Abstract
The protein family known as fp-1 provides mussel byssus with a protective outer coating and has drawn much attention for its water resistant bioadhesive properties in vitro. A new fp-l isolated from the green shell mussel Perna canaliculus (pcfp-1) reveals a composition dominated by only four amino acids: 3,4-dihydroxyphenyl-L-alanine (dopa), lysine, proline, and valine at ~20 mol % each. SDS–PAGE and MALDI-TOF mass spectrometry detected size variants at 48 and 52 kDa in preparations of purified Pcfp-1. The N-terminal sequence enabled construction of oligonucleotide primers for PCR and RACE-derived cDNAs from which the complete sequence of four variants was deduced. pcfp-1 deviates from all known homologues in other mussels in several notable respects: its mass is half, most of its sequence is represented by 75 tandem repeats of a tetrapeptide, i.e., PY*VK, in which Y* is dopa, prolines are not hydroxylated, and thiolate cysteines are clustered in homologous sequences at both the amino and carboxy termini. Amino acids in the repeat sequence show a striking resemblance to proline-rich cell wall proteins with tandemly repeated PPVYK pentapeptides [Hong, J. C., Nagao, R. T., and Key, J. L. (1987) J. Biol. Chem. 262, 8367–8376]. Cysteine plays a key role in cross-linking pcfp-1 by forming adducts with dopaquinone. Significant 5-S-cysteinyldopa and smaller amounts of 2-S-cysteinyldopa were detected in hydrolysates of the byssal threads of P. canaliculus. The cross-links could also be formed by oxidation of pcfp-1 in vitro using mushroom tyrosinase. Cysteinyldopa cross-links were present in trace amounts only in the byssus of other mussel species.
Mussel adhesive proteins have become important macro-molecular paradigms for engineering new adhesive polymers with bio-inspired attributes. Many recent developments in biotechnology (1, 2), wood and paper processing (3), and biomedical materials (4, 5) are pointedly mimicking mussel adhesive chemistry. A common theme involves introducing the 3,4-dihydroxyphenylalanine (dopa)1 functionality as well as additional flanking sequences of mussel adhesive proteins into other polymer backbones (6). Dopa, however, is revealing a chemical versatility that is more complex than first imagined (7); consequently, a better fundamental understanding of how this versatility is controlled and exploited in nature is needed.
As in other areas of biological investigation, the exploration of homologous systems often leads to unanticipated insights into structure–function relationships. This case in point compares byssal coating protein known as pcfp-1 from the green shell mussel Perna canaliculus (Gmelin) with mefp-1 from the blue mussel Mytilus edulis L. mefp-1 was first isolated from M. edulis, where, following cross-linking, it provides the holdfast with a thin but robust protective outer cuticle (8). M. edulis fp-1 is a 110 kDa protein that consists of more than 70 decapeptide repeats that collectively endow the molecule with an open extended structure (9, 10). Five of 10 residues in each decapeptide are targeted for post-translational modification: two of dopa, two of 4-trans-hydroxyproline, and one of trans-2,3-cis-3,4-dihydroxyproline (11, 12). dopa has been implicated in both cross-linking (cohesive) and interfacial (adhesive) interactions, which are proposed to include, among others, aryl couplings (13), coordination complexes with Fe3+ ions (14, 15), and complexation of metal oxides (16). Moreover, dopa chemistry is largely responsible for the technical difficulties encountered in working with extracellular matrices made of dopa-containing proteins, the foremost of which are rapid insolubilization above pH 7 and loss of antigenicity (17, 18).
Our studies with P. canaliculus fp-1 (pcfp-1) reveal that, although dopa chemistry is versatile in general, its reactivity is strongly and specifically defined by the available functional chemistry in the protein. This is especially true in the case of pcfp-1, where dopa occurs in two distinct chemical environments: in the cysteine-rich N- and C-termini and in the repetitive core sequence. While the core sequence of pcfp-1 retains features common to other mussel fp-1s, it exhibits an uncanny resemblance to the cysteine- and proline-rich proteins of the plant cell wall.
MATERIALS AND METHODS
Protein Extraction
Live green shell mussels from New Zealand were obtained from Harbor Meat & Seafood (Santa Barbara, CA) and maintained in open circulating seawater tanks at ~15 °C. For extraction of pcfp-1, fresh mussels were killed by severing the adductor muscles, following which the foot was carefully dissected and either used directly or stored at −80 °C until it was used.
Approximately 3 g of frozen mussel feet was partially thawed, cut into small pieces with a clean single-edge razor blade, and homogenized on ice with 15 mL of 5% acetic acid (v/v) and protease inhibitors (10 mg/L pepstatin and 10 mg/L leupeptin) using a frosted-glass Kontes tissue grinder. The homogenate was centrifuged at 20000g for 40 min in a refrigerated Sorval 5B centrifuge. The supernatant (~12 mL) was collected and kept in an ice-cold bath. Chilled (70%, w/v) perchloric acid (PCA) was added slowly to give a final PCA concentration of 1.4%. The mixture was stirred for 30 min on ice and centrifuged at 20000g for 30 min. The pellet was discarded, and the supernatant was dialyzed overnight at 4 °C against 4 L of 5% acetic acid using dialysis tubing with a molecular weight cutoff of 1000. The dialysates were freeze-dried and redissolved in a small volume of 5% acetic acid. This concentrated crude extract of pcfp-1 was subjected to further purification steps.
Protein Purification
Pure pcfp-1 was achieved by gel permeation chromatography using a Shodex KW-803 column (5 μm, 8 mm × 300 mm) according to ref 19. The column was equilibrated and eluted with 5% acetic acid with 0.2% TFA at a flow rate of 0.2 mL/min. For each run, a maximum volume of 200 μL of crude concentrate was loaded onto the Shodex KW-803 column. The elutants were monitored at 280 nm.
Electrophoresis
Routine electrophoresis was conducted on polyacrylamide gels (7% acrylamide and 0.2% N,N′-methylenebisacrylamide) containing 5% acetic acid with 8 M urea. The advantage of this type of PAGE for the analysis of dopa-containing proteins was previously established (20). The proteins were stained with Coomassie Blue R-250, whereas dopa was stained with nitroblue tetrazolium redox cycling (21).
