Abstract
The mouse Otx2 gene is a homeobox transcription factor required as early as gastrulation for the proper development of the head. We compared gene expression profiles in wild-type and Otx2−/− 6.5 days postcoitum embryos by using a serial analysis of gene expression assay adapted to microdissected structures. Among a broader list, the study of six genes found to be differentially expressed allows defining a role for Otx2 in the orchestration of cell movements leading to the adequate organization of the embryo before gastrulation.
Mutation of the Drosophila orthodenticle (otd) gene specifically affects the development of the most anterior segments of the fly fated to form the head (1). Numerous Otd homologues have been found in a variety of species covering most of the metazoan phyla, based on the conservation of a bicoïd class homeodomain (2). Two murine Otd homologues, Otx1 and Otx2, were characterized and shown to be also associated with rostral development. The transcription factor Otx2 is the first of these two genes to be expressed in the course of murine development [5.75 days postcoitum (dpc)]. Ubiquitously transcribed in the embryonic ectoderm and visceral endoderm before gastrulation, Otx2 is progressively restricted from 6.5 to 7.5 dpc to the anterior region of the embryo as the primitive streaks elongates. Later, Otx2 is transcribed in the cephalic region and the developing sense organs (3).
Functional studies revealed that Otx2 is required as early as gastrulation for normal development of the brain. Thus, embryos homozygous for a targeted mutation of Otx2 display an early lethal phenotype characterized by the lack of head structures anterior to rhombomere 3 (4). Further analysis by means of chimeric embryos demonstrated that Otx2 expression is sequentially necessary, in the visceral endoderm at the onset of gastrulation for neural induction, and later in the anterior neuroectoderm itself, to specify forebrain and midbrain regions (4, 5). Understanding of the mechanisms involved in the acquisition of anterior identity requires the identification of Otx2 downstream genes, among which should be the factor(s) that, in the visceral endoderm, mediate anterior neuroectoderm induction.
Large-scale analysis of gene expression should conceivably help in delineating such downstream genes. We therefore used the serial analysis of gene expression (SAGE) method, which provides quantitative gene expression profiles (6), and was recently scaled-down to make possible the analysis of microdissected structures (7). Two SAGE libraries were constructed, from wild-type (WT) and Otx2−/− embryos at early gastrulation (6.5 dpc), the stage at which Otx2 function is primarily required. Then, 140 differentially represented mRNA tags were identified, and several mRNAs were further analyzed by in situ hybridization. Our study therefore allows the identification of several Otx2 potential target genes and describes global anterior–posterior patterning defects in Otx2−/− embryos.
Materials and Methods
Embryos Genotyping, in Situ Hybridization, and 5-Bromo-4-Chloro-3-Indolyl β-d-Galactoside (X-Gal) Staining.
Embryos were obtained by mating Otx2+/− mice. Midday of the day of the vaginal plug was considered 0.5 dpc. Embryos were dissected at room temperature in PBS and processed for the generation of SAGE libraries or in situ hybridization and X-Gal staining. For SAGE libraries, the embryonic and extraembryonic portions were separately transferred into 1.5-ml polypropylene tubes. Each portion was centrifuged for 2 min at 2000 × g, frozen in liquid nitrogen, and stored at −80°C until use. Embryo genotyping was performed by PCR on the extraembryonic portion as previously described (4). X-Gal staining and whole-mount in situ hybridization of embryos were carried out as previously described by Acampora et al. (4) and Henrique et al. (8), respectively. Hybridization probes were generated from image clones by in vitro transcription using digoxygenin-UTP for RNA labeling. Embryos genotypes were established by PCR after hybridization and staining.
Generation of SAGE Libraries.
The embryonic portions of Otx2+/+ or Otx2−/− embryos harvested at 6.5 dpc were used to generate SAGE libraries. Thirty embryos were used in each case. Libraries were generated by using the SAGE adaptation for downsized extracts (SADE) method (7). Poly(A) RNAs were isolated through hybridization to oligo(dT)25 covalently bound to magnetic beads by using Dynabeads mRNA direct kit (Dynal, Great Neck, NY). Briefly, frozen embryos were pooled in 100 μl of lysis binding buffer supplemented with 2 μg glycogen (Roche Molecular Biochemicals). After extensive rinsing of the beads, first-strand cDNA was synthesized by using Superscript Moloney murine leukemia virus (M-MLV) reverse transcriptase (Life Technologies). All subsequent steps of libraries generation were performed as described by Virlon et al. (7) for microdissected samples.
DNA Sequencing and SAGE Data Analysis.
