Abstract
NO, produced from l-arginine in a reaction catalyzed by NO synthase, is an endogenous free radical with multiple functions in mammalian cells. Here, we demonstrate that endogenously produced NO can suppress c-Jun N-terminal kinase (JNK) activation in intact cells. Treatment of BV-2 murine microglial cells with IFN-γ induced endogenous NO production, concomitantly suppressing JNK1 activation. Similarly, IFN-γ induced suppression of JNK1 activation in RAW264.7 murine macrophage cells and rat alveolar macrophages. The IFN-γ-induced suppression of JNK1 activation in BV-2, RAW264.7, or rat alveolar macrophage cells was completely prevented by NG-nitro-l-arginine, a NO synthase inhibitor. Interestingly, the IFN-γ-induced suppression of JNK1 activation was not affected by 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one, an inhibitor of guanylyl cyclase. 8-Bromo-cGMP, a membrane-permeant analogue of cGMP, did not change JNK1 activation in intact cells either. In contrast, S-nitro-N-acetyl-dl-penicillamine (SNAP), a NO donor, inhibited JNK1 activity in vitro. Furthermore, a thiol reducing agent, DTT, reversed not only the in vitro inhibition of JNK1 activity by SNAP but also the in vivo suppression of JNK1 activity by IFN-γ. Substitution of serine for cysteine-116 in JNK1 abolished the inhibitory effect of IFN-γ or SNAP on JNK1 activity in vivo or in vitro, respectively. Moreover, IFN-γ enhanced endogenous S-nitrosylation of JNK1 in RAW264.7 cells. Collectively, our data suggest that endogenous NO mediates the IFN-γ-induced suppression of JNK1 activation in macrophage cells by means of a thiol-redox mechanism.
Nitric oxide (NO) is the diffusible and short-lived free radical that can be produced in mammalian cells by the family of NO synthases (NOSs), including neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS) (1, 2). Both nNOS and eNOS are constitutively expressed and their enzymatic activities are regulated by changes in intracellular concentrations of free Ca2+ (3, 4). iNOS is an enzyme whose activity is independent of the intracellular concentration of free Ca2+ and is regulated at the transcriptional level (5). Its expression can be induced in many cell types, including macrophages, by cytokines and lipopolysaccharide. iNOS, which produces high levels of NO, plays a role in cellular immune responses (6, 7). NO has been shown to function as an intracellular signaling regulator in a variety of cellular events (8). NO exerts many of its functions through S-nitrosylation of proteins (9). S-nitrosylation appears to be involved in the regulatory mechanisms of many cellular activities, including signal transduction, DNA repair, host defense, blood pressure control, and neurotransmission (9).
The intracellular signaling cascades of mitogen-activated protein kinases (MAPKs) are conserved among many eukaryotes ranging from yeast to mammals (10–12). The MAPK signaling pathway is composed of several components that include MAPK, MAPK kinase (MAP2K), and MAP2K kinase (MAP3K). When activated, MAP3K phosphorylates and stimulates MAP2K, which, in turn, stimulates MAPK (10, 13). The MAPKs are a family of serine/threonine kinases, which include the extracellular signal-regulated kinases, p38 MAPK, and the c-Jun N-terminal kinases/stress-activated protein kinases (JNKs/SAPKs). JNKs/SAPKs are strongly activated in cellular responses to various stresses such as UV radiation, heat shock, osmotic shock, DNA-damaging agents, and metabolic inhibitors as well as proinflammatory cytokines, such as tumor necrosis factor-α and IL-1β (10, 14). The upstream components of JNK/SAPK in the JNK/SAPK pathway include MAP2K such as SEK1/MKK4/JNKK1 and MKK7 and MAP3K such as MEKK1 (15–17). The JNK/SAPK pathway appears to play a role in stress-activated cellular events, including proliferation, differentiation, and apoptosis (10, 18).
We previously demonstrated that JNK/SAPK has a cysteine residue that is sensitive to thiol-modifying agents (19). Because NO is a typical endogenous thiol-reactive molecule, we investigated in the present study a possible effect of NO on JNK/SAPK. We report here that endogenous NO can suppress JNK/SAPK through a thiol-redox mechanism. The thiol-redox regulation of JNK/SAPK by NO may be important to the understanding of the mechanism by which iNOS induction and NO mediate intracellular signaling leading to many cellular functions.
Materials and Methods
Cell Culture and Transfection.
