Abstract
In many animals, establishment of the germ line depends on segregation of a specialized cytoplasm, or ‘germ plasm’, to a small number of germline precursor cells during early embryogenesis1. Germ plasm asymmetry involves targeting of RNAs and proteins to a specific region of the oocyte and/or embryo2. Here we demonstrate that germ plasm asymmetry also depends on degradation of germline proteins in non-germline (somatic) cells. We show that five CCCH finger proteins, components of the Caenorhabditis elegans germ plasm, are targeted for degradation by the novel CCCH-finger-binding protein ZIF-1. ZIF-1 is a SOCS-box protein that interacts with the E3 ubiquitin ligase subunit elongin C. Elongin C, the cullin CUL-2, the ring finger protein RBX-1 and the E2 ubiquitin conjugation enzyme UBC5 (also known as LET-70) are all required in vivo for CCCH finger protein degradation. Degradation is activated in somatic cells by the redundant CCCH finger proteins MEX-5 and MEX-6, which are counteracted in the germ line by the PAR-1 kinase. We propose that segregation of the germ plasm involves both stabilization of germline proteins in the germ line and cullin-dependent degradation in the soma.
Establishment of the germ line in C. elegans begins with a series of unequal divisions that divide successive germline blastomeres into somatic and germline daughter cells (Fig. 1a). The germ plasm contains RNA-binding proteins and RNA-rich organelles (P granules), which are maintained preferentially in germline blastomeres. Among germ plasm components are five proteins that share two CCCH fingers: PIE-1, POS-1, MEX-1 and the functionally redundant and 70% identical MEX-5 and MEX-6 (refs 3–6). CCCH fingers (a class of zinc finger) were first described in the vertebrate protein TTP/Nup475/Tis-11, where they are implicated in RNA binding7. In germline blastomeres, CCCH finger proteins are on P granules and diffuse throughout the cytoplasm. During each asymmetric division, PIE-1, POS-1 and MEX-1 segregate preferentially with the germline daughter, whereas MEX-5 and MEX-6 segregate preferentially with the somatic daughter3–6,8,9 (Fig. 1a; see also ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). After division, the CCCH finger proteins persist in the germline blastomeres at either high (PIE-1, MEX-1 and POS-1) or low (MEX-5 and MEX-6) levels. In contrast, in somatic blastomeres they do not persist beyond one (PIE-1, MEX-1 and POS-1) or two (MEX-5 and MEX-6) divisions3–6,8,9 (Fig. 1a; see also movies at ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). Earlier studies with PIE-1 demonstrated that asymmetric partitioning to germline blastomeres and turnover in somatic blastomeres are mediated by two independent mechanisms10. Turnover depends on the first CCCH finger (ZF1) in PIE-1, which targets PIE-1 for degradation specifically in somatic blastomeres10. ZF1s from PIE-1, POS-1 and MEX-1 (PIE-1ZF1, POS-1ZF1, MEX-1ZF1), and ZF2 from MEX-5 (MEX-5ZF2) are sufficient to target green fluorescent protein (GFP) for degradation in somatic blastomeres (Methods; see also ref. 10 and ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). In contrast, PIE-1ZF2, POS-1ZF2, MEX-1ZF2, MEX-5ZF1 and either finger in TTP/Nup475/Tis-11 are not sufficient to trigger soma-specific degradation10. These findings suggest that CCCH finger proteins are targeted for degradation by a mechanism that recognizes specific CCCH fingers.
Figure 1.
