Abstract
A Clostridium acetobutylicum ATCC 824 genomic library was constructed using randomly sheared DNA. Library inserts conferring increased tolerance to 1-butanol were isolated using two protocols. Protocol I utilized a single round of butanol challenges in batch culture, while protocol II, which gave clearly superior outcomes, was based on the serial transfer of stationary-phase cultures into progressively higher butanol concentrations. DNA microarray analysis made a high-resolution assessment of the dynamic process of library enrichment possible for the first time. Protocol I yielded a library insert containing the entire coding region of the gene CAC0003 (which codes for a protein of unknown function) but also several DNA fragments containing promoter regions. Protocol II enabled the successful identification of DNA fragments containing several intact genes conferring preferential growth under conditions of butanol stress. Since expression using the employed library is possible only from natural promoters, among the enriched genes, we identified 16 genes that constitute the first cistron of a transcriptional unit. These genes include four transcriptional regulators (CAC0977, CAC1463, CAC1869, and CAC2495). After subcloning plasmids carrying the CAC0003 and CAC1869 genes, strains 824(pCAC0003) and 824(pCAC1869) exhibited 13% and an 81% increases, respectively, in butanol tolerance relative to the plasmid control strain. 824(pCAC1869) consistently grew to higher cell densities in challenged and unchallenged cultures and exhibited prolonged metabolism. Our serial enrichment approach provided a more detailed understanding of the dynamic process of library enrichment under conditions of selective growth. Further characterization of the genes identified in this study will likely enhance our understanding of the complex phenotype of solvent tolerance.
Clostridium acetobutylicum ATCC 824 is important for the biological production of butanol and acetone, two solvents currently produced mainly from petrochemical feedstocks. Until the mid-1950s, C. acetobutylicum was used in the profitable ABE (acetone, butanol, and ethanol) fermentation (56). In response to increasing demands for limited petroleum resources, interest in ABE fermentation has been renewed to the point that new fermentation facilities are again becoming economically viable (15, 25).
An important part of biological research in this area is understanding and in turn ameliorating the impact of intermediate and final metabolite products on cell growth and product formation. For instance, the switch in C. acetobutylicum fermentation from the accumulation of acetic and butyric acids during the acidogenic exponential phase to acid reassimilation and butanol, acetone, and ethanol formation during the solventogenic stationary phase may be due to butyrate accumulation (37, 40, 55, 58). Before long, however, the accumulation of butanol also has a negative impact on cellular processes due primarily to the partitioning of polar solvents into the hydrophobic region of the phospholipid bilayer (43).
Fundamental work on the impact of alcohols and ketones on Escherichia coli showed that the intercalation of solvents within the lipid bilayer increases membrane fluidity (21) and also affects lipid-protein interactions integral to membrane function (22, 23). The microbial response to perturbations in membrane fluidity, or “homeoviscous adaptation,” has been studied extensively with regard to the response of E. coli to temperature upshifts, a stress phenomenon with an impact on membrane fluidity similar to that of solvent toxicity (44). Responses of various microorganisms to solvents include active solvent expulsion by molecular pumps, alterations in the composition of membrane-lipid headgroups, and the adjustment of the protein content within the cell membrane (53).
Butanol has a lower partition coefficient (0.8) than either toluene (2.5) or benzene (2.0) and is therefore predictably more toxic (43). When clostridia are exposed to butanol, the ratio of saturated to unsaturated fatty acids incorporated in the membrane lipid bilayer increases, presumably to compensate for the fluidity increase imposed by the solvent (7, 8, 28). Solvents inhibit membrane-bound ATPases, resulting in a drop in internal pH and the abolishment of the ΔpH gradient across the membrane (10, 46). Butanol also inhibits glucose uptake, thus inhibiting energy generation, which is compounded by an independent drop in intracellular ATP levels (10).