Rat alveolar macrophages were isolated from lungs of 6-wk-old male Sprague–Dawley rats. The isolated alveolar macrophages were grown in DMEM without FBS for 1 h, and then cultivated in the same medium containing 3% FBS. HEK293, BV2 murine microglial cells, and RAW264.7 murine macrophage cells were maintained in DMEM supplemented with 10% FBS in 5% CO2 at 37°C. For DNA transfections, cells were plated in 100-mm dishes at 1 × 106 cells per dish, grown overnight, and transfected with indicated expression vectors by using Lipofectamine (Life Technologies) or calcium phosphate (20).
Immunocomplex Kinase Assay.
Cultured cells were harvested and lysed in a lysis buffer (19). Cell lysates were subjected to centrifugation at 12,000 × g for 10 min at 4°C. The soluble fraction was immunoprecipitated with appropriate antibodies, and the immunopellets were assayed for enzymatic activities of indicated protein kinases, as described previously (21, 22). Phosphorylated proteins were separated by SDS/PAGE and quantified by using a Fuji BAS 2500 PhosphorImager. Glutathione S-transferase (GST)-cJun(1–79), GST-SAPKβ(K55R), and GST-SEK1(K129R) were used as substrates for JNK, SEK1, and MEKK1, respectively. Protein concentrations were determined by the Bradford method (Bio-Rad).
Measurement of NO Production.
NO production was quantified using a Griess method kit (Promega) according to the manufacturer's protocol. The Griess method measures nitrite, which is a stable breakdown product of NO.
Measurement of S-Nitrosylation.
Cells were harvested, lysed, and subjected to immunoprecipitation using mouse monoclonal anti-JNK1 antibody. The immunopellets were extensively washed five times with lysis buffer (19) and then twice with PBS. The immunopellets were incubated with 100 μM HgCl2 and 100 μM 2,3-diaminonaphthalene (DAN) for 30 min at room temperature, and then added with 1 M NaOH. The quantity of a fluorescent triazole generated from the reaction between DAN and NO released from S-nitrosylated JNK1 was monitored by a fluorometer (Perkin-Elmer HTS 7000) with an excitation wavelength of 375 nm and an emission wavelength of 450 nm (23, 24).
Luciferase Reporter Assay for c-Jun-Dependent Transcription.
Transcription-stimulating activity of c-Jun was determined using the PathDetect luciferase reporter kit (Stratagene) as described previously (19). RAW264.7 cells were transiently transfected with pFR-Luc, pFA2-c-Jun, and pFC-MEKK along with pcDNA3-βgal. After 48 h of transfection, the cells were assayed for luciferase activity by using a kit (Promega). The activity of the luciferase reporter protein in the transfected cells was normalized in reference to β-galactosidase activity in the same cells.
Results
NO Mediates IFN-γ-Induced Suppression of JNK Activation in Intact Cells.
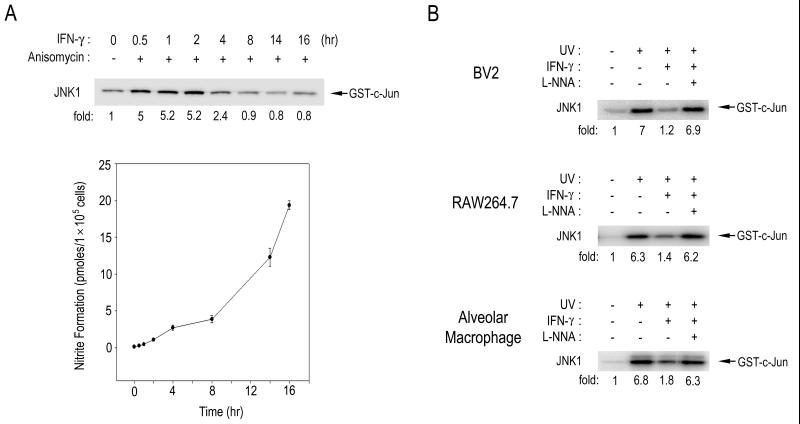
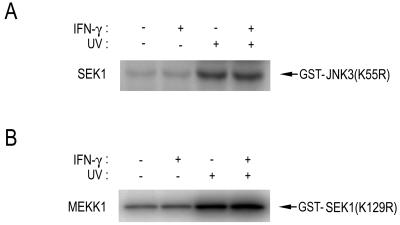
It has been well established that cytokines such as IFN-γ can induce expression of iNOS, which then produces a large amount of NO in certain cell types including macrophages and microglial cells (25, 26). To investigate a possible effect of the endogenous NO production on the JNK pathway, we treated BV-2 murine microglial cells with IFN-γ and examined the effect of INF-γ on JNK1 activation in the cells (Fig. 1). Exposure of BV-2 cells to IFN-γ resulted in an increase in NO production (Fig. 1A). Concomitantly, the IFN-γ treatment caused a decrease in the anisomycin-stimulated JNK1 activity in the cells (Fig. 1A). Immunoblot analysis using anti-JNK1 antibody indicated that