Sequencing reactions were performed on DNA minipreps by using Big Dye terminator sequencing chemistry (Applied Biosystems) and run on 377-XL Applied Biosystems automated sequencers. Sequence files were analyzed by using SAGE software (6). Assessment of significant differences between the two libraries was made by Monte–Carlo simulation analysis (9). A P value of 0.05 or less was considered significant.
Results and Discussion
Thirty embryonic portions (the extraembryonic half being used for PCR genotyping), corresponding to ≈30,000 cells, were processed for each library. A total of 27,100 and 21,443 tags were sequenced from the WT and Otx2−/− library, corresponding to 11,256 and 8,893 different tags (or transcripts), respectively. GenBank matches (known cDNAs or genes) were found for 28% of the tags from the WT library and 30% of the Otx2−/− library. The sensitivity achieved is illustrated by the fact that presence of the tags isolated from cerberus-related-1 or nodal transcripts, albeit being expressed in a subpopulation of cells (10, 11) were detected by the SAGE method (Table 1).
Table 1.
Abundance values of tags corresponding to genes involved in embryogenesis
| Tag | WT | Otx2−/− | GenBank match |
|---|---|---|---|
| TCTAATGACT | 1 | 1 | Nodal gene, a TGF-β-like gene |
| AAAGCCAAGC | 1 | 1 | Lim1, putative transcription regulator |
| CAATTCCTGA | 0 | 1 | Cerberus-related 1 (Cerr1) |
| AGGCTGAGCT | 4 | 0 | Embryonic ectoderm development protein (eed) |
| CAGAGGCAGG | 0 | 2 | Wnt-4 |
| TTCACTCTGG | 1 | 0 | Fibroblast growth factor receptor 1 (bFGF-R) |
| CTCTTGCCCC | 1 | 0 | Fibroblast growth factor 4 (FGF-4) |
| GTGCTGGAGA | 1 | 2 | Fibroblast growth factor 8 (FGF-8) |
| ATATTTATGT | 2 | 0 | Fibroblast growth factor (FGF-15) |
| CCATCTGGTG | 3 | 0 | Transforming growth factor α (TGFα) |
| TAATTGCTGT | 2 | 0 | Distal-less homeobox 1 (Dlx-1) |
| GATGCCGTGG | 1 | 3 | Gl/S-specific cyclin D3 |
| TCCCTGAGTT | 1 | 0 | E-cadherin |
| TGGAGGTGGA | 2 | 2 | Smad3 |
| AGAAAGATGC | 1 | 1 | Smad4 |
The sequence of the tag is given along with its abundance in the WT and Otx2−/− libraries, as well as the identity of the transcript from which it originates. Bold characters indicate that differential expression was confirmed by in situ hybridization.
The number of tags differentially represented (P < 0.05) in the two libraries, assessed by Monte–Carlo simulation, reached 141. Of those, 55 match to a cDNA (39%), 27 correspond to expressed sequenced tags (EST) (19%), and 59 are to date totally unknown (42%). A majority of these differentially represented tags correspond to genes expressed at high levels and taking part in basic cell functions not specifically related to development. Therefore, a selected set of genes was studied (Tables 2 and 3). Because genes that play important roles during embryogenesis, for instance transcription factors and secreted molecules can be poorly transcribed, tags bearing less significant variations but belonging to critical gene families were also taken into account and studied in more depth (Table 1).
Table 2.