Amino Acid Analysis and Protein Sequencing
The purified and alkylated pcfp-1s were hydrolyzed in 6 M HCl with 5% phenol in vacuo at 110 °C for 24 h. The samples were then flash-evaporated at 50 °C under vacuum and washed to dryness with a small volume of Milli-Q water followed by methanol. Amino acid analysis was performed according to conditions described previously with a Beckman System 6300 Auto Analyzer (21). The amino acid sequence of intact pcfp-1 was determined by automated Edman degradation on a Porton Instruments (Porton, CA) microsequencer (model 2090). Resolution of the phenylthiohydantoin (PTH) derivatives of dopa and alanine was affected by a modified gradient program (22).
Cysteine Modification
Cysteine in pcfp-1 was modified by alkylation with iodoacetate before and after reduction with dithiothreitol (23). Purified pcfp-1 was reconstituted in 50 μL of 50 mM ammonia bicarbonate. Protein was reduced by adding 5 μL of 0.2 M dithiothreitol and incubating the sample at room temperature for 1 h. pcfp-1 was alkylated with 15 μL of 500 mM iodoacetate at room temperature for 40 min in dark. Alkylated pcfp-1 with iodoacetate was subjected to amino acid analysis after hydrolysis. Cysteine was detected as carboxymethylcysteine, which runs at 9.5 min by amino acid analysis.
Isolation of Cysteinyldopa Cross-Links
Perna and Mytilus threads were hydrolyzed in 6 M HCl with 5% phenol in vacuo at 110 °C for 2 h. Hydrolysates were then flash-evaporated at 50 °C under vacuum and washed to dryness with small volume Milli-Q water followed by methanol. The flash-evaporated hydrolysate was taken up in 100 mM sodium phosphate buffer (pH 7.5), microfuged for 10 min at 15000g to remove insolubles, and applied to a pre-equilibrated phenylboronate column (Affigel Boronate, Bio-Rad). Bound ligands on the phenylboronate column were washed extensively with 10 volumes of 100 mM phosphate buffer, and desalted by 2.5 mM NH4HCO3 and Milli-Q water to facilitate subsequent amino acid analysis and electrospray ionization mass spectrometry described below. Fractions eluted with 5% acetic acid were lyophilized and subjected to a modified ninhydrin-based amino acid analysis. Pure authentic 2- and 5-S-cysteinyldopa standards were a gift and detected by amino acid analysis at 47.6 and 54.7 min, respectively.
Cross-Linking in Vitro
Approximately 1 mg of pcfp-1 was resuspended in 400 μL of 100 mM phosphate buffer (pH 7.5) and mixed with mushroom polyphenol oxidase (Sigma, T7755, ≥2000 units/mg) at an enzyme:pcfp-1 ratio of 1:10 by weight. The mixture was incubated at room temperature with constant stirring for 1 h. The reaction was stopped by adding 100 μL of 6 M HCl. The resulting mixture was hydrolyzed in 6 M HCl with 5% phenol at 110 °C for 2 h in vacuo. The hydrolysate was flash-evaporated to dryness, reconstituted in 1 mL of 50 mM phosphate buffer, and applied to the phenylboronate column. Fractions eluted with 5% acetic acid were subjected to amino acid analysis for 5-S-cysteinyldopa. A control reaction was carried out using boiled enzyme (2 min).
Molecular Cloning
Total RNA was extracted from the P. canaliculus foot using a RNase Plant Mini Kit from Qiagen (Valencia, CA). Briefly, one freshly dissected foot was used, and Qiagen’s protocols were followed after initial tissue disruption under liquid nitrogen with a mortar and pestle.
First-strand cDNA was synthesized from total RNA using Superscript II reverse transcriptase with an Adapter Primer, 5′-GGC CAC GCG TCG ACT AGT ACT (T)16-3′(Invitrogen). The product of the room temperature reaction was used in a subsequent PCR.
On the basis of the known amino acid sequence of the N-terminus of pcfp-1, VY*PNVIKPY* (Y* stands for dopa), the 3′ ends of pcfp-1 were cloned using 3′ Rapid Amplification of cDNA Ends (3′ RACE) with degenerate oligonucleotide (sense 5′-GTN TAY CCN AAY GTN ATH AAR CCN TA-3′), and an Abridged Universal Amplification Primer (antisense 5′-GGC CAC GCG TCG ACT AGT AC-3′, Invitrogen).
The PCR was carried out in 25 μL of 1× buffer B (Fisher) and 5 pmol of each primer, 5 μmol of each dNTP, 1 μL of first-strand reaction sample, and 2.5 units of Taq DNA polymerase (Fisher) for 35 cycles on a Robocycler (Stratagene). Each cycle consisted of 30 s at 94 °C, 30 s at 52 °C, and 1 min at 72 °C, with a final extension of 15 min. The PCR products were subjected to 1% agarose gel electrophoresis, purified, cloned into a PCR TA vector (TOPO TA Cloning Kit, Invitrogen), and transformed into competent Top10 cells for amplification, purification, and sequencing. The insert encodes the C-terminal sequence of pcfp-1 with a 3′-untranslated region.
To obtain the 5′ end of the cDNA of pcfp-1, the GeneRacer kit was used to obtain sequence information from full-length transcripts by a 5′ RACE. PCR was conducted with the gene specific primer (antisense 5′-GGT TTC ACA TAA GGC TTG ACG TAA GG-3′) which primes the N-terminus of pcfp-1, and a GeneRacer 5′ primer from Invitrogen.
Mass Spectrometry
The mass of pcfp-1 was determined by matrix-assisted laser desorption and ionization with time of flight (MALDI-TOF) using a PerSeptive Biosystems Voyager DE model (AB Biosystems, Foster City, CA) in the positive ion mode with delayed extraction. The MALDI matrix was prepared by dissolving sinapinic acid (10 mg/ mL) in 30 vol % acetonitrile. The purified pcfp-1 protein was dissolved in this matrix solution to give a final concentration between 1 and 10 pmol/μL. Approximately 1 μL of this solution was applied to the target plate and allowed to evaporate. The sample spots were irradiated using an N2 laser (LSI, Inc., Cambridge, MA) with a wavelength of 337 nm, a pulse width of 8 ns, and a frequency of 5 Hz. MALDI ionization generates protonated singly and doubly charged ions for the pcfp-1 protein (mostly singly charged ions) which were accelerated using an accelerating voltage of either 20 or 25 kV. Bovine serum albumin was used as molecular mass calibrant (average masses of 66 430 and 33 215 Da for mono-and diprotonated ions, respectively).