the IFN-γ treatment did not affect cellular levels of JNK1 in the BV-2 cells (data not shown). Likewise, IFN-γ suppressed the UV-induced JNK1 activation in BV-2 cells, RAW264.7 murine macrophage cells, and rat primary alveolar macrophage cells (Fig. 1B). Interestingly, NG-nitro-l-arginine (l-NNA), a NOS inhibitor, completely abolished the IFN-γ-induced suppression of the JNK1 activation in those cells. These results suggest that IFN-γ can suppress JNK1 activation in macrophages and microglial cells and that NO mediates the IFN-γ-induced suppression of JNK1 activation. Next, we examined whether IFN-γ could also suppress the activities of SEK1 and MEKK1, the upstream kinases of JNK1 (Fig. 2). Exposure of cells to UV light induced stimulation of SEK1 or MEKK1 in RAW264.7 cells. The UV-stimulated activities of SEK1 and MEKK1 were changed little, if at all, by IFN-γ treatment. These data suggest that IFN-γ, by inducing NO production, might repress the activation of JNK itself rather than activation of the upstream kinases, SEK1 and MEKK1, of JNK.
Figure 1.
IFN-γ suppresses JNK1 activation by means of the induction of NO generation in microglial and macrophage cells. (A) BV2 murine microglial cells were pretreated with IFN-γ (100 units/ml) for the indicated times and then exposed to 10 μM anisomycin for 15 min at 37°C. Enzymatic activities of JNK1 in the BV2 cells were measured by immunocomplex kinase assay using anti-JNK1 antibody. For NO measurement, 100 μl of culture medium in each plate was collected and analyzed for NO release by the Griess method. (B) BV2, RAW264.7, and rat primary alveolar macrophage cells were pretreated with IFN-γ (100 units/ml) for 16 h, and then incubated with 2 mM l-NNA for 30 min, where indicated. After the chemical treatments, the cells were exposed to UV light (60 J/m2) and further incubated for 30 min at 37°C. JNK1 activities in the cells were determined by immunocomplex kinase assay using anti-JNK1 antibody.
Figure 2.
IFN-γ does not affect either SEK1 or MEKK1 in RAW264.7 cells. RAW264.7 cells were treated with IFN-γ (100 units/ml) for 16 h. Where indicated, the cells were exposed to UV light (60 J/m2) and further incubated for 30 min at 37°C. Cell lysates were subjected to immunoprecipitation using anti-SEK1 or anti-MEKK1 antibody. The immunopellets were examined for SEK1 or MEKK1 activity by immunocomplex kinase assay using GST-SAPKβ(K55R) or GST-SEK1(K129R) as substrate.
cGMP Does Not Mediate NO-Induced Inhibition of JNK1.
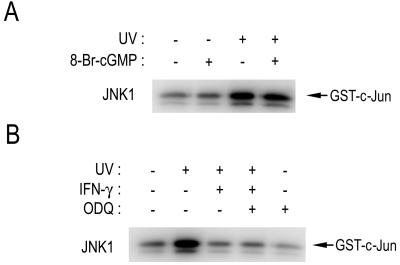
One of the well-characterized targets of NO is guanylyl cyclase, which produces cGMP (27). cGMP has been shown to mediate some of the biological functions of NO (28). We, therefore, investigated a possibility that cGMP might mediate the IFN-γ-induced suppression of JNK1 activation. First, we examined the effect of 8-bromo-cGMP, a membrane-permeant analogue of cGMP, on the UV-induced JNK1 activation (Fig. 3A). Exposure of RAW264.7 cells to UV light induced the JNK1 activation, and the UV-stimulated JNK1 activity was not affected by 8-bromo-cGMP. Second, we examined the effect of 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one (ODQ), an inhibitor of guanylyl cyclase (29), on the IFN-γ-induced suppression of JNK1 activation in RAW264.7 cells (Fig. 3B). ODQ did not affect the inhibitory effect of IFN-γ on JNK1 activation. Taken together, our results suggest that cGMP is not involved in the mechanism by which endogenous NO mediates the IFN-γ-induced suppression of JNK1 activation in RAW264.7 cells.