List of tags present at higher levels in the Otx2−/− than in the WT library
| Tag | WT | KO | KO/WT | P | GenBank match |
|---|---|---|---|---|---|
| ATAAAAAAAA | 0 | 9 | >9 | 0.004 | Peroxisomal PTS2 receptor (U69171) |
| TACGAGCACA | 1 | 9 | 9.0 | 0.018 | EST, highly similar to ubiquinol cytochrome C reductase (AA033187) |
| TTGCCTTATT | 1 | 9 | 9.0 | 0.018 | EST 331499, similar to human IP-30 protein (W13914) |
| TTCCCAGAAA | 1 | 9 | 9.0 | 0.018 | EST, similar to ATP synthase subunit D mitochondrial (D13120) |
| AGCGTGGCCT | 0 | 8 | >8 | 0.008 | AAC-11 inhibitor of apoptosis (U35846) |
| AGCCCCGCCT | 0 | 8 | >8 | 0.008 | Integrin binding protein kinase, ILK (U94479) |
| TCTCTAAATA | 2 | 16 | 8.0 | 0.001 | Cystatin B (U59807) |
| CGCTTGGCAG | 1 | 8 | 8.0 | 0.030 | Δ Proteasome subunit (U13393) |
| TTCTGTGCAG | 1 | 8 | 8.0 | 0.030 | EST J0515B06 (AU0119257) |
| TGACACCCTT | 1 | 8 | 8.0 | 0.030 | EST 633613 (AA184527) |
| TGTTCATACA | 0 | 6 | >6 | 0.018 | ESTs (AI155073; AA433547) |
| TGTAGTCTGC | 0 | 6 | >6 | 0.018 | EST, similar to human argine rich protein (AA217925) |
| CCCCTCCCAC | 0 | 6 | >6 | 0.018 | ESTs (AA871006; AI115602; W64163) |
| TTCTATGCCT | 3 | 19 | 6.3 | 0.001 | EST 493579 (AA087133) |
| TCCTTTGCCC | 0 | 5 | >5 | 0.042 | Zona pellucida (Zp-1) gene, exon 1 (U24227) |
| TGCTGAAGGA | 0 | 5 | >5 | 0.042 | Zinc finger protein Requiem (req) (U10435) |
| TGGGGTAGAT | 0 | 5 | >5 | 0.042 | Calregulin (ERp60) (M92988) |
| TGTGGGAAAA | 0 | 5 | >5 | 0.042 | EST, vanin 3 (AI118382) |
| ATAGCAAAGA | 0 | 5 | >5 | 0.042 | EST, similar to canis familiaris VIP-36 protein (AW229370) |
| AGCACAGCCT | 0 | 5 | >5 | 0.042 | EST 947464 (AI505056) |
| AGTTTGCTCA | 0 | 5 | >5 | 0.042 | Peroxisomal biogenesis factor (Pex11b) (AF093671) |
| CAGGCGGCCA | 0 | 5 | >5 | 0.042 | Catenin α 1 (X59990) |
| GAGTACTTTT | 0 | 5 | >5 | 0.042 | Purine-nucleoside phosphorylase (X56548) |
| CTGAATGACG | 2 | 11 | 5.5 | 0.012 | Nuclear RNA helicase Bat1 (AF118128) |
| GCAGAACCAG | 3 | 16 | 5.3 | 0.003 | A10 mRNA, partial cds (L21027) |
| CTGCTCCTGC | 2 | 10 | 5.0 | 0.025 | Metallothionein 2 (K02236) |
| AGTTTGTCTG | 2 | 10 | 5.0 | 0.025 | EST 1480726 (AI035276) |
| AGCGAAGTGG | 2 | 10 | 5.0 | 0.025 | EST, highly similar to human N-terminal acetyltransferase (AA068754) |
| CAACTCCAAT | 2 | 9 | 4.5 | 0.049 | Calpactin I light chain (p11) (M16465) |
| TTTTGCACAG | 2 | 9 | 4.5 | 0.049 | ESTs 765113; 947489 (J0528F02) |
| CTGAGAACTT | 2 | 9 | 4.5 | 0.049 | β-Globin gene (AA960070) |
| GGAATGCGGG | 2 | 9 | 4.5 | 0.049 | EST 1248132 (AA960070) |
| GTGACTGGGT | 5 | 21 | 4.2 | 0.001 | Mitochondrial cytochrome c oxidase subunit IV (COX IV) (M37829) |
| TTGTTGGAGG | 3 | 11 | 3.7 | 0.033 | EST (W29669) |
| TTAAATAAAA | 3 | 11 | 3.7 | 0.033 | Sid393p (AB025049) |
| GTGTTGTTTA | 5 | 14 | 2.8 | 0.037 | cdc2 mRNA for CDC2 kinase (X16461) |
| CTGTGTGTGG | 7 | 18 | 2.6 | 0.033 | Prostatic secretory glycoprotein (p12) (X06342) |
| TGAAAATTGG | 9 | 23 | 2.6 | 0.015 | COX7cl mRNA for cytochrome c oxidase VIIc (X52940) |
| GACAAGGCCA | 28 | 67 | 2.4 | 0.000 | Apolipoprotein A–I (X64262 S37) |
| ATTTCAAGAA | 9 | 21 | 2.3 | 0.022 | EST 1450276 (AI035675) |
| CACGTTGTCA | 8 | 18 | 2.3 | 0.050 | Embigin precursor (J03535) |
| AATAAACCGT | 8 | 18 | 2.3 | 0.050 | Adenine phosphoribosyltransferase (APRT) (M11310) |
| CAAAAAAAAA | 11 | 24 | 2.2 | 0.025 | EST 863707 (AA512428) |
| TGGGGAAAGG | 13 | 28 | 2.2 | 0.021 | Ribosomal protein L35a (Y16430) |
