Abstract
In this article we describe two new complete genomic sequences of Old World Arenaviruses: the Mopeia (MOP) virus and the reassortant MOP/LAS virus, clone 29, or ML29. This reassortant has the large (L) RNA from MOP virus and the small (S) RNA from Lassa (LAS) virus, Josiah strain. Recent studies showed that the ML29 virus is not pathogenic for mice, guinea pigs, or macaques, can completely protect guinea pigs from Lassa virus, and elicit vigorous cell-mediated immunity in immunized monkeys (Lukashevich, I. S., Patterson, J., Carrion, R., Moshkoff, D., Ticer, A., Zapata, J., Brasky, K., Geiger, R., Hubbard, G. B., Bryant, J., and Salvato, M. S., J Virol 79, 13934–13942, 2005). This is a molecular characterization of a reassortant virus, which has been put forward as a live attenuated vaccine candidate against Lassa Fever. Sequence analysis of this reassortant virus revealed 5 non-conservative amino acid substitutions that distinguished it from the parental LAS and MOP viruses. Three substitutions were found outside the conserved RNA-dependent RNA polymerase (RdRp) motifs. A fourth substitution was located between the glycoprotein (GPC)-cleavage site and the putative fusion peptide of GP2. The nucleocapsid protein (NP) contained a fifth substitution in the carboxyl-terminal region of the protein. Two mutations were found within each non-coding terminus of the L segment and one mutation was located in the 3′ non-coding region of the S segment of the MOP/LAS virus. ML29 mutations in its genomic termini may have implications for the genetic stability and replication efficiency of ML29 reassortant.
Keywords: Arenavirus, Lassa virus, Mopeia virus, Bi-segmented genome, Reassortant virus
Introduction
Mopeia (MOP) virus, once known as Mozambique virus, was discovered in 1977 [1] and is a negativestrand RNA virus in the Arenaviridae family, related to other Old World arenaviruses like Lassa (LAS), Mobala, Ippy, and lymphocytic choriomeningitis virus (LCMV) [2]. It is carried by the African multi-mammate rat, Mastomys natalensis, widely spread in sub-Saharan Africa [3]. In contrast to its West African relative, LAS virus, MOP virus causes no lethal disease in guinea pigs or primates and can be used to vaccinate against LAS virus [4].
ML29 virus was generated in cell culture by segment reassortment between the AN20410 strain ofMOPvirus and the Josiah strain of LAS virus. It has the large (L) RNAfromMOPvirus and the small (S)RNAfrom LAS virus [5, 6]. ML29 has a small plaque phenotype similar to MOP virus and is non-pathogenic for newborn mice similar to LAS virus. The ML29 clone, encoding the replication machinery of MOP and the antigenic determinants of LAS virus was chosen for further characterization as a potential vaccine. Our recent studies showed that guinea pigs vaccinated with ML29 were fully protected from lethal challenge with LAS virus and protected animals did not manifest any clinical or biochemical abnormalities [7]. TheML29 does not cause any detectable clinical manifestations in immunized monkeys and shares epitopes with LAS virus that are responsible for eliciting a fully protective response to LAS virus [7]. Earlier studies with rhesus macaques had also shown the efficacy ofMOPas a vaccine against LAS virus in which the vaccinated animals developed a mild fever and a brief and low-titer viremia [8]. In strain 13 guinea pigs MOP vaccination protected animals against the death. However, in contrast to ML29-vaccinated animals, MOP-protected guinea pigs had functional liver alterations after LAS challenge [7]. Hybridization of RNA from human PBMC infected with ML29 and MOP revealed differences between the two viruses in DNA array analysis. The ML29 gene expression profile was more similar to the profile of normal cells than to MOP-infected cells (Lukashevich, I. S., and Salvato, M. S., in press).