Improvements to solvent tolerance and titers in C. acetobutylicum are not limited to the metabolic engineering of membrane functions. The overexpression of groESL, a class I stress response operon, resulted in increased solvent tolerance and solvent titers 33% higher than those of a plasmid control strain (50), indicating a denaturing effect of solvents on functional enzymes and a role for stress proteins in solvent tolerance. Secondly, knockout of the metabolic intermediate enzyme butyrate kinase by chromosomal insertion resulted in a strain that produced butanol titers of 225 mM (1.97% [vol/vol]) (19). These observations underscore the potential complexity of solvent toxicity and warrant a genomic approach to discovering genes that impart solvent tolerance.
Screening of genomic libraries has been used to identify phenotype-specific genes (16, 18, 24). Genomic libraries can be generated from fragmented genomic DNA ligated into a suitable expression vector. Altered gene expression as a result of cloned fragments has the potential to confer a selective growth advantage under applied solvent stress. Plating of stressed cultures and subsequent colony isolation and sequencing have traditionally been the methods for identifying selected gene fragments (41). More recently, E. coli genomic-library DNA was hybridized to DNA microarrays in order to identify genes enriched upon selective growth in the presence of the antimicrobial agent Pine-Sol (18).
In this work, we identified elements of a genomic library enriched by preferential growth under conditions of butanol stress. First, a genomic library was generated, and its coverage was estimated by hybridization to C. acetobutylicum DNA microarrays. Next, the genomic library was used to electrotransform C. acetobutylicum, and library-bearing cultures were subjected to two stress selection protocols in order to identify gene fragments conferring preferential growth under conditions of butanol stress. To visualize the dynamics of library enrichment over the course of a challenge experiment, plasmid library inserts were amplified by PCR, fluorescently labeled, and hybridized on DNA microarrays. Enriched genomic inserts containing whole genes were then subcloned back into wild-type C. acetobutylicum in order to characterize the growth and solvent tolerance properties conferred by the enriched genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and PCR primers.
One Shot Top10 E. coli (Invitrogen, Carlsbad, CA) was used for library construction. E. coli ER2275(pAN1) (31) was used to methylate plasmid DNA. C. acetobutylicum ATCC 824 was received from the American Type Culture Collection (ATCC, Manassas, VA). The pIMP1 vector (32) was utilized for generating the pLib1 genomic library, and forward (5′-ACACCACGTAGTTATTGGGAGGTC) and reverse (5′-GCTGCAAGGCGATTAAGTTGGGTA) PCR primers flanking the library insertion site were used for PCR amplification.
Growth conditions.
E. coli cultures were grown aerobically at 37°C in liquid Luria-Bertani (LB) medium or on solid LB agar plates supplemented with ampicillin (100 μg/ml) as necessary. C. acetobutylicum ATCC 824 was revived from −85°C storage by plating onto 2× yeast-tryptone-glucose (pH 5.8) solid medium under anaerobic conditions at 37°C. Individual colonies that were at least 5 days old were transferred into 10-ml tubes of liquid clostridial growth medium (CGM) (containing 80g/liter glucose and buffered with 30 mM sodium acetate unless otherwise noted) (54), heat shocked at 70 to 80°C for 10 min to induce germination, and grown overnight to the mid-exponential growth phase (absorbance at 600 nm [A600] of ∼0.6). Flask cultures were then inoculated (1:100 [vol/vol]) with mid-exponential-phase cells. Recombinant strains were supplemented with erythromycin (Em) (40 μg/ml for plates and 100 μg/ml for liquid cultures).
Analytical methods.
Cell growth was monitored by reading the A600 using a Thermo Spectronic (Rochester, NY) Biomate3 spectrophotometer. Culture supernatants were analyzed for glucose, acetate, butyrate, butanol, acetone, ethanol, and acetoin using a Waters (Milford, MA) high-performance liquid chromatograph (50).
DNA isolation and manipulation.
Plasmid DNA was isolated from E. coli using Hurricane plasmid miniprep and maxiprep kits (Gerard Biotech, Oxford, OH). Genomic and recombinant-plasmid DNAs from C. acetobutylicum were isolated as described previously (20). Genomic and plasmid DNAs were fragmented using a Branson Sonifier 150 (Branson Ultrasonics, Danbury, CT) fitted with a 3/8-inch tip. DNA for sonication was diluted to 100 μg/ml in Tris-EDTA buffer (10 mM Tris-HCl and 1 mM EDTA [pH 8.0]) and sonicated on ice for three 5-s pulses with a 1-min rest between pulses. Power settings were adjusted to achieve the desired fragment length, which was confirmed on a 0.7% agarose gel or using an Agilent (Palo Alto, CA) 2100 Bioanalyzer with a DNA 1000 or DNA 7500 LabChip.