Positive ion TOF ESI-MS was performed on a PE Sciex QStar quadrupole/time-of-flight tandem mass spectrometer. Samples were injected with a syringe pump at 5 μL/min, and typical settings were a capillary voltage of 3.5 kV, a cone tension of 45 V, and a collision voltage of 10 V for MS. Cross-link candidates were further analyzed by MS/ MS using argon gas and a collision voltage of 30 V for collision-induced decomposition on the same instrument. Structure was deduced from decomposition-induced fragment ion masses.
RESULTS
Like the fp-1s of other marine mussels, pcfp-1 was isolated by a procedure based on perchloric acid extraction (18) (Figure 1A). pcfp-1 was effectively separated from the remaining soluble contaminants by gel permeation chromatography (Figure 1B). Protein from the peak fractions was subjected to standard automated Edman degradation for sequencing and to acid hydrolysis for amino acid composition. An N-terminal sequence of 12 amino acids was obtained (Table 1). The first three residues (Valdopa-Pro) were homogeneous, after which two residues were detected in each cycle with major and minor components at a ratio of ~4:3. Continuing the sequence further into the N-terminus was quickly limited by the heterogeneity and frequency of poorly cleaved Pro residues.
Figure 1.
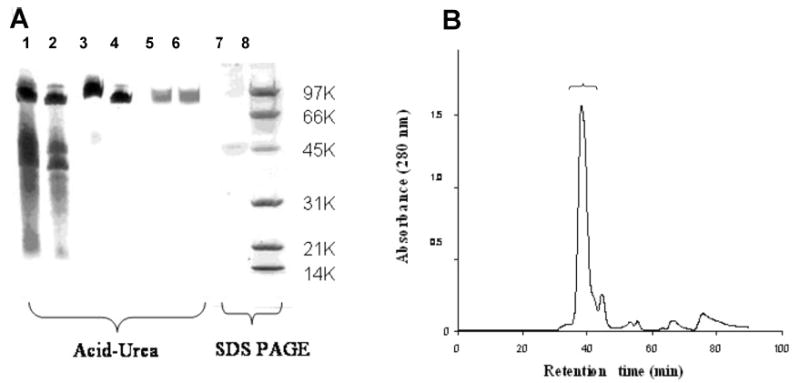
Purification of pcfp-1. (A) Polyacrylamide gel electrophoresis of key steps in purification of pcfp-1. Lanes 1 and 2 are loaded with 40 and 20 μL of crude PCA extract of pcfp-1, respectively, and stained with Coomasie Blue R-250. Lanes 3 and 4 are loaded with 40 and 20 μL of crude extract of pcfp-1, respectively, and stained for dopa with NBT in glycinate. Lanes 5 and 6, which correspond to fractions 39 and 40 (each 40 μL), respectively, are loaded with pcfp-1 purified by chromatography on a Shodex KW-803 column. Lanes 1–6 show 7% acid–urea PAGE. Lane 7 contains 20 μL of purified pcfp-1 run on 15% SDS–PAGE. Lane 8 contains the low-molecular weight markers: 5 μg each of phosphorylase b (97 400), bovine serum albumin (66 200), ovalbumin (45 000), carbonic anhydrase (31 000), soybean trypsin inhibitor (21 500), and lysozyme (14 400). (B) Gel permeation chromatography of pcfp-1 crude extract with a Shodex KW-803 column. The bracketed peak contains purified pcfp-1.
Table 1.
Amino Acid Composition of pcfp-1 in Mole Percent (residues per 100 residues)a
| amino acid | purified (mol %) | purified (mol/mol) | predicted (mol/mol) |
|---|---|---|---|
| Hyp | 0 | – | – |
| Asx | 3.0 | 12 | 11 |
| Thr | 1.5 | 6 | 3 |
| Ser | 1.9 | 7 | 6 |
| Glx | 1.1 | 4 | 0 |
| Pro | 22.5 | 90 | 91 |
| Gly | 3.8 | 15 | 11 |
| Ala | 1.9 | 8 | 3 |
| Cys | 0.6 | 5 | 13 |
| Val | 17.8 | 74 | 79 |
| Met | 0 | 0 | 0 |
| Ile | 0.5 | 1 | 3 |
| Leu | 0.9 | 4 | 4 |
| dopa | 17.5 | 72 | – |
| Tyr | 3.0 | 13 | 87 |
| Phe | 0.5 | 1 | 0 |
| His | 0.7 | 1 | 1 |
| Hyl | 1.1 | 5 | – |
| Lys | 23.3 | 97 | 101 |
| Arg | 0.6 | 2 | 1 |
| total | 100 | 397 | 414 |
The mole per mole composition of the second column assumes an average mass of 49.7 kDa; the predicted composition is based on pcfp-1/1.
The amino acid composition of purified pcfp-1 revealed primarily four amino acids: Pro, Val, Lys, and dopa each at levels of approximately 20–23 mol % (Table 2). There was no detectable 4-trans-hydroxyproline, and cystine was detected at ~0.6 mol %.
Table 2.
Edman Sequencing of the N-Terminus of pcfp-1 (10 cycles)
| cycle | major amino acid | minor amino acid |
|---|---|---|
| 1 | Val | – |
| 2 | dopa | – |
| 3 | Pro | – |
| 4 | Asn | Lys |
| 5 | Val | Lys |
| 6 | Ile | Pro |
| 7 | Lys | dopa |
| 8 | Pro | Val |
| 9 | dopa | Lys |
| 10 | Ala | Pro |
Peak HPLC fractions were also subjected to MALDI-TOF mass spectrometry (Figure 2). This revealed a pair of monoprotonated doublets at 52 712/51 446 and 47 989/ 46 712, which appear to correspond to a separated pair of bands on SDS–PAGE, but moved as a single band on acid urea (Figure 1A).