All arenaviruses have similar biochemistry and morphology. They are enveloped and the typical Arenavirus genome consists of two single-stranded RNA molecules, a large RNA (L, 7.2 kb), and a small RNA (S, 3.4 kb) [9]. Each of the RNA segments has an ambisense coding strategy, and genes are separated by a non-coding intergenic region: on its 5′ end, the S genome segment encodes a surface glycoprotein precursor (GPC) that undergoes cleavage-maturation to become structural glycoproteins GP-1 and GP-2, and on its 3′ end the S segment encodes the nucleocapsid protein (NP). The large (L) segment encodes a small zinc-binding (Z) protein on its 5′ end, and it encodes the viral RNA-dependent RNA polymerase (RdRp or L protein) on its 3′ end.
Here we describe and compare the L and S RNA sequence of MOP virus AN20410 (GenBank acc. No. NC_006574, NC_006575) and the L and S RNA sequences of the reassortant MOP/LAS virus, clone 29 (GenBank acc. No. NC_006572, NC_006573). We speculate on the impact of changes in the genomic termini on virus replication and on the possible adaptive changes in the viral polymerase that could allow it to transcribe heterologous termini.
Materials and methods
Viruses a2nd cells
MOP virus strain AN20410, isolated from Mastomys natalensis in Mozambique, was kindly provided by Dr. G. van der Groen (Institute of Tropical Medicine, Antwerpen, Belgium) to Dr. I. S. Lukashevich and had three passages on Vero E6 cells, before cloning and sequencing. Mopeia/Lassa reassortants were generated by Dr. I. S. Lukashevich, Vasiuchkov A. D., Stel’makh T. A., Scheslenok E. P., and Shabanov A. G. (Department of Special Pathogens, Byelorussian Research Institute of Epidemiology and Microbiology, Minsk, Belarus) as previously described [5]. The MOP/LAS clone 29 or ML29 was isolated from a stock of reassortants, had been three times plaque-purified and propagated one passage in BHK-21 and seven passages in Vero E6 cells (C1008 ATCC, Cat. #CRL-1586, Lot #1618471, 3-22-01 and Lot #3546415, 2-2-04). The seventh passage constituted the master seed stock for vaccine work and molecular characterization. ML29 and MOP viruses were grown on Vero E6 cells, cultured in EME Media with 2% fetal bovine serum (FBS), 1 × antibiotic and antimycotic (Invitrogen Corporation, USA, Cat. No. 15240) at 37°C in a 5% CO2 atmosphere. The culture supernatant was changed every 24 h after infection, and at 72 h after infection maximum viral yield was determined by plaque assay. TheML29 genotype was initially identified by dot-blot hybridization to strand specific probes [5], by RT-PCR (Fig. 1), and also confirmed by sequence analysis (the current publication).
Fig. 1.
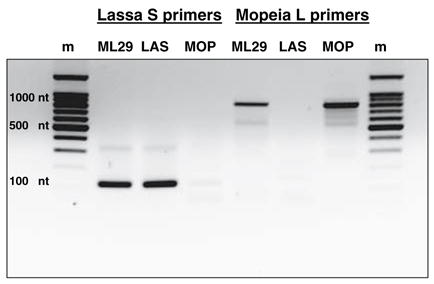
Genotyping of the ML29 virus. RNA from cells infected with ML29 or with parental viruses, MOP, or LAS was reverse transcribed and amplified with MOP L- or with LAS S-specific primers; m, markers, 100 nt DNA ladder
Plaque titration was done on Vero E6 cells (same ATCC Cat. No. as above but Lot F-15030-1999). These cells were used specifically for plaque assay and needed 5 rather than 1 min of trypsin exposure. Under crystal violet staining ML29 generated clear plaques, similar to those generated by LAS virus. MOP virus (AN20410) generated dark-staining plaques in this Vero cell line. Using this method it was possible to distinguish between the two viruses by plaque morphology. The Vero cells and the viral strains were all mycoplasma-free.