Figure 3.
cGMP does not mediate NO-induced suppression of JNK activation in intact cells. (A) RAW264.7 cells were pretreated with 100 μM 8-bromo-cGMP (8-Br-cGMP) for 30 min, and then irradiated with 60 J/m2 UV light, followed by an additional 1-h incubation at 37°C. (B) RAW264.7 cells were pretreated with IFN-γ (100 units/ml) for 16 h, and then treated with 100 μM ODQ for 30 min. After the chemical treatments, the cells were exposed to 60 J/m2 UV light, then incubated further for 1 h at 37°C. (A and B) The cells were harvested, lysed, and analyzed for JNK1 activity by immunocomplex kinase assay using anti-JNK1 antibody.
In Vitro Effect of SNAP on MEKK1, SEK1, or JNK1 Activity.
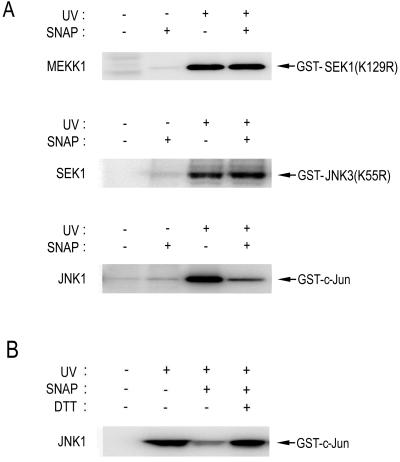
As shown in Fig. 1, the IFN-γ treatment suppressed JNK1 activity, but not the activities of upstream kinases, including SEK1 and MEKK1, in intact cells. We, therefore, investigated the possibility that NO might directly act on and inhibit JNK1. To test this possibility, we examined the in vitro effect of S-nitro-N-acetyl-dl-penicillamine (SNAP) on JNK1 and its upstream kinases SEK1 and MEKK1 (Fig. 4A). SNAP is a NO donor. SNAP inhibited JNK1, but not MEKK1 or SEK1, in vitro. These data suggest that JNK may be a major target protein of NO in the JNK signaling pathway.
Figure 4.
In vitro effect of SNAP on JNK1, SEK1, or MEKK1 activity. HEK293 cells were irradiated with 60 J/m2 UV light and then incubated for 1 h at 37°C. The cell lysates were subjected to immunoprecipitation using anti-MEKK1, anti-SEK1, or anti-JNK1 antibody, as indicated. In A, the resultant immunopellets were treated with 100 μM SNAP for 20 min on ice, and then examined for the indicated protein kinase activities by immunocomplex kinase assay. In B, the resultant immunopellets were exposed to 100 μM SNAP for 20 min on ice, and then incubated with 10 mM DTT for 20 min on ice. JNK1 activity in the immunopellets was determined by immunocomplex kinase assay.
Many lines of evidence point to NO as a modulator of the functions of a variety of proteins using a redox mechanism (9). Furthermore, we previously demonstrated that JNK1 could be inhibited by thiol-oxidizing agents and that it contains a cysteine residue critical for thiol-redox regulation (19). Therefore, it seems possible that NO may inhibit JNK1 through a thiol-redox mechanism. To test this possibility, we examined whether a thiol reducing reagent, DTT, could reverse the inhibitory action of SNAP on JNK1 (Fig. 4B). SNAP inhibited an enzymatic activity of activated JNK1 in vitro, and the in vitro inhibition of JNK1 activity by SNAP was completely blocked by DTT. These data suggest that NO may regulate JNK1 by means of a thiol-redox mechanism.
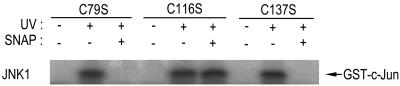
Cys116 of JNK1 Is Important in the Thiol-Redox Regulation by NO.
To better understand the thiol-redox regulation of JNK1 by NO, we examined the sensitivity of the JNK1 mutants C79S, C116S, and C137S to SNAP (Fig. 5). The cysteine residues at the 79th, 116th, and 137th amino acid positions in JNK1 are those conserved among the JNK isoforms, but not in the extracellular signal-regulated kinases or p38 MAPK. We transiently transfected HEK293 cells with each JNK1 mutant construct and then treated the transfected cells with UV light. The UV irradiation induced the activation of JNK1(C79S), JNK1(C116S), and JNK1(C137S) (Fig. 5). SNAP inhibited the enzymatic activity of JNK1(C79S) and JNK1(C137S) in vitro. Interestingly, JNK1(C116S) was resistant to the in vitro inhibition by SNAP. Thus, our data suggest that Cys116 of JNK1 may be critical in the thiol-redox mechanism by which SNAP suppresses JNK1 activation. These results are consistent with our previous data that demonstrated that Cys116 of JNK1 is the redox-sensitive cysteine residue (19).