Figure 2.
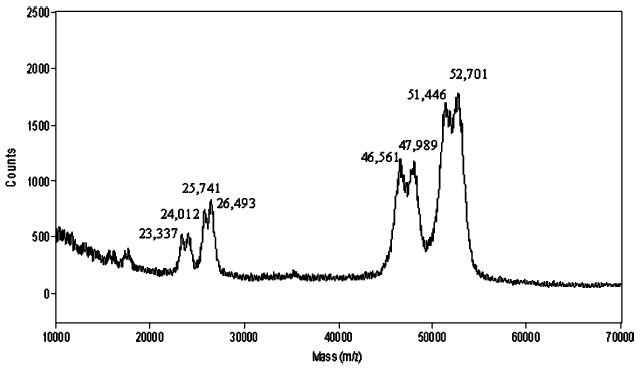
MALDI-TOF mass spectrum of purified pcfp-1. Peak clusters from right to left are [M + H]+, [M + 2H]2+, and [M + 3H]3+. Delayed extraction (200 ns) in positive ion mode with an accelerating voltage of 25 000 V, a grid voltage at 93%, and a guide wire voltage at 0.1%. The spectrum represents an average of 254 scans.
Given their repetitive structure, a tendency to oxidize at neutral pH, and a large number of Lys-Pro linkages, pcfp-1 posed insurmountable challenges to traditional sequencing by peptide mapping. Consequently, a molecular strategy for deriving cDNA-deduced sequences was devised on the basis of degenerate oligonucleotides designed from the N-terminus which was unique before degenerating into more than 75 tandem high-fidelity repeats of the core sequence (Figure 3). Four related clones, apparently reflecting the observed variants of pcfp-1, were amplified, cloned, and sequenced. The sequence of one of these, pcfp-1/1, is shown in Figure 3. The others are given in the Supporting Information. Evident is a signal peptide sequence that is 18 residues long. Cleavage between S-18 and V-19 is predicted by “SIGNALP” (EXPASY), and is consistent with the observed N-terminus in pcfp-1/1. Sandwiched between N- and C-terminal Cys-rich sequences each ~45 residues in length is a highly repetitive core structure that consists almost entirely of PYVK tetrapeptide repeats. Only two thematic deviations occur: (1) three tetrapeptides with P → T (two) or Y → C substitutions (one) and (2) 13 repeats of an abbreviated variant, PK, from which the core amino acids YV are absent.
Figure 3.
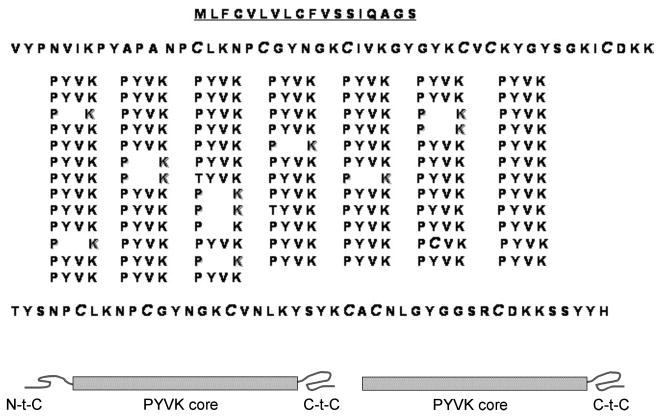
Complete sequence of pcfp-1 deduced from cDNA (variant pcfp-1/1). Schematics reflect the main difference between pcfp-1/1 (left) and truncated variant pcfp-1/3 (right).
In all, four different sequence variants were found that correlate reasonably well with the four mass variants in the mass spectrum of Figure 2. Pcfp-1/1 and pcfp-1/2, with a calculated mass of ~50 kDa, differ by a single PYVK tetrapeptide (mass of 505 Da). Pcfp-1/3 and pcfp-1/4, in contrast, both lack the Cys-rich sequence at the amino terminus and are lower by 45–46 kDa with the latter being 16 residues or four tetrapeptides (~2 kDa) shorter than the former. The peaks in Figure 2 have higher observed masses than the cDNA-deduced ones in part because of the extensive hydroxylation of Tyr to dopa. Variant pcfp-1/1, for example, is the largest of the variant clones at 49 666 Da. If it corresponds to the observed mature protein mass of 52.7 kDa (Figure 2), then the mass difference between the two amounts to ~3 kDa. Since pcfp-1/1 contains 75 repeats of the PYVK tetrapeptide, an increase in mass of 1.2 kDa would be anticipated if all these Tyr residues were hydroxylated to dopa. An additional 224 Da would be gained if all the Tyr residues in the termini were hydroxylated to dopa. This still leaves room (1.8 kDa) for other modifications, which are unknown at this time.
The presence of thiolate cysteines (five to seven residues per protein) in pcfp-1 is unprecedented for an fp-1 as well as for known fps in general. The Cys residues in the EGF domains of mefp-2 (Mytilus) are all disulfide-bonded (24). Reductive alkylation of pcfp-1 with DTT and iodoacetate followed by hydrolysis and amino acid analysis showed that total cysteine (detected as carboxymethylcysteine) is present at 2.7 mol %. Approximately half of this (52.2 ± 5%; n = 3) can be alkylated prior to reduction by DTT. As deduced from cDNA, all Cys residues but one are sequestered to the N- and C-terminal ends of pcfp-1 in sequences that are more than 60% identical (Figure 3).
Since dopa is oxidized to dopaquinone, and the latter reacts readily with free cysteine in vitro (25, 26), the fate of cysteinyl groups in pcfp-1 seemed to be a plausible target for further investigation. Hydrolysates of byssal threads were screened for condensation products of dopa and cysteine. Positive identification of cysteinyldopas is based on four criteria: run time in amino acid analysis compared with known standards, binding to phenylboronate (which is specific for catechols), ESI mass spectrometry, and fragmentation by MS/MS. Both 2- and 5-S-cysteinyldopa were detected in acid hydrolysates of byssal threads; however, the 5-S isomer is 10 times more abundant, reaching levels of >1 mol % in threads. Cysteinyldopa isomers are separated well by ion exchange used in amino acid analysis with elution times of 47.6 min (2-S) and 54.7 min (5-S) (Figure 4).