Virus concentration and RNA extraction
For all sequencing work, virus was first concentrated from Vero cell culture supernatants. Around 15 ml of cell culture supernatant collected at 72 h were concentrated 100 times using Amicon Ultra 100,000 MWCO centrifugal filter devices (Cat. No. UFC910024). RNA was extracted from 150 μl of concentrated supernatant using the QIAamp viral RNA mini kit (Qiagen Cat.No. 52904).
cDNA synthesis and amplification of viral sequences
For reverse transcription of the full-length L and S viral RNA of viruses we used Superscript III first strand synthesis system from Invitrogen (Cat. No. 18080-051). Mix 1 was incubated with MopRT primer (Table 1) in a 50-μl volume at 65°C for 5 min. The mix contained 7 μl (equivalent of 5.1 × 107 PFU) of ML29 or MOP virus RNA, 1 μl of 1 μM MopRT primer, 5 μl of 10 μM dNTP’s, 38 μl DEPC water bringing the total volume to 50 μl. The mixture was quickly chilled on ice for 5 min. Meanwhile “cDNA synthesis mix” had been made: 10 μl 10 × RT buffer, 20 μl 25 mM MgCl2, 10 μl 0.1 M DTT, 5 μl RNase OUT, and 5 μl Superscript III RT, at a total volume of 50 μl. After 5 min of incubation on ice, Mix 1, and the cDNA synthesis mix were combined together and incubated for 1 h at 50°C and 15 min at 85°C. Then 2 μl RNase H were added and incubated for 20 min at 37°C. The synthesized cDNAs were stored at −20°C or processed immediately for PCR.
Table 1.
Primers for RT and PCR amplification
| Primer name | Sequence 5′-3′ | Length | Position |
|---|---|---|---|
| MopRT | CGCACAGTGGATCCTAGGC | 19 | L 7271–7253 |
| ML29RT | CGCACCGGGGATCCTAGGC | 19 | L 7271–7253 |
| LassaS3NTR | CGCACAGTGGATCCTAGGCTATTGGATTG | 29 | S 3402–3374 |
| MOP0/20-5p | GTGGATCCTAGGCTTTTTGG | 20 | L 7–26 |
| LAS5/25-5p | ACCGGGGATCCTAGGCATTT | 20 | L 2–23 |
| UNi20Mop | CGCACAGTGGATCCTAGGCA | 20 | L 7271–7252 |
| MVP1 | CTTATGGTGGAAGTCTGAAACAAAGGCCATT | 31 | L 5221–5251 |
| MVP2 | ATCTGATCTGTCAACCTATCTGGAGTTTC | 29 | L 5021–5049 |
| LasS3NTR | CGCACAGTGGATCCTAGGCTATTGGATTG | 29 | S 3402–3374 |
For amplifying the L segment for both viruses we used Expand High Fidelity PCR System (Cat. No. 1 732 641) from Roche Diagnostics GmbH. Primers used for PCR were: MopRT, for reverse transcription, and MOP0/20-5p and LAS5/25-5p, for amplification (Table 1). The thermo cycler was programed for 95°C for 2 min one cycle, 95°C for 30 s, 55°C for 1 min, 72°C for 3 min, repeated for 45 cycles, then 72°C for 10 min, and finally 4°C. Amplicon bands (Fig. 2A) were extracted from gels, purified by QIAquick gel extraction Kit from QIAGENMDUSA(Cat. No. 28704) and used for direct sequencing or for cloning. For amplifying the remaining 3′ ends of MOP and ML29 viruses we used two different primer sets which overlapped the sequenced areas, UNi20Mop, MVP1, and UNi20Mop/MVP2 (Table 1). The thermo cycler was programed for 95°C, 2 min one cycle: 95°C for 30 s, 55°C for 1min, 7°C for 2 min, repeating for 45 cycles, then 72°C for 10 min, and ending with 4°C. Amplicon bands were again extracted from gels, purified by the QIAquick Kit, and used for sequencing or cloning (Fig. 2B).
Fig. 2.