| TTTGCCGGCA | 27 | 58 | 2.1 | 0.001 | Ubiquitin (X51703) |
| AATAAAGTGG | 16 | 33 | 2.1 | 0.016 | Cyclophilin (M60456) |
| AATGCCCTCA | 65 | 130 | 2.0 | 0.000 | Acidic ribosomal phosphoprotein P1 (U29402) |
| GAGAGGGCAA | 84 | 162 | 1.9 | 0.000 | Ribosomal protein S12 (X15962) |
| TGAAATGAAC | 23 | 44 | 1.9 | 0.010 | EST 475993 (AA049909) |
| GTTCCAAAGC | 24 | 45 | 1.9 | 0.011 | Hexokinase 1 (J05277) |
| CGTATTACCT | 27 | 49 | 1.8 | 0.010 | REX-3 (AP051347) |
| CCGCGAGGCC | 42 | 75 | 1.8 | 0.003 | Ribosomal protein S26 (RPS26) (U67770) |
| TTCCAGGCCC | 20 | 35 | 1.8 | 0.029 | Apolipoprotein E (M12414) |
| AAAGCCCGGA | 79 | 138 | 1.7 | 0.001 | Ribosomal protein S8 (X73829) |
| GCATTGCCAA | 62 | 107 | 1.7 | 0.001 | J1 protein, yeast ribosomal protein L3 homologue (Y00225) |
| AGCAAGCAGG | 45 | 77 | 1.7 | 0.003 | Cytoplasmic beta-actin (MI2481) |
| CTCCTTGTCA | 23 | 38 | 1.7 | 0.048 | Phosphatidylethanolamine binding protein (U43206) |
| AGAAAAAAAA | 36 | 57 | 1.6 | 0.027 | Skeletal muscle chloride channel (X62897) |
| AAACAACCCA | 37 | 56 | 1.5 | 0.045 | Mitochondrial gene for cytochrome b (X57779) |
| AAGCGGGCTT | 51 | 76 | 1.5 | 0.020 | EST, highly similar to human ribosomal 60S protein L34 (AA574826) |
| GGGGTTTACC | 76 | 110 | 1.4 | 0.012 | Ribosomal protein S16 (M11408) |
| AATGATGAGG | 79 | 105 | 1.3 | 0.040 | Heat shock protein 70 cognate (M19141) |
Each tag is given its sequence, its abundance in both libraries (WT and KO), the ratio value (KO/WT), the P value calculated by Monte–Carlo simulations, and its GenBank match. Only tags displaying GenBank matches are shown. The complete list of tags obtained in WT and Otx2−/− will be available at http://www.dsv.cea.fr/thema/get/sade.html.
Table 3.
List of tags present at lower levels in the Otx2−/− than in the WT library
| Tag | WT | KO | WT/KO | P | GenBank match |
|---|---|---|---|---|---|
| CTCCTGCCCC | 9 | 0 | >9 | 0.01 | ClpP protease (AJ005253) |
| CTAGAGGAAA | 9 | 1 | 9.0 | 0.03 | EST 1005223 (AA606898) |
| ATGCCGCCCC | 8 | 1 | 8.0 | 0.04 | Nonmuscle tropomyosin 5 (X53753) |
| GTCGTGACAG | 8 | 1 | 8.0 | 0.04 | p68 RNA helicase (X65627) |
| TTCAATTTAA | 8 | 1 | 8.0 | 0.04 | ESTs (AI047965; AA967648) |
| CAACAAAGGT | 7 | 0 | >7 | 0.02 | HSP60 protein (X53584) |
| ATAGTTGCTA | 6 | 0 | >6 | 0.03 | EST 577496 (AA139307) |
| ACAGAGATGA | 5 | 0 | >5 | 0.05 | ESTs, similar to human hypothetical protein (Q15004) |
| ACCATCCTGA | 13 | 3 | 4.3 | 0.01 | rbm3 (AB016424) |
| TCTTGTGAAA | 12 | 3 | 4.0 | 0.02 | EST 597651 (AI447986) |
| AGCTGAAGGT | 21 | 6 | 3.5 | 0.01 | Suil (Z50159) |
| ACCTTTGCAT | 10 | 3 | 3.3 | 0.05 | Protein arginine methyltransferase (Carm1) (AF117887) |
| AAGAAGCCAC | 46 | 19 | 2.4 | 0.00 | Pyrubate kinase M (D38379) |
| TGATTCCCTC | 48 | 21 | 2.3 | 0.00 | External transcribed spacer (X56974) |
| CACATCTCAA | 50 | 24 | 2.1 | 0.00 | 24.6-kDa protein (M93980) |
| TTGTTACAGA | 33 | 19 | 1.7 | 0.04 | EST J1008D06 (AU041730) |
| TCTACCATTT | 40 | 24 | 1.7 | 0.04 | EST, highly similar to human 40S ribosomal protein (AI931810) |
| TCTCTTCCCA | 90 | 56 | 1.6 | 0.01 | Ribosomal protein S4 (Rps4) (M73436) |
| TGATGCCCTC | 466 | 306 | 1.5 | 0.00 | α-Galactosidase A (U34071) |
| TGACTCCCTC | 1118 | 955 | 1.2 | 0.00 | B2 repetitive sequence (M31447) |
| TGACGCCCTC | 461 | 394 | 1.2 | 0.02 | Pelle-like protein kinase (AF103876) |
The content of each column is similar to that described in Table 2. Additional data will be available at http://www.dsv.cea.fr/thema/get/sade.html.