Library construction.
C. acetobutylicum genomic DNA was fragmented by sonication at a power setting of 7 to generate an average fragment size of ∼3 kb, as determined by agarose gel electrophoresis. Damaged termini were repaired to generate blunt ends by combining 10 μg of sonicated DNA with 10 units of T4 DNA polymerase (New England Biolabs, Ipswich, MA), 10 μl of bovine serum albumin (100 μg/ml), 10 μl of New England Biolabs buffer 2 (100 mM Tris-HCl, 100 mM MgCl2, 500 mM NaCl, 1 mM dithiothreitol [pH 7.9]), and 5 μl of 2.5 mM deoxynucleoside triphosphates in a total volume of 100 μl and incubating the mixture for 2 h at 15°C. After purification using a GFX column (GE Healthcare Biosciences Corp., Piscataway, NJ), blunt-ended DNA fragments were ligated by T4 DNA ligase into the pIMP1 vector that had been linearized by digestion with PvuII and dephosphorylated using calf intestinal phosphatase according to the manufacturer's specifications (New England Biolabs). Ligated DNA was then transformed into Top10 competent E. coli cells to generate ∼10,000 transformant colonies over multiple transformations. Individual transformant colonies were combined by the application of 5 ml of liquid LB medium to each transformant plate, followed by a 2-h incubation with gentle mixing. After incubation, the liquid LB medium was recovered from each plate and used to inoculate 500 ml of LB medium with 100 μg/ml ampicillin and grown overnight, and frozen stocks and a maxiprep of the plasmid library were generated as described above. Prior to electroporation of competent C. acetobutylicum cells, plasmid library DNA was methylated in E. coli ER2275(pAN1) to prevent digestion by a membrane-bound endonuclease (31).
Butanol challenges, library enrichment, and metabolic properties of enriched cultures.
Two challenge protocols were employed in this study. Protocol I was based on methods described previously by Gill et al. for the enrichment of an E. coli genomic library (18). Briefly, methylated plasmid library DNA was used to transform C. acetobutylicum as described previously (32), yielding a transformation efficiency of 500 CFU per microgram (6 × 104 total transformants). The transformed C. acetobutylicum [pLib1] culture was grown to early exponential phase (A600 of ∼0.6), split into four 50-ml aliquots, and used to inoculate (1:10 [vol/vol]) four challenged cultures (450 ml of CGM containing 100 μg/ml Em and 0%, 0.6%, 1.2%, or 1.5% [vol/vol] butanol after inoculation). Challenged cultures were monitored for growth and product profiles, and plasmid DNA was isolated from each of the four challenged cultures at the onset of stationary phase.
For protocol II, transformations were conducted as described above but with a voltage setting of 2.0 kV, resulting in an increase in the transformation efficiency to 5 × 104 CFU/μg (8 × 105 total transformants). When the transformed cultures reached an A600 of 0.6, 50-μl aliquots of the frozen stock were prepared using CGM containing 15% glycerol. The desired numbers of frozen stocks were subsequently thawed, used to inoculate individual 10-ml test tubes of CGM, and incubated overnight. At an A600 of 0.6, the test tubes were used to inoculate 100 ml of CGM (buffered with 30 mM acetate) containing 100 μg/ml Em and 0.5% butanol. Butanol-challenged cultures were incubated until they reached stationary phase, at which point 1 ml was used as an inoculum for new 100-ml flasks containing progressively higher butanol concentrations. This process was continued until the stationary-phase A600 was reduced to less than 2.0, at which point the inoculum volume was increased to 5 ml and transfers again continued until no growth was observed within 48 h of inoculation. Plasmid DNA was isolated from the unchallenged C. acetobutylicum [pLib1] inoculum as well as from each challenged culture at the onset of the stationary phase. Supernatant samples were taken from each challenged culture to determine end-point metabolite levels. Isolated plasmid DNA was used for DNA microarray analysis and also to transform E. coli to generate sufficient material for the sequencing of individual genomic inserts. Finally, test tubes with 10 ml of CGM containing 100 g/liter glucose and 100 μg/ml Em were inoculated in duplicate from each challenge flask, grown for 168 to 216 h, and sampled to determine the solvent production capabilities of cultures enriched in fragments conferring tolerance to butanol.