Gel electrophoresis of amplified and purified L and S segments. A, 5′ portion (5.3 kilobases) of L segments; B, 3′ portion (2.1 kilobases) of L segments; C, full length S segments
Using the same strategy as described above, we used two different primers sets to generate full S-segments, one for MOP virus strain AN20410 and one for LAS virus Josiah strain. For the S segment of MOP virus we used primers called Mop0/20-5p and MopRT. The Cycler program was 94°C for 2 min and (94°C for 30 s, 50°C for 45 s, and 72°C for 3 min) repeating for 45 cycles, then 7 min at 72°C and 4°C, to finish. For the S segment of ML29 we used LassaS3NTR and LAS5/25-5p primers (Table 1). The Cycler program was 94°C for 2 min and (94°C 30 s, 57°C 45 s, 72°C 3 min) repeating for 45 cycles, then 7 min at 72°C and 4°C to finish. Amplicon bands were again extracted from gels and purified by the QIAquick Kit (Fig. 2C).
Cloning the sequencing
Cloning used the Topo XL-PCR cloning kit from Invitrogen (Cat. No. 45-0008), according to the manufacturer’s instructions. Clones were analyzed by EcoRI digestion and correct clones were used for sequencing. We finally obtained overlapping clones of 5.3 and 2.1 kilobases for the L segments of both viruses. We did not succeed to clone the full S-segments and generated three overlapping clones for each virus to complete sequence of S segments.
Sequence was generated by direct sequencing of PCR fragments and by sequencing cloned fragments. Final results were obtained from two independent clones from both strands or from two independent PCR reactions and reconfirmed by the use of walking primers (sequences are available upon request). For verification of 5′ and 3′ end nucleotide sequences of MOP and ML29 viruses PCR amplification reactions contained MopRT and ML29RT primers. DNA amplicons and cloned fragments were used for sequencing. The sequences were analyzed using Vector NTI Advance 9.0 software from InforMax (Invitrogen).
Results
In the present paper we describe genomic sequences of two viruses: MOP virus (AN 20410) and MOP/LAS reassortant, ML29. The complete nucleotide sequences of these viruses were submitted to Genbank. The sequence of the other parental virus, LAS-Josiah, has been published previously [10–12]. Here we discuss the relationship of the reassortant to the parental viruses and the implications of the few nucleotide changes detected in the progeny virus.
The genotype of ML29 virus was confirmed by RT/PCR amplification with MOP-L- and LAS-S-specific primers (Fig. 1). Genome segments of ML29 were amplified and purified for cloning and sequencing as showed in Fig. 2. The L RNAs of both viruses, ML29 and MOP, are 7271 nucleotides in length. Similar to the parental MOP virus, the ML29 L RNA has 60% homology with LAS-Josiah L RNA, and 57% amino acid homology for both L and Z proteins. The ML29 virus L segment differs from the MOP sequence by 12 nucleotides (Table 2). Mutations in both segments were mostly A to G and U to C transitions. Most of them seem to be derived from viral RdRp “mistakes” during MOP/LAS replication in tissue culture after isolation of the primary ML29 virus clones. Transversions A6C, U8G and A7264C, U7266G were located at 5′ and 3′ termini of the L RNA and contributed to the stability of the predictable secondary structure. The 5′ and 3′ terminal conservative regions form specific “panhandle” structures involved in the replication and transcription (see below).
Table 2.