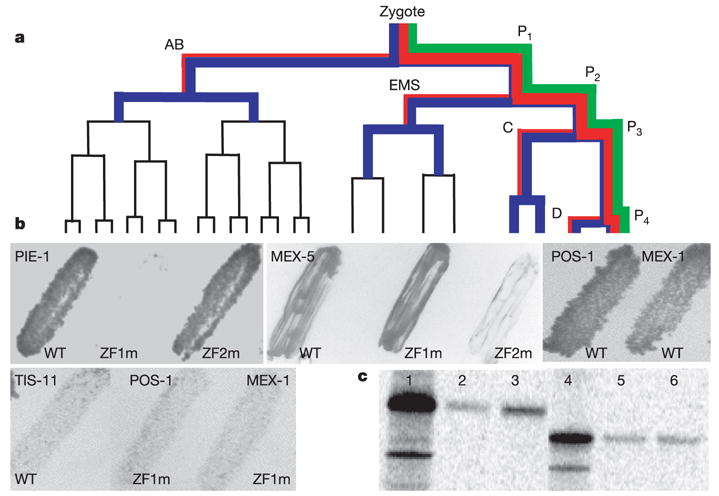
ZIF-1 interacts with CCCH fingers targeted for degradation. a, Early embryonic lineage. P1 to P4 are the germline blastomeres. AB, EMS, C and D are the somatic blastomeres. PAR-1 (green), PIE-1 (red) and MEX-5 (blue) segregations are indicated; line thickness indicates relative levels. b, Yeast transformed with ZIF-1 and finger domains from indicated proteins. ZF1m and ZF2m are SSCH instead of CCCH. Growth indicates interaction. WT, wild type. c, Labelled PIE-1 proteins (full-length, lanes 1–3; ZF1 deleted, lanes 4–6) synthesized in rabbit reticulolysates and incubated with E. coli-synthesized GST (lanes 2 and 5) or GST–ZIF-1 (lanes 3 and 6) on Sepharose beads. Bound proteins were eluted and run on SDS–PAGE. 100% of input (lanes 1 and 4) was loaded on GST column; 50% of bound was loaded on gel.
To identify trans-acting factors in this process, we performed a yeast two-hybrid screen for proteins that bind to the PIE-1 fingers (Methods). The screen identified ten isolates of F59B2.6, a new protein that we named ZIF-1 (zinc finger interacting factor 1). In the two-hybrid assay, ZIF-1 interacted with the zinc finger domains of PIE-1, POS-1, MEX-1 and MEX-5, but not TIS-11 (Fig. 1b). The interaction was blocked by mutations in the finger required for degradation, but not by mutations in the other finger (Fig. 1b). Glutathione S-transferase (GST) pull-down assays confirmed the specificity of the ZIF-1–PIE-1 interaction (Fig. 1c). We conclude that ZIF-1 binds to CCCH finger motifs targeted for degradation in somatic lineages.
To determine the in vivo function of ZIF-1, zif-1 activity was reduced in embryos by RNA-mediated interference (RNAi)11. Unlike wild-type embryos, zif-1(RNAi) embryos maintained PIE-1, POS-1, MEX-1, MEX-5 and MEX-6 in somatic blastomeres (Fig. 2a–c and data not shown; see also ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). zif-1(RNAi) embryos also failed to degrade GFP::PIE-1ZF1 and GFP::MEX-5ZF2 fusions in somatic lineages (ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). Consistent with ZIF-1 acting by binding to CCCH fingers, zif-1(RNAi) did not affect the localization of a GFP::PIE-1 fusion lacking ZF1 (Fig. 2d) and did not affect the distribution of germline-enriched factors that do not contain CCCH fingers (PAR-2 and PGL-1; Fig. 2e). Furthermore, zif-1(RNAi) did not disrupt asymmetric partitioning in germline blastomeres: CCCH finger proteins were still preferentially segregated to the germline (PIE-1, POS-1, MEX-1) or somatic (MEX-5) daughter cell at each asymmetric division (Fig. 2a, b, g and data not shown; see also ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). pie-1 RNA, which in the wild type is maintained preferentially in germline blastomeres, was also not affected in zif-1(RNAi) embryos (Fig. 2f). Finally, zif-1(RNAi) did not block turnover of OMA-1, a CCCH finger protein degraded in all cells after the one-cell stage12 (data not shown). We conclude that ZIF-1 is not generally required for protein degradation or asymmetric partitioning in germline blastomeres, but is essential for soma-specific degradation of CCCH finger proteins. zif-1(RNAi) embryos die during embryogenesis (Methods), suggesting that perdurance of CCCH finger proteins in somatic lineages interferes with embryonic development.
Figure 2.