Figure 4.
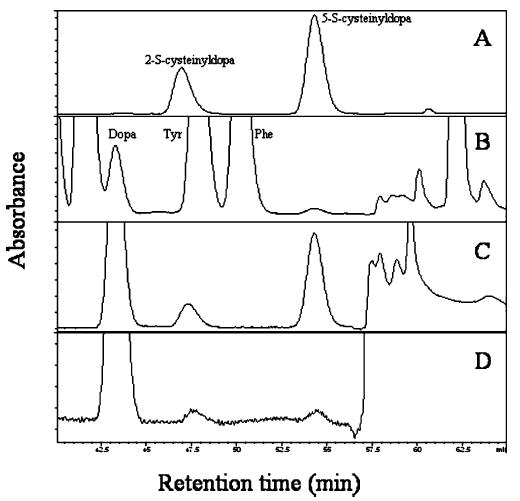
Amino acid analysis of cysteinyldopa cross-link content: (A) authentic 2- and 5-S-cysteinyldopa, (B) Perna byssal thread hydrolysate (hydrolysis for 2 h at 110 °C), (C) byssal thread hydrolysate after phenyl boronate chromatography, and (D) hydrolysate of purified pcfp-1 after in vitro oxidation with mushroom tyrosinase post-phenyl boronate chromatography. Absorbance is measured at 570 nm for ninhydrin.
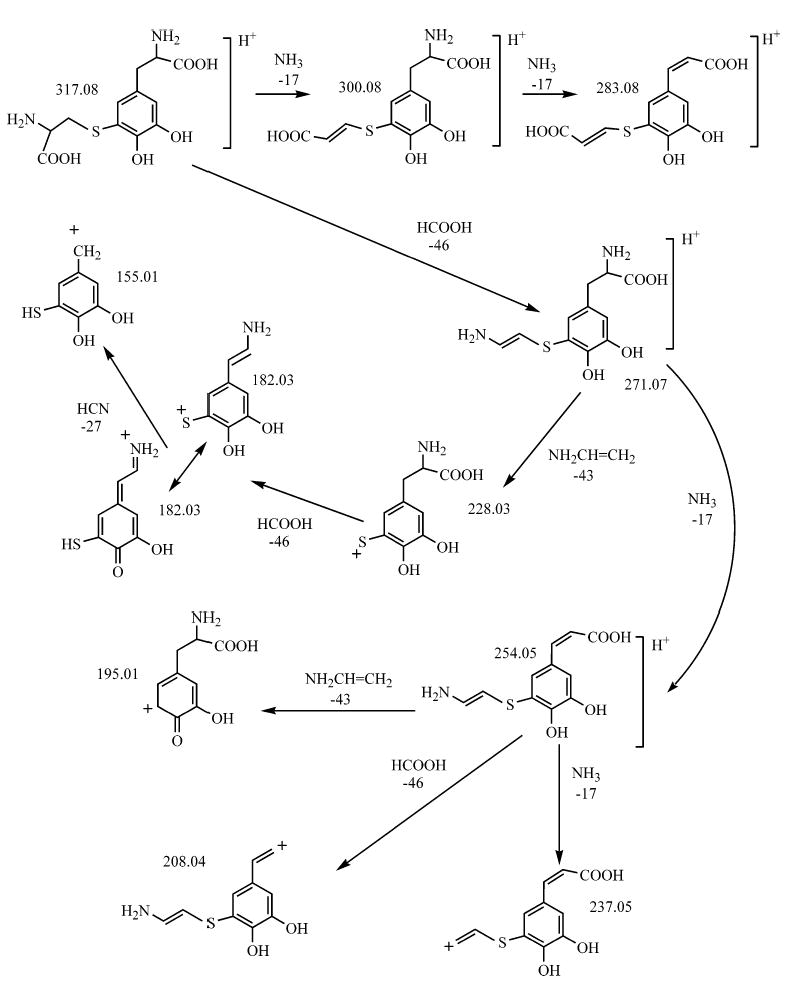
Both cysteinyldopa isomers were effectively captured from hydrolysates by phenyl boronate affinity chromatography, and fractions eluted by 5% acetic acid were subjected to ESI mass spectrometry which usually gave the protonated (317.1 Da) and sodiated (339.1 Da) masses for both the standards and the protein-derived cross-links (the relative abundance of the sodiated form reflecting Na+ copurified with the cross-links) (Figure 5). The protonated forms of both standard cysteinyldopa and byssus-derived compounds subjected to MS/MS produce similar spectra and exhibit fragmentation that is consistent with earlier fast atom bombardment (FAB) studies on methylated and pentafluoropropionylated 2-S and 5-S isomers (27). The proposed structures of these ions are Scheme 1 illustrated in Scheme 1. It is noteworthy that the 2- and 5-S isomers can be readily distinguished by the relative abundance of the 182 and 228 Da ions in the MS/MS of 2-S-cysteinyldopa (Figure 5).
Figure 5.
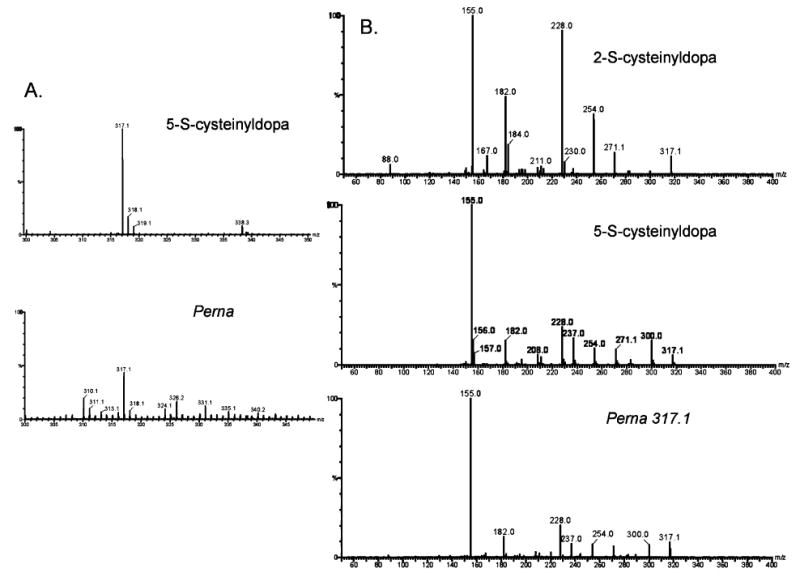
ESI mass spectrometry of the monoprotonated form of 5-S-cysteinyldopa. Authentic 5-S-cysteinyldopa and a compound isolated from hydrolyzed P. canaliculus threads by phenyl boronate chromatography (A). CID and MS/MS of the m/z 317 peak from 2-S and 5-S standards and Perna phenyl boronate-derived fractions, respectively (B).