Mutations in genomic segments of attenuated MOP/LAS reassortant (clone ML29)
| Nucleotide | L RNA position | ORF/NCR | Nucleotide | S RNA position | ORF/NCR |
|---|---|---|---|---|---|
| A/C | 6 | 5′ | A/G | 869 | GPC |
| U/G | 8 | 5′ | U/C | 1366 | GPC |
| C/U | 810 | RdRp (L) | G/A | 1842 | NP |
| G/A | 1423 | RdRp (L) | G/U | 1849 | NP |
| U/C | 3319 | RdRp (L) | U/C | 2785 | NP |
| U/C | 3519 | RdRp (L) | G/A | 3328 | 3′ |
| U/C | 3664 | RdRp (L) | |||
| G/A | 4002 | RdRp (L) | |||
| U/C | 4177 | RdRp (L) | |||
| A/U | 4665 | RdRp (L) | |||
| A/C | 7264 | 3′ | |||
| U/G | 7266 | 3′ | |||
| Amino acid | Position | RdRp | Amino acid | Position | GPC/NP |
| Y/N | 851 | RdRp (L) | K/E | 272 | GPC (GP2) |
| R/G | 1233 | RdRp (L) | A/D | 485 | NP |
| D/N | 2136 | RdRp (L) | N/S | 173 | NP |
ORF, open reading frame; NCR, 5′ and 3′ non-coding regions; RdRp, RNA-dependent RNA polymerase (L protein); GPC, glycoprotein precursor; NP, nucleocapsid protein. Non-conservative amino acid substitutions are underlined
The L RNA encodes Z and L protein (RdRp). The Z protein is 103 amino acids long with a molecular weight of 11,646 kDa for both viruses, and the L protein is 2238 amino acids for both viruses with a molecular weight of 256,187 kDa for MOP virus, and 256,336 kDa for ML29. Biochemical parameters of predicted L proteins of both viruses were very close and similar with those described for LAS L protein [11]. Mutations in the coding region of the L RNA resulted in three non-conservative amino acid substitutions at positions 851 (Tyr → Asn), 1233 (Arg → Gly), and 2136 (Asp → Asn), as shown in Fig. 3. Only the R1233G mutation was located within the putative RdRp domain and mapped between motif A, Asp-X(2)-Lys-Trp, and motif B, Gly-X(5)-Ser (11). These motifs represent stretches of highly conserved sequences that are linked by less conserved variable regions [11].
Fig. 3.
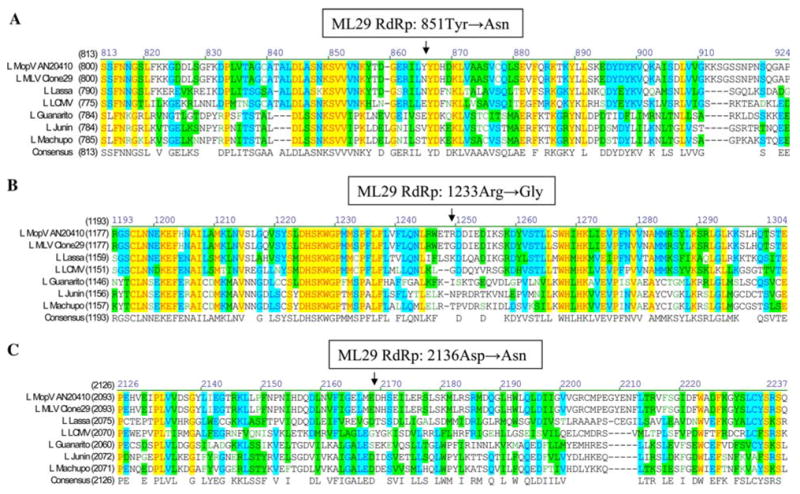
Location of amino acid substitutions detected in RdRp (L protein) of ML29 reassortant. Mutations in the coding region of the L RNA resulted in 3 non-conservative amino acid substitutions located outside the highly conserved motifs of at positions: A, 851 (Tyr → Asn); B, 1233 (Arg → Gly); C 2136 (Asp → Asn). GenBank accession numbers: MOP AN20410, NC_006574; ML29 reassortant, NC_006572; LAS-Josiah, NC_004297; LCMV-ARM, DQ361066; Guanarito, NC_005082; Junin, NC_005080; Machupo, NC_005079
This paper describes the first MOP-AN20410 virus L RNA sequence, the S genomic segments of two other MOP strains have already been published. MOP virus AN20410 S RNA sequence has a 3427 nucleotide length and is eight nucleotides longer than MOP virus AN21366 strain (access. No. M33879) [13] and 43 nucleotides longer than the Mozambique strain (access. No. DQ328874) [14] with 79% nucleotide homology. ML29 S RNA is exactly the same length, 3402 nucleotides, as LAS-Josiah strain.