MIC assays.
For MIC assays of freshly inoculated cultures, butanol was added to CGM containing 100 μg/ml Em to generate a dilution series of 0 to 2% butanol, and 9.9 ml was aliquoted into individual test tubes. Multiple colonies per strain were then inoculated into butanol-free medium and grown to the mid-exponential growth phase (A600 of ∼0.6), and 100 μl was used to inoculate the butanol dilution series in quadruplicate per strain (i.e., four biological replicates per strain). Challenge tubes were then incubated for 12 h and assayed for growth (A600), and biological replicate values were averaged. For MIC assays of actively growing cultures, 400 ml of CGM with 100 μg/ml Em was grown to an A600 of 1.0 in duplicate for each strain. Ten milliliters of culture was then aliquoted into individual test tubes containing butanol to generate a final concentration of 0 to 2%, with four independent test tubes per challenge level (i.e., four technical replicates for each of two biological replicates per strain). Challenge tubes were sampled for growth (A600) after 6, 12, and 18 h of incubation, and replicate values were averaged at each challenge level and sample time.
From each MIC assay, percent tolerance relative to unchallenged cultures was estimated at each challenge level and sample time as follows:
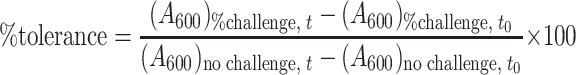 |
To compare butanol tolerance between strains, the area under the percent tolerance (Area%T) curve of each strain was estimated using the trapezoid rule. Finally, the relative tolerance (% RT) of strains bearing plasmid library inserts compared to the 824(pIMP1) plasmid control strain was estimated as follows:
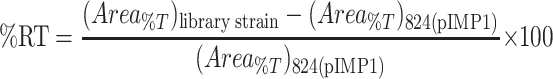 |
C. acetobutylicum DNA microarrays.
The design and use of DNA microarrays employed in this study have been described previously (4). Briefly, primers were designed to generate a PCR product for each gene that is not homologous to the rest of the genome. Individual PCR products for ∼95% of the genes in the C. acetobutylicum genome were then generated, purified, spotted onto Corning UltraGAPS slides, and UV cross-linked at 300 mJ.
Microarray analysis was employed to asses nonspecific hybridization of the vector used for library generation (pIMP1) to DNA microarray spots by hybridizing 1 μg of sonicated, Cy3-labeled pIMP1 vector against 1 μg of sonicated, Cy5-labeled C. acetobutylicum genomic DNA. While the genomic DNA channel showed signal-to-noise ratios greater than 3 for over 85% of all genes, signal intensities in the Cy3 channel were at background levels for all but three genes, CAC0706, an endo-1,4-β-glucanase; CAC3153, an uncharacterized protein; and CAC3428, a 6Fe-6S prismane cluster-containing protein (35), which showed considerable nonspecific hybridization with the pIMP1 shuttle vector. These genes were therefore discarded during the DNA microarray analysis of library enrichment.
Microarray analysis of library enrichment.
For protocol I challenge experiments, wild-type challenge culture plasmid DNA was sonicated at a power setting of 3 to generate an average fragment length of 1.0 kb, as determined by agarose gel electrophoresis. Amino-allyl dUTP (aa-dUTP) was incorporated by random hexamer-primed extension of 2 μg of the sonicated template using a Klenow fragment (New England Biolabs) (39). Cyanine dyes (Cy3 and Cy5) were then coupled to the aa-dUTP, and unincorporated dyes were removed using a QIAGEN (Valencia, CA) PCR purification kit as described previously (4). Upon hybridization of butanol-enriched pLib1 plasmid DNA to DNA microarrays, more genes showed signal intensities above the background (3,895 genes) than with similarly labeled plasmid library DNA isolated from E. coli and used to estimate the library coverage (2,981 genes) (see Results). This superfluous signal was due to highly active, membrane-bound nucleases of C. acetobutylicum that digest genomic DNA upon cell lysis (27), which in turn generates fluorescent signal not attributable to genomic-library inserts upon hybridization to DNA microarrays.