Scheme 1.
The abundance of cysteinyldopa cross-links in Perna threads is an order of magnitude greater than that in three species of Mytilus (Figure 6). To ascertain whether these cross-links can also form in vitro using purified pcfp-1, the protein was oxidized by tyrosinase at pH 7.5, and aliquots were removed at different intervals, hydrolyzed, and analyzed for dopa and cysteinyldopa content by amino acid analysis. Both the 2- and 5-S-cysteinyldopa cross-link isomers were detectable at 1 h (Figure 4D).
Figure 6.
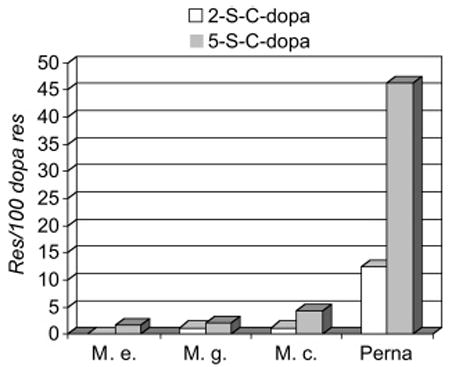
Comparing the cysteinyldopa content of byssal threads from Perna with three species of Mytilus. Cross-link content is expressed as the mole percent of the dopa content.
DISCUSSION
Given their role as a protective coating of byssal threads (28), it seems peculiar that the fp-1s of M. edulis and P. canaliculus should be so different. Pcfp-1 exhibits a smaller mass (50 kDa in pcfp1 vs 110 kDa in mefp-1), a shorter consensus repeat (PYVK in pcfp-1 vs AKPSYPPTYK in mefp1), no hydroxylation of Pro to Hyp (pcfp-1) in contrast to 100% hydroxylation of –PP– (mefp-1), and 13 Cys residues compared to none in mefp-1. There are also some similarities: Tyr is largely hydroxylated to dopa in both, Pro and Lys are prominent in both (usually in Lys-Pro sequences), and Tyr, Pro, and Lys positions are conserved with high fidelity in both cores (Tables 2 and 3). Consensus repeats of pcfp-1 also resemble features of other fp-1s. The consensus repeat of Aulacomya is seven amino acids long and ends in VK (29), while in Trichomya, it is five amino acids long (SYYPK), including three of the same amino acids (30). The most striking match, however, is not with any mussel protein but with a soybean proline-rich protein (PRP) of the cell wall, which contains 43 PPVYK repeats (31). Undoubtedly, pcfp-1 and PRP sequences were independently evolved, but it seems extraordinary that both serve as protective coatings and are closely coupled to oxidative cross-linking (32, 33). The resemblance is further enhanced with the recent report of hybrid PRPs with cysteine-rich C-terminal domains (34).
Table 3.
Comparison of Consensus Repeat Sequences in fp-1 Proteins from Various Musselsa
| Species | consensus | (repeats) | reference |
|---|---|---|---|
| Perna canaliculus | PYVK | (72) | present |
| Aulacomya ater | AGYGGVK | (29) | |
| Trichomya hirsute | SYYPK | (30) | |
| Modiolus modiolus | SSYYPK | (30) | |
| Choromytilus chores | AKPSYPTGYKPPVK | (29) | |
| Mytilus edulis | AKPSYPPTYK ---- | (71) | (40) |
| Mytilus galloprovincialis | AKPSYPPTYK ---- | (85) | (41) |
| Glycine max | PPVYK | (43) | (31) |
The consensus repeat number (#) is not precisely known for proteins known only from Edman sequencing.
Since dopa in mefp-1 has been associated with the high-affinity binding of metal ions such as Fe3+ in vitro and in situ (14, 15), we assumed that it might serve the same role in pcfp-1. However, the low metal content of Perna threads vis-à-vis Mytilus (ref 35 and unpublished observations) as well as the significant Cys thiolate content in pcfp-1 suggested that Perna threads utilize at least some of the dopa by another pathway, that is, the formation of cysteinyldopa cross-links following oxidation of dopa to dopaquinone. This pathway was confirmed by the presence of 5-S-cysteinyldopa in hydrolyzed Perna threads. The demonstration that Perna preferentially exploits such cross-links is evident from a comparison of the dopa and cysteinyldopa content in threads from P. canaliculus and three species of Mytilus [M. edulis, Mytilus galloprovincialis, and Mytilus californianus (Figure 6)]. Both 2- and 5-S-cysteinyldopa are present at levels at least 1 order of magnitude higher in Perna threads than in threads from the other mussels. Nothing in the present results can distinguish whether the cysteinyldopas occur as intra-or intermolecular cross-links in the native structure. In all likelihood, both modes of cross-linking occur.
5-S-Cysteinyldopa has become important as a urinary marker for melanoma (36). It is formed from free dopa and cysteine, which are precursors of the phaeomelanins. 5-S-Cysteinyldopa cross-links have been detected in serum albumin, alcohol dehydrogenase, and gluten formed in vitro by the action of tyrosinase (37, 38). Its detection in the byssal threads of Perna, however, represents the first case of its natural occurrence as a protein cross-link. Given that the nucleophilic attack of an o-quinone by cysteine is 5 × 104 times faster than that by the ε-amino group of lysine (39), it seems to have been chosen well for a material like byssus that needs to set rapidly from secreted soluble proteins. Perhaps the more intriguing question is why the reaction is not more widely practiced in mussels.