The SRNAof ML29 was derived from LAS virus and differs in sequence by 6 mutations (Fig. 4). These mutations resulted in three amino acid substitutions. The first non-conservative substitution, K272E, was located in the amino-terminal region of GP2 glycoprotein between GPC cleavage motif and the putative fusion peptide of arenaviruses [15]. This region is very conserved and all strains of LAS virus sequenced so far had amino acid substitutions only within the KDT tri-peptide (Fig. 4). The second amino acid substitution was homologous, N173S, and mapped in the central region of NP. The second non-conservative substitution, A485DA is located in the carboxyl-terminal region of NP. The S segment of ML29 also had a G3328A mutation in the 3′ non-coding region (Table 1).
Fig. 4.
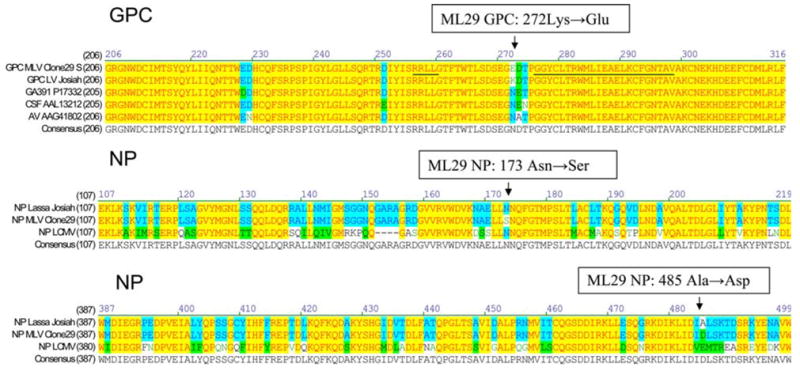
Location of amino acids substitutions in GPC and NP proteins of ML29 reassortant. A non-conservative substitution, K272E, is located in the amino-terminal region of GP2 glycoprotein between GPC cleavage motif (RRLL, underlined) and the putative fusion peptide of arenaviruses (GGYCLTRWMLIEAELKCFGNTAV, underlined). A conservative mutation, N173S, is mapped to the central region of NP protein. A non-conservative substitution, A485D, is located in the carboxyl-terminal region of NP protein. GenBank accession numbers: ML29 reassortant, NC_006573; LAS-Josiah, NC_004296; LAS-GA391, X52400; LAS-CSF, AF333969; LASAV, AF246121; LCMV-ARM, NC_004294
Discussion
It is most likely that the attenuated phenotype of ML29 is associated with MOP L RNA. The mutations that were introduced in the L segment of MOP/LAS virus during LAS-MOP co-cultivation and/or during subsequent selection steps can additionally contribute to the MOP/LAS phenotype. Reassortment experiments on LCMV [18] and Pichinde virus [19] strains indicate that virulence of pathogenic strains of these viruses is associated with the L segment.
Amongst the six mutations detected in the S segment, the A869G mutation resulted in a substitution Lys to Glu at position 272 within the amino-terminal region of GP2 glycoprotein. This region is very conserved among arenaviruses and a stretch of 24 amino acids at positions 276–299 (numbering according to the sequence of GPC, LAS-Josiah) seems to be involved in fusion activity between cellular membrane and viral surface. The fusion domain also overlaps with a human T-helper cell epitope, which is highly conserved in all LAS-LCMV strains, and shows more than 90% similarity in all New World arenaviruses of clade B [20]. The K272E substitution is located between the fusion domain and the RRLL motif, the cellular subtilase SKI-1/S1P cleavage site [15]. Interestingly, all LAS strains analyzed so far had mutations in this spot between amino acid residues 272–274, KDT for LAS-Josiah GP2 (Fig. 4). Substitution Lys to Glu introduces two negatively charged R groups in positions 272–273 (Asp-Glu) distinguishing the MOP/LAS reassortant from other LAS virus strains so far analyzed.