For protocol II challenge experiments, plasmid library inserts were amplified by PCR using plasmid-specific primer regions flanking the library insert site, yielding effective amplification of genomic-library DNA template and pIMP1 vector template but without nonspecific amplification of C. acetobutylicum genomic DNA (see Fig. S1A in the supplemental material). When intended for hybridization to DNA microarrays, PCR was conducted in the presence of 275 μM dATP, 180 μM dGTP, 180 μM dCTP, 90 μM dTTP, and 180 μM aa-dUTP, and PCR products were purified using a Microcon YM-30 column (Millipore, Billerica, MA) and washed twice with 450 μl distilled water according to manufacturer's specifications. Cyanine dyes were then coupled to the aa-dUTP, and unincorporated dyes were removed using a QIAGEN PCR purification kit as described previously (4). To test for PCR bias on the representation of library inserts, DNA microarrays were used to compare the products of PCR amplification to the original library (see Fig. S1B and C in the supplemental material). The amount of data scatter increased with the number of PCR cycles used for insert amplification; therefore, 20 PCR cycles were used to generate sufficient product for microarray hybridization without biasing library representation.
Gene fragment enrichment was determined by averaging background-subtracted and nonspecific-hybridization-subtracted signal intensities for the three replicate microarray spots per gene and ranking in decreasing order of average signal intensity.
Microarray data accession number.
DNA microarray data were deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under the series accession number GSE6289.
RESULTS
Genomic-library characterization.
Approximately 10,000 E. coli transformants containing, on average, a 1.5-kb genomic DNA fragment (based on the sequencing of 22 library clones) inserted into the pIMP1 shuttle vector were combined to form the genomic library pLib1. To estimate the coverage across the C. acetobutylicum genome, 4.2 μg of purified library plasmid DNA was fragmented and labeled with Cy3 as described above and hybridized on DNA microarrays against 1 μg of fragmented, Cy5-labeled C. acetobutylicum genomic DNA (4.2 μg of labeled library DNA was hybridized in order to have approximately 1 μg of hybridized genomic-insert DNA and 3.2 μg of pIMP1 vector). Signal intensities for 73% of all genes had signal-to-noise ratios greater than 3, which is indicative of statistically significant signal above background. Therefore, the pLib1 library is estimated to contain approximately 73% of the genes of the C. acetobutylicum genome.
Library enrichment at discrete butanol challenge levels (protocol I).
C. acetobutylicum transformed with methylated pLib1 DNA was grown to the exponential phase and used to inoculate CGM containing 100 μg/ml Em and ca. 0, 0.6, 1.2, and 1.5% (vol/vol) butanol. A dose-dependent butanol inhibition of cell growth and metabolite production was observed (see Fig. S2A to E in the supplemental material). Plasmid DNA was isolated from individual colonies of the culture challenged with 1.56% butanol for sequencing of genomic inserts (see Table S1 in the supplemental material). One of the sequenced inserts (sequence 1, found in 6 of 23 sequenced plasmids) contained a complete, intact gene (intergenic region, open reading frame [ORF], and terminator region of CAC0003, a small conserved protein ortholog of the uncharacterized protein YAAA in Bacillus subtilis). Other isolated library inserts contained promoter elements and partial gene fragments that potentially titrate regulatory proteins detrimental to a tolerant phenotype (33, 48, 49) or produce aptamer-like transcripts that function to improve solvent tolerance (11). However, library inserts isolated using protocol I failed to identify many genes imparting a solvent-tolerant phenotype, and therefore, a more robust library enrichment protocol was developed.
Increasing enrichment through stationary-phase transfers (protocol II).