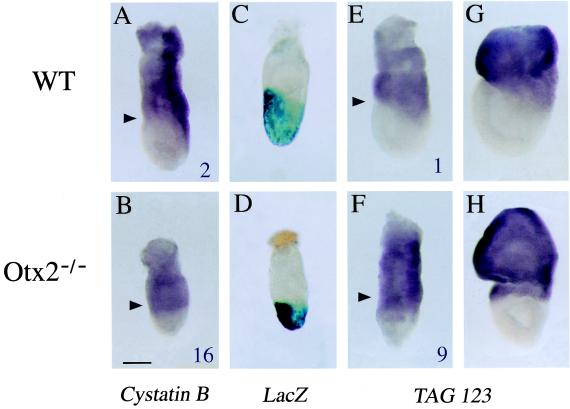
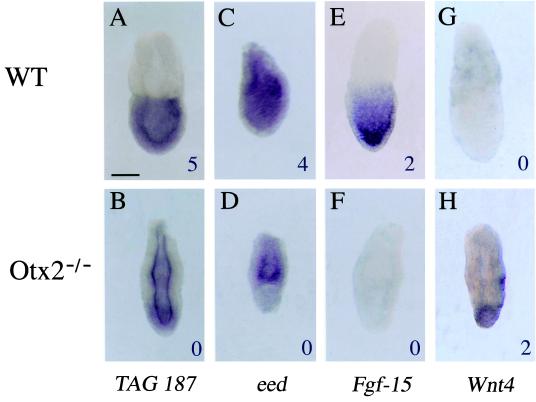
To provide a potential link of these data to the Otx2−/− phenotype, whole mount in situ hybridization (8) was performed on WT and mutant embryos at 6.5 dpc. Six tags corresponding to genes predicted by SAGE to be differentially expressed between both types of embryos were confirmed by using this technique: (i) tag 123 and 15, which corresponds to an EST (331499) and to the cystatin B gene, respectively, and were both detected at higher levels in the mutant than in the WT library (Table 2); (ii) tag 187, which match to several ESTs, all highly similar to a human hypothetical protein (Q15004), and was present at a lower level in the mutant than in the WT library; (iii) tags corresponding to Wnt4, Fgf-15, and eed (embryonic ectoderm development) mRNAs (Table 1). The mRNA known through EST 331499, which is similar to a human interferon-induced protein of unknown function (12), and that encoding the protease inhibitor cystatin B (13), display comparable spatial expression patterns (Fig. 1). In WT embryos, they are expressed in the extraembryonic visceral endoderm and in the embryonic posterior proximal part where the primitive streak forms (Fig. 1 A, E, and G). In mutant embryos, their expression domain is wider and form a ring encompassing the entire proximal embryonic region at the expense of the normal asymmetrical localization (Fig. 1 B, F, and H). Considering that the SAGE data were obtained from the embryonic portion, this extended distribution agrees with the fact that tags for both transcripts were much more abundant in the mutant than in the WT embryos. Fig. 1 also shows that the distribution of mRNAs for EST 331499 and cystatin B is strikingly complementary to the lacZ expression domain, which reflects sites for Otx2 transcription. Thus, these two mRNAs locate in cells of the visceral endoderm not expressing Otx2 and irrespective of the embryonic–extraembryonic boundary of the underlying ectoderm. Their altered distribution in mutant embryos suggests that Otx2 is indirectly necessary for the accurate regionalization of the visceral endoderm. On the contrary, modifications in the expression profiles for tags 187, eed, Wnt4, and Fgf-15 (Fig. 2) are associated to the embryonic ectoderm layer. Tag 187 was found in ESTs that show sequence similarity with a hypothetical human protein isolated from an immature myeloid cell line (14). The gene is expressed throughout the embryonic ectoderm (Fig. 2A). As expected from the SAGE data (WT count 5, Otx2−/− count 0), this expression decreases in Otx2−/− embryos without complete disappearance, suggesting that Otx2 is necessary for its correct transcription (Fig. 2B). A more striking difference was found regarding eed transcription, which is normally ubiquitous at 6.5 dpc. Eed is the mouse homologue of Drosophila extra sex combs gene, a known repressor of homeotic genes. In mouse, it has been shown to play a role in the formation of the antero-posterior axis at gastrulation (ref. 15; Fig. 2C). SAGE analysis counted four times the eed tag in the embryonic portion of WT embryos but never in the mutants (Table 1). This result is confirmed in the in situ experiments in which little or no transcription is found in the embryonic region of Otx2−/− embryos (Fig. 2D). Conversely, eed expression in the extraembryonic portion is not affected. Hence, eed