A non-conservative, Ala to Asp substitution, was found at the position 485 outside of a zinc finger-like domain [21] located in the carboxyl-terminal region of arenavirus NP. A homologous, N173S, amino acid substitution was located in the middle-region of NP and seemed to be not involved in NP–RNA interaction. The non-coding region preceding the NP open reading frame contained a G3328A nucleotide mutation. Is not clear how S RNA-derived mutations contribute to the MOP/LAS attenuated phenotype. The data on protective activity clearly indicates that major LAS virus antigens encoded by the S segment of the MOP/LAS reassortant induce protective and partially cross-reactive immune responses [7].
Models for Arenavirus replication seek to explain the maintenance of one copy of each genome segment in each virion particle. One idea [22], is that the genomic termini preferentially associates such that the 3′ end of the S RNA anneals to the 5′ end of the L RNA and the 3′ end of the L RNA preferentially anneals to the 5′ end of the S RNA. Preferential annealing of an L segment to an S segment would result in the preferential packaging of L/S complexes over L/L or S/S complexes. The simple prediction that the heterologous termini pair with lower negative free energy than the homologous termini would insure preferential annealing of heterologous segments. We predicted the secondary structure for the first and last 27 nucleotides of ML29 by genetic algorithm [23]. The ML2 virus has mutations in its genomic termini that render its MOP segment more similar to LAS virus at these termini. As seen in Fig. 5, these mutations improve the ability of L (MOP) and S (LAS) segments to associate in comparison to the parental viruses.
Fig. 5.
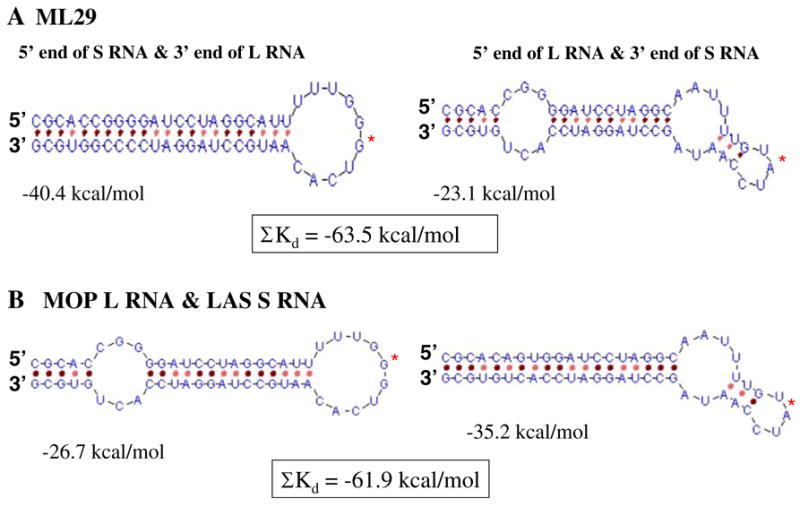
The terminal RNA secondary structure predicted by a genetic algorithm (http://wwwmgs.bionet.nsc.ru/mgs/programs/2dstructrna). The fusion area is marked by a red star. Total energy for terminal structures of ML29 RNA and terminal structures formed by MOP L and LAS S segments are boxed
Acknowledgments
The authors wish to thanks Dr. Robert Powell for excellent assistance in BSL3 laboratory, Dr. C. David Pauza for comprehensive discussion of results and Dr. Lisa Sadzewicz for clone sequencing assistance. This work was supported by NIH grants R21-AI52367, RO1-AI52367 and Middle Atlantic Regional Center of Excellence (MARCE) in Biodefense and Emerging Infectious Diseases Research to Dr. Igor S. Lukashevich.
Footnotes
The nucleotide sequence data for the of the S and L RNAs of the ML29 and Mopeia virus strains reported in this paper have been submitted to the GenBank nucleotide sequence database and have been assigned the accession numbers: NC_006572-75.