Figure 5.
Cys116 of JNK1 is critical for the NO-mediated thiol-redox regulation of JNK1. HEK293 cells were transfected with a plasmid vector expressing HA-JNK1(C79S), HA-JNK1(C116S), or HA-JNK1(C137S). After 50 h of transfection, the cells were exposed to UV light (60 J/m2) and then incubated further for 1 h at 37°C. The cell lysates were subjected to immunoprecipitation using mouse monoclonal anti-HA antibody. The immunopellets were treated with 100 μM SNAP for 20 min on ice, washed twice with PBS solution, and then examined for JNK1 activity by immunocomplex kinase assay.
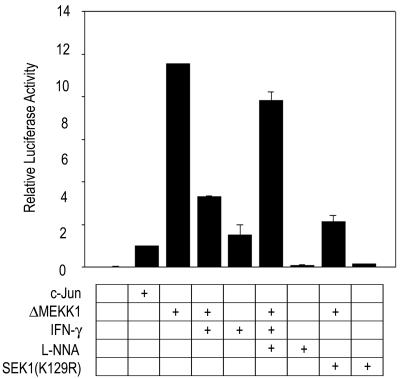
IFN-γ Suppresses the JNK-Mediated c-Jun Activation by Induction of NO Production.
One of the immediate downstream signals of JNK is the phosphorylation of c-Jun, resulting in an enhancement of the transcription stimulating activity of c-Jun (30). We, therefore, tested whether the JNK inhibition by the IFN-γ-driven NO production could result in a suppression of the JNK-dependent transcription stimulating activity of c-Jun in RAW264.7 cells. The transcriptional activity of c-Jun was measured using a luciferase reporter gene assay (Fig. 6). Overexpression of active MEKK1, ΔMEKK1, resulted in an increase in the c-Jun-mediated luciferase reporter activity, and the ΔMEKK1-stimulated luciferase activity was inhibited by coexpressed SEK1(K129R), a dominant-negative mutant of SEK1. These results suggest that ΔMEKK1 induced the c-Jun-mediated luciferase reporter activity through the MEKK1-SEK1-JNK pathway. IFN-γ completely abolished the ΔMEKK1-induced stimulation of the c-Jun-mediated luciferase activity in transfected RAW264.7 cells. Furthermore, l-NNA reversed the inhibitory effect of IFN-γ on the ΔMEKK1-stimulated transactivating activity of c-Jun. Thus, these results suggest that IFN-γ-induced endogenous NO suppresses the JNK pathway-dependent stimulation of the transactivating activity of c-Jun in RAW264.7 cells.
Figure 6.
IFN-γ suppresses the c-Jun-mediated luciferase reporter activity through an induction of NO production. RAW264.7 cells were transiently transfected with luciferase reporter gene, c-Jun, ΔMEKK1, and SEK1(K129R) constructs, as indicated. pcDNA3-β-galactosidase was also included in all transfections. After 24 h of transfection, the cells were exposed to 100 units/ml IFN-γ for 16 h at 37°C, then incubated with 2 mM l-NNA for 6 h. The cells were harvested and assayed for luciferase activity. Luciferase activity was normalized to β-galactosidase activity.
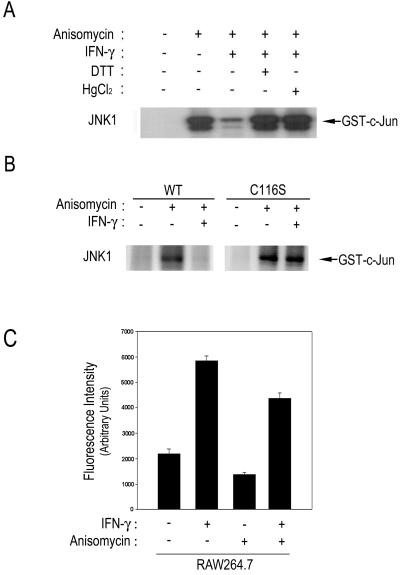
In Vivo S-Nitrosylation of JNK1.