expression in the embryonic half requires presence of Otx2. With regards to Fgf-15 (16), in situ experiments revealed that it is expressed in all embryonic ectoderm cells of WT 6.5 dpc embryos, perhaps even in a graded pattern (the cells localized at the anterior pole being more strongly stained) (Fig. 2E). However, no expression was detectable in mutant embryos, which agrees with our SAGE data and raises the possibility that transcription of Fgf-15 cannot be achieved in the absence of Otx2. Wnt4, a secreted molecule involved in sexual differentiation and expressed in the developing spinal cord and kidneys (17, 18), is normally not transcribed during gastrulation. As expected, the corresponding tag could not be found in the WT library. Interestingly, its tag was counted twice in the mutant library (Table 1), and in situ hybridization shows a clearly distinguishable signal at the distal tip of Otx2−/− embryos (Fig. 2H). Thus, Otx2−/− embryos display an ectopic expression of Wnt4 during gastrulation.
Figure 1.
Expression of mRNAs for cystatin B and tag 123 in WT and Otx2−/− embryos at 6.5 and 7.5 dpc, and comparison to the Otx2 transcription domain. Values at the lower right hand corner of A, B, E, and F show tag abundance in the two libraries. The black arrowheads point to the embryonic–extraembryonic junction. (A and E) Expression of mRNAs for cystatin B and tag 123 in 6.5 dpc WT embryos. (B and F) Expression of mRNAs for cystatin B and tag 123 in 6.5 dpc Otx2−/− embryos. (G and H) Expression of mRNA for tag 123 in 7.5 dpc embryos. (C and D) β-galactosidase staining of Otx2+/− and Otx2−/− embryos carrying a LacZ reporter gene in the Otx2 locus. (Scale bar: 100 μm.)
Figure 2.
Expression of mRNAs for tag 187 (Q15004), eed, Fgf-15, and Wnt4 in WT and Otx2−/− 6.5 dpc embryos. (Upper) Expression in WT embryos. (Lower) Expression in Otx2−/− embryos. Values at the lower right-hand corners of each picture give the abundance of the corresponding tag in either WT or Otx2−/− libraries. (Scale bar: 100 μm.)
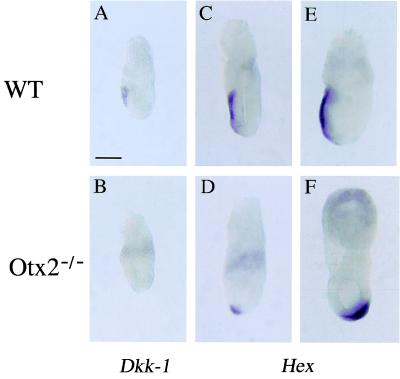
The extent of the anomalies observed in both the epiblast and visceral endoderm lead us to think that Otx2 mutant embryos suffered global antero-posterior patterning defects. Thus, to gain deeper insights into the understanding of the Otx2 phenotype at gastrulation, the two recently described marker genes Dickkopf-1 (Dkk-1) (19) and Hex (20) were also tested. Dkk-1 is a member of a family of secreted proteins and is involved in head induction. It is expressed at 6.5 dpc in the anterior visceral endoderm (AVE) (ref. 21; Fig. 3A) and believed to be the head organizer in mouse. Dkk-1 transcription is abolished in the visceral endoderm of 6.5 dpc mutant embryos (Fig. 3B). This could account for the loss of head structures in Otx2−/− embryos. Expression of the Hex homeobox gene displays anterior asymmetry before gastrulation. Hex-expressing cells are found at the distal tip of the visceral endoderm at 5.5 dpc, and subsequently migrate to the AVE (ref. 20; Fig. 3 C and E). Our results show that, in the Otx2−/− mutant embryos, AVE precursor cells are specified. Indeed, Hex mRNA is expressed at the distal tip in these embryos, albeit the expected anterior migration is impeded (Fig. 2D). This leads, in Otx2−/− 7.5 dpc embryos, to the ectopic confinement of Hex-expressing cells to the region where the node is normally located (Fig. 3F).