References
- 1.Wulff H, McIntosh BM, Hamner DB, Johnson KM. Bull World Health Organ. 1977;55:441–444. [PMC free article] [PubMed] [Google Scholar]
- 2.Salvato MS, Clegg JCS, Buchmeier MJ, Charrel RN, Gonzalez J-P, Lukashevich IS, Rico-Hesse R, Romanowski V. In: Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Elsevier/Academic Press; London: 2005. pp. 725–733. [Google Scholar]
- 3.Johnson KM, Taylor P, Elliott LH, Tomori O. Am J Trop Med Hyg. 1981;30:1291–1293. doi: 10.4269/ajtmh.1981.30.1291. [DOI] [PubMed] [Google Scholar]
- 4.Walker DH, Johnson KM, Lange JV, Gardner JJ, Kiley MP, McCormick JB. J Infect Dis. 1982;146:360–368. doi: 10.1093/infdis/146.3.360. [DOI] [PubMed] [Google Scholar]
- 5.Lukashevich IS, Vasiuchkov AD, Stel’makh TA, Scheslenok EP, Shabanov AG. Vopr Virusol. 1991;36:146–150. [PubMed] [Google Scholar]
- 6.Lukashevich IS. Virology. 1992;188:600–605. doi: 10.1016/0042-6822(92)90514-p. [DOI] [PubMed] [Google Scholar]
- 7.Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, Brasky K, Geiger R, Hubbard GB, Bryant J, Salvato MS. J Virol. 2005;79:13934–13942. doi: 10.1128/JVI.79.22.13934-13942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher-Hoch SP, McCormick JB, Auperin D, Brown BG, Castor M, Perez G, Ruo S, Conaty A, Brammer L, Bauer S. Proc Natl Acad Sci USA. 1989;86:317–321. doi: 10.1073/pnas.86.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clegg JCS, Bowen MD, Buchmeier MJ, Gonzalez J-P, Lukashevich IS, Peters CJ, RicoHesse R, V Romanowski. In: Seventh Report of the International Committee on Taxonomy of Viruses. van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniliff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editors. Academic Press; San Diego: 2000. pp. 633–640. [Google Scholar]
- 10.Auperin DD, McCormick JB. Virology. 1989;168:421–425. doi: 10.1016/0042-6822(89)90287-0. [DOI] [PubMed] [Google Scholar]
- 11.Lukashevich IS, Djavani M, Shapiro K, Sanchez A, Ravkov E, Nichol ST, Salvato MS. J Gen Virol. 1997;78( Pt 3):547–551. doi: 10.1099/0022-1317-78-3-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djavani M, Lukashevich IS, Sanchez A, Nichol ST, Salvato MS. Virology. 1997;235:414–418. doi: 10.1006/viro.1997.8722. [DOI] [PubMed] [Google Scholar]
- 13.Wilson SM, Clegg JC. Virology. 1991;180:543–552. doi: 10.1016/0042-6822(91)90068-m. [DOI] [PubMed] [Google Scholar]
- 14.Emonet S, Lemasson JJ, Gonzalez JP, de Lamballerie X, Charrel RN. Virology. 2006;350(2):251–257. doi: 10.1016/j.virol.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Basak A, Chretien M, Seidah NG. FEBS Lett. 2002;514:333–339. doi: 10.1016/s0014-5793(02)02394-3. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Oldstone MB. J Virol. 1985;55:704–709. doi: 10.1128/jvi.55.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harnish DG, Dimock K, Bishop DH, Rawls WE. J Virol. 1983;46:638–641. doi: 10.1128/jvi.46.2.638-641.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meulen J, Badusche M, Satoguina J, Strecker T, Lenz O, Loeliger C, Sakho M, Koulemou K, Koivogui L, Hoerauf A. Virology. 2004;321:134–143. doi: 10.1016/j.virol.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Tortorici MA, Ghiringhelli PD, Lozano ME, Albarino CG, Romanowski V. J Gen Virol. 2001;82:121–128. doi: 10.1099/0022-1317-82-1-121. [DOI] [PubMed] [Google Scholar]
- 22.Salvato MS. The Arenaviridae. Plenum Press; New York: 1993. [Google Scholar]
- 23.Titov II, Ivanisenko VA, Arna NAKG. Predicting 2D structure of RNA by genetic algorithm. 2000 http://wwwmgs.bionet.nsc.ru/mgs/programs/2dstructrna/