A perusal of the entire pcfp-1 sequence reveals that 14 dopa (or Tyr) residues lie outside the repetitive core in the N- and C-terminal domains. These exhibit more sequence variability than the PYV consensus of the core region. GYX, for example, where X is G, S, N, or K, occurs six of 14 times in the terminal domains. If these dopas were more readily oxidized to quinones than the dopa in PYV sequences, then cross-linking might be restricted initially to the terminal domains. This raises the intriguing possibility that variants with both terminal domains intact (pcfp-1/1 and -1/2) might function in chain propagation, whereas those without N-terminal domains (pcfp-1/3 and -1/4) would serve as chain terminators in the self-assembly of pcfp-1.
Supplementary Material
Acknowledgments
2- and 5-S-Cysteinyldopa were generously donated by Drs. K. Wakamatsu and S. Ito of Fujita-Gakuen University (Toyoake, Japan). We thank Drs. Chengjun Sun and James Pavlovich for their assistance with phenylboronate columns and mass spectrometry, respectively.
Abbreviations
- dopa
3,4-dihydroxyphenylalanine
- ESI
electrospray ionization
- MALDI-TOF
matrix-assisted laser desorption ionization with time of flight
- mefp-1
M. edulis foot protein 1
- pcfp-1
P. canaliculus foot protein 1
- PCA
perchloric acid
- PAGE
polyacrylamide gel electrophoresis
- MS/MS
tandem mass spectrometry
Footnotes
This research was supported by a grant from the National Institutes of Health (R01 DE015415).
GenBank accession numbers AY960603 (fp1-1), AY960604 (fp1-2), AY960605 (fp1-3), and AY960606 (fp1-4).
SUPPORTING INFORMATION AVAILABLE
Complete cDNA-deduced sequences of all pcfp-1 variants. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Xu C, Xu K, Gu H, Zheng R, Liu H, Zhang X, Guo Z, Xu B. Dopamine as a robust anchor to immobilize functional molecules on the iron oxide shell of magnetic nanoparticles. J Am Chem Soc. 2004;126:9938–9939. doi: 10.1021/ja0464802. [DOI] [PubMed] [Google Scholar]
- 2.Burdine L, Gillette TG, Lin HJ, Kodadek T. Periodate-triggered cross-linking of DOPA-containing peptide–protein complexes. J Am Chem Soc. 2004;126:1142–1143. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- 3.Li K, Geng X, Simonsen J, Karchesy J. Novel wood adhesives from condensed tannins and polyethylenimine. Int J Adhes Adhes. 2004;24:327–333. [Google Scholar]
- 4.Dalsin JL, Hu BH, Lee BP, Messersmith PB. Mussel adhesive protein mimetic polymers for the preparation of nonfouling surfaces. J Am Chem Soc. 2003;125:4253–4258. doi: 10.1021/ja0284963. [DOI] [PubMed] [Google Scholar]
- 5.Huang K, Lee BP, Ingram DR, Messersmith PB. Synthesis and characterization of self-assembling block copolymers containing bioadhesive end groups. Biomacromolecules. 2002;3:397–406. doi: 10.1021/bm015650p. [DOI] [PubMed] [Google Scholar]
- 6.Deming TJ. Mussel byssus and biomolecular materials. Curr Opin Chem Biol. 1999;3:100–105. doi: 10.1016/s1367-5931(99)80018-0. [DOI] [PubMed] [Google Scholar]
- 7.Waite JH. Adhesion a la moule. Integr Comp Biol. 2002;42:1172–1180. doi: 10.1093/icb/42.6.1172. [DOI] [PubMed] [Google Scholar]
- 8.Benedict CV, Waite JH. Location and analysis of byssal structural proteins of Mytilus edulis. J Morphol. 1986;189:261–270. doi: 10.1002/jmor.1051890207. [DOI] [PubMed] [Google Scholar]
- 9.Deacon MP, Davis SS, Waite JH, Harding SE. Structure and mucoadhesion properties of mussel glue protein in dilute solution. Biochemistry. 1998;37:14108–141112. doi: 10.1021/bi9806192. [DOI] [PubMed] [Google Scholar]
- 10.Haemers S, van der Leeden MC, Frens G. Coil dimensions of the mussel adhesive protein Mefp-1. Biomaterials. 2005;26:1231–1236. doi: 10.1016/j.biomaterials.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 11.Taylor SW, Waite JH, Ross MM, Shabanowitz J, Hunt DF. trans-2,3-cis-3,4-Dihydroxyproline in the tandemly repeated consensus decapeptides of an adhesive protein from Mytilus edulis. J Am Chem Soc. 1994;116:10803–10804. [Google Scholar]
- 12.Waite JH. Evidence for a repeating 3,4-dihydroxy-phenylalanine and hydroxyproline containing decapeptide in the adhesive protein of the mussel Mytilus edulis. J Biol Chem. 1983;258:2911–2915. [PubMed] [Google Scholar]
- 13.McDowell LM, Burzio LA, Waite JH, Schaefer J. Rotational echo double resonance detection of cross-links formed in mussel byssus under high-flow stress. J Biol Chem. 1999;274:20293–20295. doi: 10.1074/jbc.274.29.20293. [DOI] [PubMed] [Google Scholar]
- 14.Sever MJ, Weisser JT, Monahan J, Srinivasan S, Wilker JJ. Metal mediated cross-linking in the generation of a marine-mussel adhesive. Angew Chem. 2004;43:448–450. doi: 10.1002/anie.200352759. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SW, Chase DB, Emptage MH, Nelson MJ, Waite JH. Ferric ion complexes of a DOPA-containing adhesive protein from Mytilus edulis. Inorg Chem. 1996;35:7572– 7577. [Google Scholar]
- 16.Dalsin JL, Lin L, Tosatti S, Vörös J, Textor M, Messersmith PM. Protein Resistance of Titanium Oxide Surfaces Modified by Biologically Inspired mPEG-DOPA. Langmuir. 2005;21:640–646. doi: 10.1021/la048626g. [DOI] [PubMed] [Google Scholar]