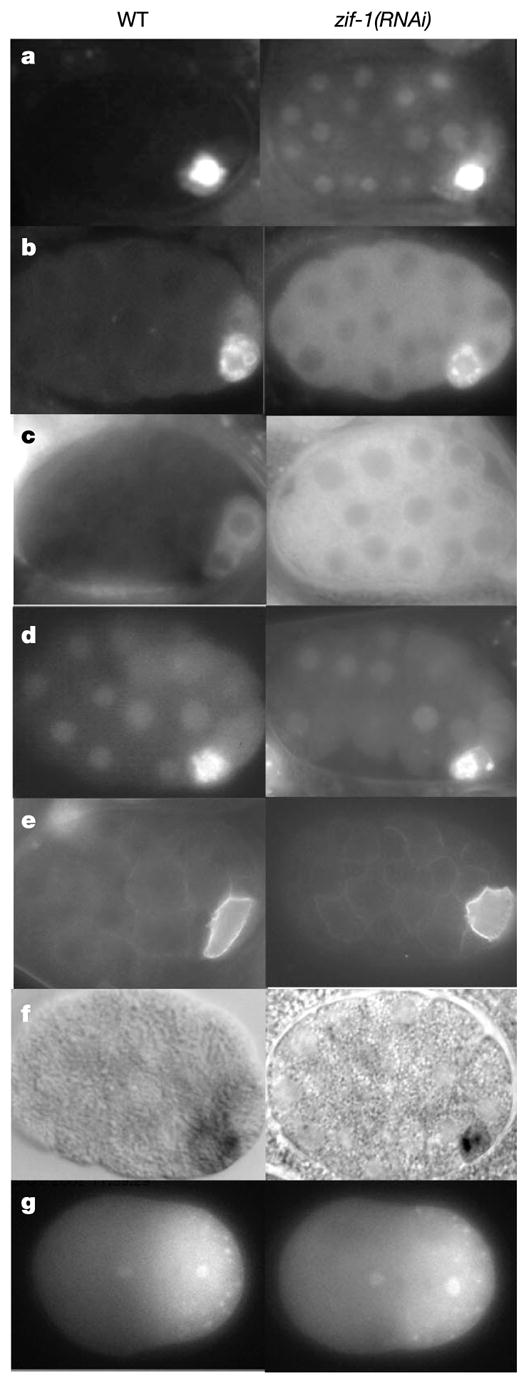
Depletion of ZIF-1 blocks degradation of CCCH finger proteins in somatic cells, but does not affect other soma–germline asymmetries. a–f, 28-cell wild-type and zif-1(RNAi) embryos expressing PIE-1::GFP (a), POS-1 (b, immunofluorescence5), GFP::MEX-5 (c), GFP::PIE-1 with ZF1 deleted (d), GFP::PAR-2 (e) and pie-1 RNA (f, in situ hybridization27). Measurement of GFP fluorescence (IP lab software) in ABa (soma) confirmed that PIE-1::GFP levels are higher in zif-1(RNAi) compared with wild type (t-test; P = 0.009), but do not change significantly in P2 (germ line) (t-test; P = 0.25, N = 10 four-cell embryos for each genotype). g, One-cell wild-type and zif-1(RNAi) embryos expressing a PIE-1::GFP fusion. In later stages, the zif-1(RNAi) embryo failed to degrade PIE-1::GFP in somatic blastomeres (see ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003), confirming the efficient inactivation of zif-1.
ZIF-1 does not share obvious homology with other proteins in the databases. To investigate the mechanism of action of ZIF-1, we conducted another yeast two-hybrid screen to identify potential ZIF-1 interactors (Methods). As expected, we recovered several isolates of PIE-1, POS-1, MEX-1, MEX-5 and MEX-6. In addition, we recovered two isolates of Y82E9BR.15. Sequence analysis of Y82E9BR.15 revealed that it encodes the C. elegans homologue of vertebrate elongin C (Supplementary Information). In human cells, elongin C complexes with the ubiquitin-like protein elongin B, the cullin Cul2 and the RING finger protein Rbx1 to form a multi-subunit E3 ubiquitin ligase (ECS ligase13). When bound to the substrate-recruitment subunit von Hippel-Lindau (VHL), ECS ligase targets hypoxia inducible factor (HIF1α) for ubiquitination and subsequent degradation by the proteasome. Elongin C binds to VHL through a conserved motif (SOCS box) located near the carboxy terminus of VHL. Mutations in this sequence in von Hippel-Lindau patients prevent binding of VHL to elongin C and lead to abnormal stabilization of HIF1α14. ZIF-1 contains a potential SOCS box in its C terminus (Fig. 3). Mutation of a conserved cysteine in the SOCS box blocked ZIF-1’s ability to interact with elongin C in the yeast two-hybrid assay, without affecting binding to PIE-1 (Fig. 4a). This finding suggests that, similar to VHL, ZIF-1 may function as a substrate-recruitment subunit for an ECS ligase (Fig. 4b). If so, elongin C, CUL-2, RBX-1 and the associated E2 ubiquitin-conjugating enzyme UBC5 should all be required for ZF1-dependent degradation. Complete elimination of these proteins interferes with cell divisions15 (J. Liu, S. Vasudevan and T. Kipreos, submitted; and C.D., unpublished work); however, partial depletion by RNAi yields multicellular embryos that can be examined for CCCH finger degradation. In all cases, we found that degradation of PIE-1::GFP and GFP::PIE-1ZF1 fusions was blocked in these embryos (Fig. 4c and data not shown). This block was not due to general loss of embryonic polarity or cell viability, as the distribution of GFP::PGL-1 was unaffected (Fig. 4c). Depletion of another cullin (CUL-3) or another E2 (UBC-12) did not interfere with GFP::PIE-1ZF1 degradation (Supplementary Information), confirming the specific involvement of CUL-2 and UBC5. We conclude that elongin C, CUL-2, RBX-1 and UBC5 are required with ZIF-1 to degrade CCCH finger proteins in somatic cells.