Figure 3.
Expression of mRNAs for Dkk-1 and Hex in WT and Otx2−/− embryos at 6.5 and 7.5 dpc. (A and B) Expression of Dkk-1 mRNA at 6.5 dpc in WT and Otx2−/− embryos, respectively. (C and D) Expression of Hex mRNA at 6.5 dpc in WT and Otx2−/− embryos, respectively. (E and F) Same as C and D at 7.5 dpc. (Scale bar: 100 μm.)
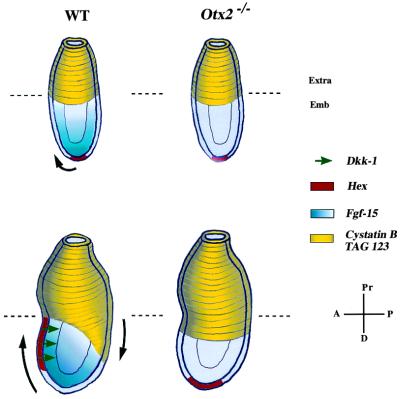
Taken together, the studies of cystatin B, tag 123, and Hex expression patterns suggest that the abnormalities presented by the mutant embryos are probably because of the defective migratory properties of the visceral endoderm tissue as a whole. This may result specifically in the mislocalization of the cells fated to form the AVE, leading to an ineffective head organizer. This critical movement could perhaps be a prerequisite for the expression of the head inductor Dkk-1. Its absence in Otx2−/− embryos supports this hypothesis. Because it has been shown that cerberus-related (10) is not required for murine development, the targeted disruption of Dkk-1 will be of great relevance for the understanding of the Otx2 phenotype.
We also found several members of the Wnt/β-catenin pathway to be affected (21). For instance, mRNA levels for integrin binding protein kinase (a kinase highly homologous to human ILK) and α-catenin are heavily up-regulated in Otx2 mutant embryos (Table 2). Overexpression of ILK could lead to the indirect depletion of β-catenin, by means of GSK3β (glycogen synthase kinase 3β) (22). The loss of β-catenin could be compensated by up-regulation of α-catenin because these two molecules are partially functionally redundant (23). Given the determining role of the Wnt/β-catenin in the formation of the Spemann organizer, it would not be surprising that Otx2 intervenes in this signaling cascade for anterior patterning. Most interesting is the complete loss of expression of Fgf-15 found in Otx2 homozygous embryos. Expressed in a distal to proximal gradient in the epiblast of WT embryos, Fgf-15 seems to parallel the expression of another secreted molecule and global regulator of antero-posterior patterning: cripto (24). It is worth noting that their expression domains are symmetric and complementary. Cripto was shown to be required for the conversion of the proximal-distal axis into the antero-posterior axis through a dialogue between the hypoblast and the epiblast. Thus, Fgf-15 could also mediate such a function by instrumenting underlying cells of the distal endoderm to shift anteriorly. Conversely, this pattern could also reflect an Otx2-mediated inductive signal emanating from the visceral endoderm toward the epiblast (Fig. 4). A similar hypothesis can be suggested for the mRNA corresponding to tag 187 (Q15004), which is much more expressed in WT than in Otx2−/− embryos in the embryonic ectoderm. Deciphering the function of this mRNA, as well as identification of the Fgf-15 partners, could lead to a deeper molecular understanding of neural induction and morphogenetic movements during gastrulation. Nevertheless, it becomes more and more apparent that Otx2 plays a pivotal role in very early development, perhaps as soon as 5.5 dpc or earlier in the global control of antero-posterior patterning through modulation of morphogenetic movements. It is noteworthy that the inactivation of Otx2 entails the double knockouts of Dkk-1 and Fgf-15 in the visceral endoderm and epiblast, respectively, in gastrulating mouse embryos.
Figure 4.