As described above, the IFN-γ-driven production of endogenous NO resulted in inhibition of JNK activity through the thiol-redox mechanism in macrophage cells. We then examined whether the IFN-γ-induced suppression of the JNK1 activation in RAW264.7 cells could be reversed by the thiol-reducing agent DTT (Fig. 7A). Exposure of RAW264.7 cells to anisomycin induced JNK1 activation, whereas IFN-γ pretreatment suppressed the anisomycin-stimulated JNK1 activity in the cells. The IFN-γ-induced suppression of the JNK1 activation was abolished in the presence of DTT. Moreover, HgCl2 also reversed the IFN-γ-induced suppression of JNK1 activation, suggesting that IFN-γ treatment might result in a modification of a cysteine residue(s) on JNK1 (Fig. 7A). Next, we examined the effect of IFN-γ on the enzymatic activities of wild-type JNK1 and JNK1(C116S) in RAW264.7 cells. Anisomycin induced stimulation of both JNK1 and JNK1(C116S) activities in the cells (Fig. 7B). IFN-γ pretreatment prevented the anisomycin-induced activation of wild-type JNK1, but not of JNK1(C116S). This result is in good agreement with the in vitro effect of SNAP on JNK1(C116S) mutant as shown in Fig. 5. Taken together, these data suggest that NO-mediated thiol-redox regulation is the primary mechanism by which IFN-γ suppresses JNK1 activation in RAW264.7 cells.
Figure 7.
INF-γ enhances in vivo S-nitrosylation of JNK1. (A) RAW264.7 cells were pretreated with 100 units/ml of IFN-γ for 16 h, and then treated with 10 μg/ml anisomycin for 15 min at 37°C. The cell lysates were subjected to immunoprecipitation using mouse monoclonal anti-JNK1 antibody. The immunopellets were incubated either with 10 mM DTT for 20 min on ice or with 100 μM HgCl2 for 30 min at room temperature, where indicated. JNK1 activities in the immunopellets were determined by immunocomplex kinase assay. (B) RAW264.7 cells were transiently transfected with pcDNA3 expressing wild-type JNK1 or JNK1(C116) mutant. After 30 h of transfection, the cells were pretreated with 100 units/ml IFN-γ for 16 h, and then treated with 10 μg/ml anisomycin for 15 min at 37°C. The cell lysates were determined for JNK1 activity by immunocomplex kinase assay using anti-JNK1 antibody. (C) RAW264.7 cells were pretreated with 100 units/ml of IFN-γ for 16 h, and then treated with 10 μg/ml anisomycin for 15 min at 37°C. The cell lysates were subjected to immunoprecipitation using mouse monoclonal anti-JNK1 antibody. The JNK1 immunoprecipitates were extensively washed with washing buffer (20 mM Hepes, pH 7.4) (the purity of JNK1 in the immunoprecipitates: >95%). The JNK1 immunoprecipitates were then reacted with 100 μM HgCl2 and 100 μM DAN for 30 min at room temperature, followed by addition of 1 M NaOH. NO released from S-nitrosylated JNK1 was quantified by measuring the fluorescence intensity of the reaction product using a fluorimeter with an excitation wavelength of 375 nm and an emission wavelength of 450 nm.
Next, we investigated whether IFN-γ-induced endogenous NO could modify endogenous JNK1 in RAW264.7 cells by S-nitrosylation. We analyzed protein S-nitrosylation by fluorometry using DAN (23, 24). RAW264.7 cells were pretreated with IFN-γ for 16 h to induce the production of endogenous NO and then exposed to anisomycin for 15 min where indicated (Fig. 7C). JNK1 was then isolated by immunoprecipitation. The JNK1 immunoprecipitates were extensively washed with washing buffer to remove nonspecific intracellular S-nitrosothiols. The JNK1 immunoprecipitates were then incubated with 100 μM HgCl2 and 200 μM DAN, which can be converted to a fluorescent triazole when reacting with NO (23, 24). Reaction between DAN and NO released from S-nitrosylated JNK1 was detected by fluorimetry. Our data showed that the IFN-γ pretreatment increased the in vivo S-nitrosylation of JNK1 in either untreated or anisomycin-treated cells (Fig. 7C).
Discussion
There has been evidence that the JNK/SAPK signaling pathway can be either positively or negatively regulated by NO, depending on cell types, types of NO donors, or the presence of other stimuli (31–36). The molecular mechanisms by which NO modulates the JNK/SAPK pathway are not yet understood, however. It is noteworthy that NO is a typical endogenous thiol-reactive molecule and that JNK/SAPK can be regulated by a thiol-redox mechanism (19). We, therefore, investigated in this study whether endogenous NO could modulate the JNK/SAPK pathway by means of thiol-redox regulation. We demonstrate here that endogenously produced NO can negatively regulate the JNK/SAPK pathway by means of JNK/SAPK inhibition in intact cells. Furthermore, IFN-γ, a strong inducer of iNOS in macrophages, enhances S-nitrosylation of JNK1 in RAW264.7 cells.