Figure 3.
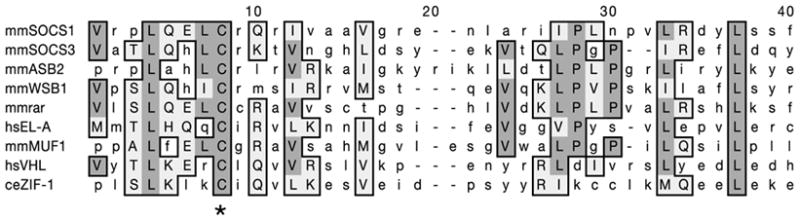
ZIF-1 is a SOCS-box protein. Alignment of the ZIF-1 SOCS box with SOCS boxes from other elongin C-interacting proteins13. Asterisk indicates cysteine mutated in Fig. 4a. ce, C. elegans; hs, Homo sapiens; mm, Mus musculus.
Figure 4.
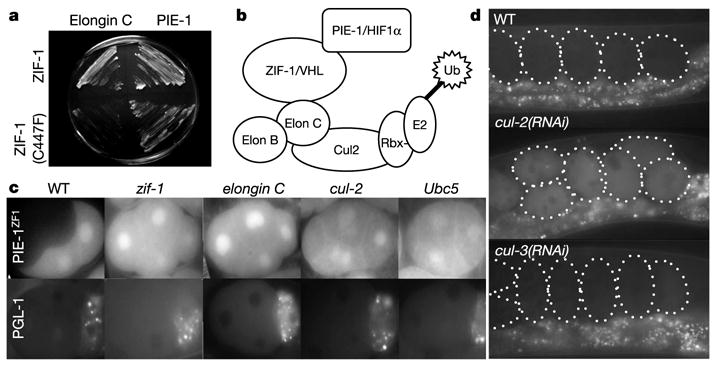
ZIF-1 may function as a substrate-recruitment factor for an elongin C/CUL-2 E3 ubiquitin ligase. a, Yeasts expressing ZIF-1, or ZIF-1 with a SOCS mutation, and elongin C or PIE-1 as indicated. Growth indicates interaction. b, Diagram after ref. 28 depicting ECS ligase with its associated E2, substrate-recruitment subunits (ZIF-1/VHL) and substrates (PIE-1/HIF1α). c, Four-cell embryos expressing GFP::PIE-1ZF1 or GFP::PGL-1 and partially depleted by RNAi for the indicated genes. Visualization of PGL-1 and GFP::PIE-1ZF1 in the same cul-2(RNAi) embryos confirmed that partial cul-2 depletion blocks degradation without affecting overall polarity (data not shown). d, Live embryos (outlined) expressing GFP::ZIF-1, driven by the pie-1 promoter (Methods), in wild-type mothers or mothers exposed to cul-2 or cul-3 double-stranded RNA. Punctate fluorescence below the embryos is gut autofluorescence from the mother.
F box proteins—the substrate recruitment subunits for the SCF class of ubiquitin ligases—are unstable in vivo and are themselves subject to ubiquitination in a SCF-dependent manner16–18. Similarly, we found that a GFP::ZIF-1 fusion is unstable in vivo, and is stabilized when CUL-2, but not CUL-3, is inactivated (Fig. 4d; see also Supplementary Information). These observations are consistent with ZIF-1 acting as a substrate recruitment factor for a CUL-2-containing E3 ligase (Fig. 4b); however, biochemical studies will be needed to confirm this hypothesis.