Defective formation of the antero-posterior axis in early gastrulating Otx2−/− embryos. Conversion of the proximal-distal axis into the antero-posterior axis begins before gastrulation. Cells of the distal visceral endoderm undergo an oriented movement toward the future anterior pole of the embryo as illustrated by the Hex expression domain (shown in red). Conversely, cells of the extraembryonic endoderm expressing cystatin B and tag 123 appear to converge to the future posterior pole (shown in yellow). Black arrows symbolize this movement. In WT embryos, the anterior pole is also marked by the expression of Dkk-1. In the ectoderm layer, Fgf-15 expression forms a gradient distributed along the proximal-distal axis before gastrulation, then along the antero-posterior axis at 6.5 dpc. In Otx2−/− embryos, the oriented movement of the cells from the visceral endoderm is abolished, resulting in the ectopic localization of the Hex expression domain as well as the accumulation of the cystatin B and tag 123-expressing cells at the embryonic–extraembryonic junction. Formation of the head organizer is also impaired, as assessed by the loss of expression of the head inductor Dkk-1. In addition, the ectodermal layer is affected, as shown by the absence of Fgf-15 expression. Hence, Otx2 is required for global cellular movements in the visceral endoderm, as well as for the proper orientation of the antero-posterior axis before gastrulation. Extra, extraembryonic region; Emb, embryonic region; A, anterior; P, posterior; Pr, proximal; D, distal. Embryos at the top are pregastrulating embryos. Embryos at the bottom are 6.5 dpc embryos.
In conclusion, the application of SAGE to the understanding of the Otx2 phenotype at gastrulation delivered extensive information. It allowed to identify transcripts whose regulation is modified in the absence of the Otx2 protein. Because SAGE provides such considerable amounts of data, a systematic functional screening needs to be set up to readily have access to the tags of primary interest. In the near future, confrontation of independent SAGE libraries performed at the same embryonic stage but over distinct mutations will permit the unraveling of genetic cascades and a better discernment on the direct effects of each mutation.
Acknowledgments
We thank Dr. J. C. Aude for updating the SADE web site with mouse embryos data. This work was supported by grants from the European commission Biotech CT-96-0378, the Association Francaise contre les Myopathies, and the Centre National de la Recherche Scientifique.
Abbreviations
- otd
orthodenticle
- dpc
days postcoitum
- SAGE
serial analysis of gene expression
- WT
wild type
- EST
expressed sequence tag
- AVE
anterior visceral endoderm
- Dkk-1
Dickkopf-1
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011513398.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011513398
References
- 1.Cohen S, Jurgens G. Nature (London) 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 2.Klein W H, Li X. Biochem Biophys Res Commun. 1999;258:229–233. doi: 10.1006/bbrc.1999.0449. [DOI] [PubMed] [Google Scholar]
- 3.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 4.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brûlet P. Development (Cambridge, UK) 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 5.Rhinn M, Dierich A, Shawlot W, Behringer R R, Le Meur M, Ang S-L. Development (Cambridge, UK) 1998;125:845–856. doi: 10.1242/dev.125.5.845. [DOI] [PubMed] [Google Scholar]
- 6.Velculescu V, Zhang L, Vogelstein B, Kinzler K. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Virlon B, Cheval L, Buhler J-M, Billon E, Doucet A, Elalouf J-M. Proc Natl Acad Sci USA. 1999;96:15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Nature (London) 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 10.Belo J A, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, De Robertis E M. Mech Dev. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- 11.Varlet I, Collignon J, Robertson E. Development (Cambridge, UK) 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- 12.Pennacchio L A, Myers R M. Genome Res. 1996;6:1103–1109. doi: 10.1101/gr.6.11.1103. [DOI] [PubMed] [Google Scholar]
- 13.Luster A, Weinshank R, Feinman R, Ravetch J. J Biol Chem. 1998;263:12036–12043. [PubMed] [Google Scholar]
- 14.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. DNA Res. 1995;2:199–210. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 15.Shumacher A, Faust C, Magnuson T. Nature (London) 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 16.McWhirter J, Goulding M, Weiner J, Chun J, Murre C. Development (Cambridge, UK) 1997;124:3221–3232. doi: 10.1242/dev.124.17.3221. [DOI] [PubMed] [Google Scholar]
- 17.Stark K, Vainio S, Vassileva G, McMahon A P. Nature (London) 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 18.Vainio S, Heikkila M, Kispert A, Chin N, McMahon A. Nature (London) 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 19.Glinka A, Wu W, Delius H, Monaghan A P, Blumenstock C, Niehrs C. Nature (London) 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 20.Thomas P Q, Brown A, Beddington R. Development (Cambridge, UK) 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- 21.Moon R, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 22.Novak A, Hsu S-C, Leung-Hagesteijn C, Raveda G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vleminckx K, Kemler R. BioEssays. 1999;21:211–220. doi: 10.1002/(SICI)1521-1878(199903)21:3<211::AID-BIES5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 24.Ding J, Yang L, Yan Y-T, Chen A, Desai N, Wynshaw-Boris A, Shen M M. Nature (London) 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]