Evidence has been accumulating that NO modulates the biological functions of many intracellular signaling proteins by S-nitrosylation (37, 38). Those proteins include N-methyl-d-aspartate receptor, type I adenylyl cyclase, protein kinase C, NF-κB, AP-1, Ras, and caspase-3 (39–47). We show now that IFN-γ-induced production of endogenous NO results in an increase in S-nitrosylated JNK1 in RAW264.7 cells, and that the S-nitrosylation is involved in the mechanism underlying the JNK1 inhibition by NO. Our data using site-directed mutagenesis imply that S-nitrosylation might occur at Cys116 of the enzyme, which has been previously shown to be a critical cysteine residue for the thiol-redox regulation of JNK1 (19).
IFN-γ has been shown to elicit NO generation in macrophages and other cell types through iNOS induction (48). We demonstrated that IFN-γ induced the suppression of JNK activation in macrophages and microglial cells and that the inhibitory effect of IFN-γ on the JNK pathway was blocked by l-NNA, a NOS inhibitor. Furthermore, IFN-γ treatment enhanced the S-nitrosylation of JNK1 in intact cells. Collectively, these findings strongly suggest that IFN-γ functions as a natural inhibitory signal for the JNK/SAPK pathway in macrophages and other iNOS-inducible cells, and that NO mediates the IFN-γ-induced suppression of JNK/SAPK through thiol-redox regulation. The negative regulation of JNK/SAPK by IFN-γ highlights a function of IFN-γ that may be important to the understanding of the molecular mechanisms by which IFN-γ exerts its multiple functions in many iNOS-inducible cells.
Acknowledgments
We thank Drs. R. J. Davis, J. Woodgett, and L. I. Zon for providing cDNA clones; Dr. V. Bocchini for BV-2 murine microglial cells; and Dr. G. Hoschek for critical reading of the manuscript. This work was supported by the Creative Research Initiatives Program of the Korean Ministry of Science and Technology.
Abbreviations
- NOS
NO synthase
- nNOS
neuronal NOS
- eNOS
endothelial NOS
- iNOS
inducible NOS
- MAPK
mitogen-activated protein kinase
- JNK
c-Jun N-terminal kinase
- SAPK
stress-activated protein kinase
- GST
glutathione S-transferase
- ODQ
1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one
- SNAP
S-nitro-N-acetyl-dl-penicillamine
- DAN
2,3-diaminonaphthalene
- l-NNA
NG-nitro-l-arginine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 14022.
References
- 1.Marletta M A. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C, Xie Q W. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 3.Bredt D S, Hwang P M, Glatt C E, Lowenstein C, Reed R R, Snyder S H. Nature (London) 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 4.Busse R, Mulsch A. FEBS Lett. 1990;265:133–136. doi: 10.1016/0014-5793(90)80902-u. [DOI] [PubMed] [Google Scholar]
- 5.Galea E, Feinstein D L, Reis D J. Proc Natl Acad Sci USA. 1992;89:10945–10949. doi: 10.1073/pnas.89.22.10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan C. J Clin Invest. 1997;100:2417–2423. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacMicking J, Xie Q W, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 8.Ignarro L J. Biochem Pharmacol. 1991;41:485–490. doi: 10.1016/0006-2952(91)90618-f. [DOI] [PubMed] [Google Scholar]
- 9.Stamler J S. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 10.Ip Y T, Davis R J. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 11.Su B, Karin M. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 12.Davis R J. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer H J, Weber M J. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minden A, Karin M. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 15.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Nature (London) 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Nature (London) 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 17.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. Proc Natl Acad Sci USA. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minden A, Lin A, Claret F X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 19.Park H S, Park E, Kim M S, Ahn K, Kim I Y, Choi E-J. J Biol Chem. 2000;275:2527–2531. doi: 10.1074/jbc.275.4.2527. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim J, Lee H, Park J, Kim H, Choi E-J. Nature (London) 1996;381:804–806. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 22.Shim J, Park H S, Kim M J, Park J, Park E, Cho S G, Eom S J, Lee H W, Joe C O, Choi E-J. J Biol Chem. 2000;275:14107–14111. doi: 10.1074/jbc.275.19.14107. [DOI] [PubMed] [Google Scholar]