ZIF-1-dependent degradation is restricted to somatic cells, as zif-1(RNAi) stabilizes PIE-1::GFP and GFP::PIE-1ZF1 levels in somatic cells without changing levels in germ cells10 (Fig. 2; see also ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003). We found that this specificity is dependent on MEX-5 and MEX-6, the redundant CCCH finger proteins that accumulate transiently in somatic blastomeres (Fig. 1a). In mex-5(RNAi);mex-6(RNAi) embryos, GFP::PIE-1ZF1 remained stable in all blastomeres, indicating that MEX-5 and MEX-6 are required for ZIF-1-dependent degradation (Fig. 5). MEX-5 and MEX-6 levels are kept low in germline blastomeres by PAR-1, a serine threonine kinase that segregates with the germ lineage (Fig. 1a) and is essential for asymmetric divisions19. In par-1(RNAi) embryos, MEX-5 and MEX-6 are present at high levels in all early blastomeres6 and degradation of GFP::PIE-1ZF1 was activated in all cells (Fig. 5). This ubiquitous degradation requires MEX-5 and MEX-6, as well as ZIF-1 and CUL-2 (Fig. 5). Consistent with MEX-5 and MEX-6 triggering CCCH finger protein degradation, heat-shock-induced expression of MEX-5 in somatic blastomeres of mex-5(zu199); mex-6(RNAi);pie-1(RNAi) embryos correlates with loss of MEX-1 expression in those cells6. We conclude that high levels of MEX-5 and MEX-6 activate ZIF-1-dependent degradation in somatic blastomeres, and that PAR-1 blocks ZIF-1-dependent degradation in germline blastomeres, at least in part by keeping MEX-5 and MEX-6 levels low (Fig. 5b). Our data indicate that MEX-5 and MEX-6 are both activators and targets of ZIF-1-dependent degradation. We do not know whether MEX-5 and MEX-6 activate ZIF-1-dependent degradation directly, through their binding to ZIF-1, for example, or indirectly by regulating another factor. PAR-1, MEX-5 and MEX-6 are also required for the initial asymmetric partitioning of PIE-1, POS-1 and MEX-1 in germline blastomeres6, but the mechanism involved in that process is not known, and is distinct from the one described here as it does not require ZIF-1 (Fig. 2g) or PIE-1ZF1 (Fig. 2d; see also ref. 10).
Figure 5.
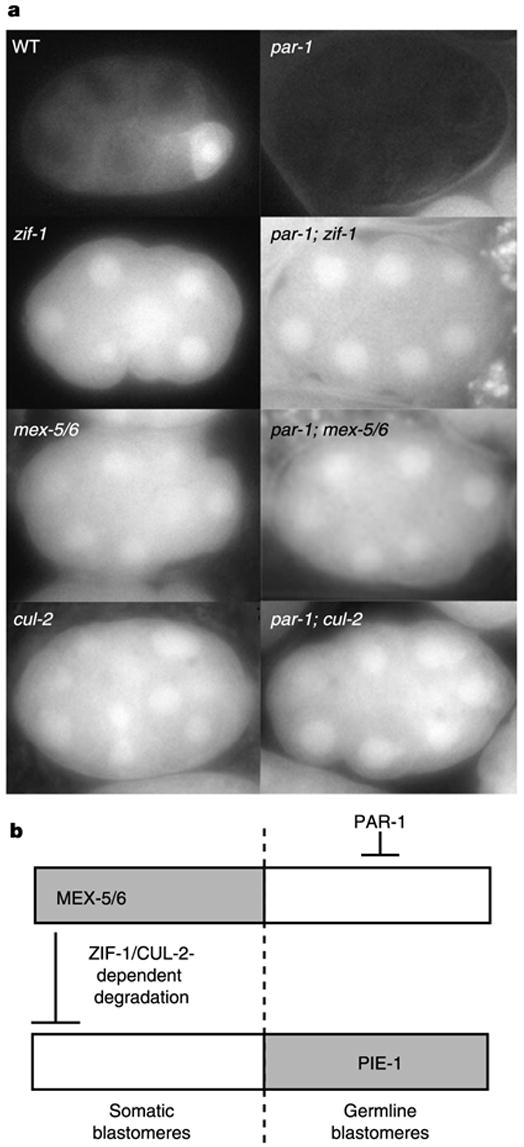
PAR-1 inhibits, and MEX-5 and MEX-6 activate, ZIF-1-dependent degradation. a, 12/15-cell embryos expressing GFP::PIE-1ZF1 with genes inactivated by RNAi. GFP::PIE-1ZF1 is degraded in somatic blastomeres in wild type, and in all cells in par-1(RNAi) embryos. Ubiquitous degradation in par-1(RNAi) requires ZIF-1, MEX-5, MEX-6 and CUL-2. Similar results were obtained with PIE-1::GFP (Supplementary Information). b, Schematic summarizing the epistatic relationships deduced from a and ref. 6.