- 17.Anderson KE, Waite JH. Immunolocalization of Dpfp1, a byssal protein of the zebra mussel Dreissena polymorpha. J Exp Biol. 2000;203:3065–3076. doi: 10.1242/jeb.203.20.3065. [DOI] [PubMed] [Google Scholar]
- 18.Robinson MW, Colhoun LM, Fairweather I, Brennan GP, Waite JH. Development of the vitellaria of the liver fluke Fasciola hepatica in the rat host. Parasitology. 2001;123:509–518. doi: 10.1017/s0031182001008630. [DOI] [PubMed] [Google Scholar]
- 19.Ohkawa K, Nishida A, Yamamoto H, Waite JH. A glycosylated precursor protein from the green mussel Perna viridis with modified Dopa side-chains. Biofouling. 2004;20:101–105. doi: 10.1080/08927010410001681246. [DOI] [PubMed] [Google Scholar]
- 20.Waite JH, Benedict CV. Assay of DOPA in invertebrate structural proteins. Methods Enzymol. 1984;107:397–413. doi: 10.1016/0076-6879(84)07028-2. [DOI] [PubMed] [Google Scholar]
- 21.Paz M, Flückinger R, Boak A, Kagan HM, Gallop PM. Specific detection of quinoproteins by redox-cycling staining. J Biol Chem. 1991;266:689–692. [PubMed] [Google Scholar]
- 22.Waite JH. Detection of peptidyl-DOPA by amino acid analysis and microsequencing techniques. Anal Biochem. 1991;192:429–433. doi: 10.1016/0003-2697(91)90560-g. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JG, Matthews JM, Ward LD, Simpson RJ. Disruption of the disulfide bonds of recombinant murine interleukin-6 induces formation of a partially unfolded state. Biochemistry. 1997;36:2380–2389. doi: 10.1021/bi962164r. [DOI] [PubMed] [Google Scholar]
- 24.Rzepecki LM, Hansen KM, Waite JH. Characterization of a cystine-rich polyphenolic protein family from the blue mussel Mytilus edulis L. Biol Bull. 1992;183:123–137. doi: 10.2307/1542413. [DOI] [PubMed] [Google Scholar]
- 25.Ito S, Prota G. A facile one-step synthesis of cysteinyldopas using mushroom tyrosinase. Experientia. 1977;33:1118–1119. doi: 10.1007/BF01946005. [DOI] [PubMed] [Google Scholar]
- 26.Roston S. Reaction of the sulfhydryl group with an oxidation product of β-3,4-dihydroxyphenylalanine. J Biol Chem. 1960;235:1002–1006. [PubMed] [Google Scholar]
- 27.Agrup G, Hansson C, Rorsman H, Rosengren AM, Rosengren E. Mass spectrometric analysis of enzymatically synthesized cysteinyldopa isomers. Commun Dep Anat, Univ Lund, Swed. 1976;5:1–17. [Google Scholar]
- 28.Holten-Andersen N, Slack N, Zok F, Waite JH. Nano-mechanical Investigation of the byssal cuticle, a protective coating of a bio-elastomer. Mater Res Soc Symp Proc. 2005;841:R3.7.1/Y3.7.1. [Google Scholar]
- 29.Burzio LA, Saez C, Pardo J, Waite JH, Burzio LO. The adhesive protein of Choromytilus chorus and Aulacomya ater: A proline-rich and a glycine-rich polyphenolic protein. Biochim Biophys Acta. 2000;1479:315–320. doi: 10.1016/s0167-4838(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 30.Rzepecki LM, Chin SS, Waite JH, Lavin MF. Molecular diversity of marine glues: Polyphenolic proteins from five mussel species. Mol Mar Biol Biotechnol. 1991;1:78–88. [PubMed] [Google Scholar]
- 31.Hong JC, Nagao RT, Key JL. Characterization of a proline-rich cell wall protein gene family of soybean: A comparative analysis. J Biol Chem. 1987;262:8367–8376. [PubMed] [Google Scholar]
- 32.Schmidt JS, Lindstrom JT, Vodkin LO. Genetic length polymorphisms create size variation variation in proline-rich proteins of the cell wall. Plant J. 1994;6:177–186. doi: 10.1046/j.1365-313x.1994.6020177.x. [DOI] [PubMed] [Google Scholar]
- 33.Bradley DJ, Kjellbom P, Lamb CJ. Eliicitor-induced and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- 34.Josè-Estanyol M, Puigdomènech P. Plant cell wall glycoproteins and their genes. Plant Physiol Biochem. 2001;38:97–108. [Google Scholar]
- 35.Sun CJ, Waite JH. Mapping chemical gradients within and along a fibrous structural tissue: Mussel byssal threads. J Biol Chem. 2005 doi: 10.1074/jbc.M508674200. (in press) [DOI] [PubMed] [Google Scholar]
- 36.Agrup G, Agrup P, Andersson T, Hafstrom L, Hansson C, Jacobsson S, Jonsson PE, Rorsman H, Rosengren AM, Rosengren E. Five years experience of 5-S-cysteinyl-dopa in melanoma diagnosis. Acta Derm-Venereol. 1979;59:381–388. [PubMed] [Google Scholar]
- 37.Ito S, Kato T, Shinpo K, Fujita K. Oxidation of tyrosine residues in proteins by tyrosinase. Detection of protein bonded 3,4-dihydroxyphenylalanine and 5-S-cysteinylDopa. Biochem J. 1984;222:407–411. doi: 10.1042/bj2220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takasaki S, Kawakishi S. Formation of protein-bound 3,4-dihydroxyphenylalanine and 5-S-cysteinyl-3,4-dihydroxy-phenylalanine as new cross-linkers in gluten. J Agric Food Chem. 1997;45:3472–3475. [Google Scholar]
- 39.Dryhurst G, Kadish KM, Scheller F, Renneberg R. Biological Electrochemistry. Vol. 1. Academic Press; New York: 1982. pp. 138–139. [Google Scholar]
- 40.Filpula DR, Lee SM, Link RP, Strausberg SL, Strausberg RL. Structural and functional repetition in a marine mussel adhesive protein. Biotechnol Prog. 1990;6:171–177. doi: 10.1021/bp00003a001. [DOI] [PubMed] [Google Scholar]
- 41.Inoue K, Odo S. The adhesive protein cDNA of Mytilus galloprovincialis encodes decapeptide repeats but no hexapeptide motif. Biol Bull. 1994;186:349–355. doi: 10.2307/1542281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