- 23.Wink D A, Grisham M B, Mitchell J B, Ford P C. Methods Enzymol. 1996;268:12–31. doi: 10.1016/s0076-6879(96)68006-9. [DOI] [PubMed] [Google Scholar]
- 24.Miles A M, Wink D A, Cook J C, Grisham M B. Methods Enzymol. 1996;268:105–120. doi: 10.1016/s0076-6879(96)68013-6. [DOI] [PubMed] [Google Scholar]
- 25.Xie Q W, Whisnant R, Nathan C. J Exp Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D, Coleclough C, Cao L, Hu X, Sun S, Whitaker J N. J Neuroimmunol. 1998;89:122–130. doi: 10.1016/s0165-5728(98)00124-6. [DOI] [PubMed] [Google Scholar]
- 27.Arnold W P, Mittal C K, Katsuki S, Murad F. Proc Natl Acad Sci USA. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popescu L M, Panoiu C, Hinescu M, Nutu O. Eur J Pharmacol. 1985;107:393–394. doi: 10.1016/0014-2999(85)90269-9. [DOI] [PubMed] [Google Scholar]
- 29.Brunner F, Stessel H, Kukovetz W R. FEBS Lett. 1995;376:262–266. doi: 10.1016/0014-5793(95)01297-x. [DOI] [PubMed] [Google Scholar]
- 30.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 31.Park D S, Stefanis L, Yan C Y I, Farinelli S E, Greene L A. J Biol Chem. 1996;271:21898–21902. doi: 10.1074/jbc.271.36.21898. [DOI] [PubMed] [Google Scholar]
- 32.Lo Y Y C, Wong J M S, Cruz T F. J Biol Chem. 1996;271:15703–15707. doi: 10.1074/jbc.271.26.15703. [DOI] [PubMed] [Google Scholar]
- 33.Kim H, Shim J, Han P L, Choi E-J. Biochemistry. 1997;36:13677–13681. doi: 10.1021/bi970837f. [DOI] [PubMed] [Google Scholar]
- 34.Wang D, Yu X, Brecher P. J Biol Chem. 1998;273:33027–33034. doi: 10.1074/jbc.273.49.33027. [DOI] [PubMed] [Google Scholar]
- 35.Lander H M, Jacovina A T, Davis R J, Tauras J M. J Biol Chem. 1996;271:19705–19709. doi: 10.1074/jbc.271.33.19705. [DOI] [PubMed] [Google Scholar]
- 36.Jun C D, Oh C D, Kwak H J, Pae H O, Yoo J C, Choi B M, Chun J S, Park R K, Chung H T. J Immunol. 1999;162:3395–3401. [PubMed] [Google Scholar]
- 37.Stamler J S, Simon D I, Osborne J A, Mullins M E, Jaraki O, Michel T, Singel D J, Loscalzo J. Proc Natl Acad Sci USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamler J S, Hausladen A. Nat Struct Biol. 1998;5:247–249. doi: 10.1038/nsb0498-247. [DOI] [PubMed] [Google Scholar]
- 39.Choi Y B, Tenneti L, Le D A, Ortiz J, Bai G, Chen H S, Lipton S A. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 40.Duhe R J, Nielsen M D, Dittman A H, Villacres E C, Choi E J, Storm D R. J Biol Chem. 1994;269:7290–7296. [PubMed] [Google Scholar]
- 41.Gopalakrishna R, Chen Z H, Gundimeda U. J Biol Chem. 1993;268:27180–27185. [PubMed] [Google Scholar]
- 42.DelaTorre A, Schroeder R A, Kuo P C. Biochem Biophys Res Commun. 1997;238:703–706. doi: 10.1006/bbrc.1997.7279. [DOI] [PubMed] [Google Scholar]
- 43.DelaTorre A, Schroeder R A, Bartlett S T, Kuo P C. Surgery. 1998;124:137–141. [PubMed] [Google Scholar]
- 44.Tabuchi A, Sano K, Oh E, Tsuchiya T, Tsuda M. FEBS Lett. 1994;351:123–127. doi: 10.1016/0014-5793(94)00839-6. [DOI] [PubMed] [Google Scholar]
- 45.Lander H M, Ogiste J S, Pearce S F, Levi R, Novogrodsky A. J Biol Chem. 1995;270:7017–7020. doi: 10.1074/jbc.270.13.7017. [DOI] [PubMed] [Google Scholar]
- 46.Lander H M, Hajjar D P, Hempstead B L, Mirza U A, Chait B T, Campbell S, Quilliam L A. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 47.Mannick J B, Hausladen A, Liu L, Hess D T, Zeng M, Miao Q X, Kane L S, Gow A J, Stamler J S. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 48.Walker G, Pfeilschifter J, Kunz D. J Biol Chem. 1997;272:16679–16687. doi: 10.1074/jbc.272.26.16679. [DOI] [PubMed] [Google Scholar]