Our findings demonstrate that restriction of CCCH finger proteins to the germ line requires degradation in somatic lineages. Instability outside of the germ plasm has been observed for other germ plasm components, including P granules in C. elegans20, Oskar protein in Drosophila21 and Vasa protein in zebrafish22. Consistent with our findings, stabilization of Oskar requires PAR-1 (ref. 21). We propose that degradation in the soma coupled with PAR-1-dependent protection in the germ line is a commonly used mechanism to restrict germ plasm components to the germ lineage. Our studies implicate the ECS family of E3 ubiquitin ligases13 in the degradation of CCCH finger proteins. The recent identification of Gustavus, a SOCS-box protein required for Vasa localization in Drosophila23, raises the possibility that ECS E3 ligases also regulate other germ plasm components.
Methods
GFP fusions
GFP fusions were driven by pie-1 promoter (and 3′ UTR)24, which is active during oogenesis (maternal expression). To determine which MEX-5 finger is sufficient for degradation, MEX-5ZF1 (QPPNYKTRLCMMHASGIKPCDMGARCKFAHGLKELRAT) and MEX-5ZF2 (PNNKYKTKLCKNFARGGTGFCPYGLRCEFVHPTDKEFQN) were fused singly to GFP. MEX-5ZF2 (ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003), but not MEX-5ZF1 (data not shown), was sufficient to target GFP for degradation. Localization dynamics were analysed by time-lapse microscopy as in ref. 9. Movies of protein localization in embryos can be viewed at ftp://ftp.wormbase.org/pub/wormbase/datasets/derenzo_2003.
Yeast two hybrid
PIE-1ZF1–ZF2 (amino acids 92–217) or ZIF-1 (F59B2.6) was fused to GAL-4 DNA-binding domain (pAS1-CYH1) and transformed into yeast PJ69-4A. Caernorhabditis elegans Gal4 activation domain library (a gift from Z. Zhao and B. Horvitz) was used as prey using standard procedures25. Approximately 600,000 transformants were screened in each screen.
RNAi
RNA interference was performed using the feeding method26. For zif-1(RNAi), Escherichia coli expressing zif-1 double-stranded RNA was plated on IPTG-containing plates for 4 h, before adding L4 hermaphrodites. Embryos were examined 27 h later; 95–100% of embryos had mislocalized PIE-1::GFP and 80–100% did not hatch (embryonic lethality).
Supplementary Material
Acknowledgments
We dedicate this study to the memory of D. Nathans. We thank E. Kipreos for critical reading of the manuscript; Z. Zhao and B. Horvitz for the yeast two-hybrid library; Y. Kohara for the POS-1 antibody; B. Aman and J. Berg for many discussions about CCCH fingers; and A. Cuenca for help with the movies. This work was supported by a NIH grant and by the Steve and Michelle Kirsch Foundation.
Footnotes
Competing interests statement The authors declare that they have no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 2.Kloc M, et al. RNA localization and germ cell determination in Xenopus. Int Rev Cytol. 2001;203:63–91. doi: 10.1016/s0074-7696(01)03004-2. [DOI] [PubMed] [Google Scholar]
- 3.Guedes S, Priess JR. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- 4.Mello CC, et al. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 5.Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 7.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 8.Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- 9.Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Detwiler MR, Reuben M, Li X, Rogers E, Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1:187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- 13.Kile BT, et al. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 14.Kamura T, et al. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H, et al. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nature Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 17.Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirbelauer C, et al. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 20.Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- 21.Riechmann V, Gutierrez GJ, Filardo P, Nebreda AR, Ephrussi A. Par-1 regulates stability of the posterior determinant Oskar by phosphorylation. Nature Cell Biol. 2002;4:337–342. doi: 10.1038/ncb782. [DOI] [PubMed] [Google Scholar]
- 22.Wolke U, Weidinger G, Koprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289–294. doi: 10.1016/s0960-9822(02)00679-6. [DOI] [PubMed] [Google Scholar]
- 23.Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–876. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- 24.Strome S, et al. Spindle dynamics and the role of gamma-tubulin in early C. elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 26.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 27.Seydoux G, Fire A. Whole-mount in situ hybridization for the detection of RNA in Caenorhabditis elegans embryos. Methods Cell Biol. 1995;48:323–337. doi: 10.1016/s0091-679x(08)61394-1. [DOI] [PubMed] [Google Scholar]
- 28.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