To more stringently enrich solvent-tolerance-conferring genetic elements, selection protocol II was developed. This protocol is based on stationary-phase transfers to increase the number of generations of selective growth coupled with PCR amplification of plasmid library inserts in order to circumvent the contaminating fragmented genomic DNA. Stationary-phase culture transfers were utilized to select for insert-bearing cells capable of delaying transitional and stationary-phase phenomena and thus exhibiting prolonged vegetative growth under butanol stress. These cells would then have a greater propensity to repopulate freshly inoculated media with increasing butanol concentrations.
Results from two independent butanol challenge experiments using stationary-phase transfers are shown in Table S2 in the supplemental material. For comparison, the corresponding values for the single-flask challenges conducted using protocol I are also shown. With the gradual selection of protocol II, challenged cultures were better able to cope with increasing butanol levels, resulting in greater product formation and glucose consumption than those with only a single bolus of butanol stress (protocol I).
Our data suggest that butanol stress promotes C. acetobutylicum degeneration—loss of the pSOL1 megaplasmid necessary for both sporulation and solvent production (14). By transfer 10 to 11, individual colonies plated from both biological replicates rapidly changed colony morphology, which is indicative of degeneracy (13, 57). Additional evidence of strain degeneration is shown in the last two columns of Table S2 in the supplemental material. By the 10th transfer of replicate 2 and the 12th transfer of replicate 1, butanol production of outgrowth tubes decreased considerably. Following the 14th transfer for both biological replicate experiments, outgrowth tubes not only resumed butanol production but also exceeded the production level of early transfers.
The pIMP1 plasmid control strain was subjected to the same stationary-phase selection protocol (protocol II) used for the enrichment of library cultures. Biological replicate plasmid control cultures had a prolonged lag phase (>24 h) at all challenge levels and were unable to grow above 1.3% (vol/vol) butanol (transfer 10) (see Table S2 in the supplemental material), consistent with previously observed values for C. acetobutylicum butanol tolerance (8, 36, 51, 52).
Direct visualization of gene fragment enrichment.
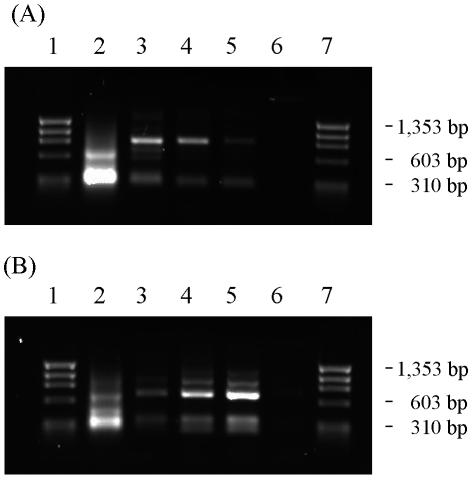
Figure 1A and B show PCR-amplified insert DNA from the 1st, 4th, 7th, 10th, and 13th transfers (lanes 2 to 6, respectively) of biological replicate challenges 1 (Fig. 1A) and 2 (Fig. 1B). The data show strong selection for a subset of the initially diverse population of chromosomal inserts. In the earliest challenged cultures, a smear of insert sizes was visible, with two broad bands of ∼300 and 600 bp, respectively (lane 2). This smear corresponds to inserts that were smaller than the average insert size of the original E. coli pLib1 library (∼1 kb) (see Fig. S1A in the supplemental material), perhaps reflecting a preferential uptake of smaller plasmids by C. acetobutylicum. Significant gene enrichment was evident as early as the fourth transfer (Fig. 1, lane 3), and a consistent set of bands persisted after the seventh transfer (lanes 4 to 5). Transfer 13 (lane 6) showed relatively less PCR product, a result of low final cell densities and therefore low miniprep yields.
FIG. 1.
Enrichment of tolerance gene fragments in (A) biological replicate 1 and (B) biological replicate 2 of stationary-phase transfer challenges (protocol II). Each lane contains the products of 20 PCR cycles using 0.1 μl of C. acetobutylicum pLib1 minipreps of the indicated challenge culture. Lanes 1 and 7, φX174-HaeIII standard; lanes 2, 3, 4, 5, and 6, PCR products from the 1st, 4th, 7th, 10th, and 13th stationary-phase transfers of the indicated replicate challenge flask.
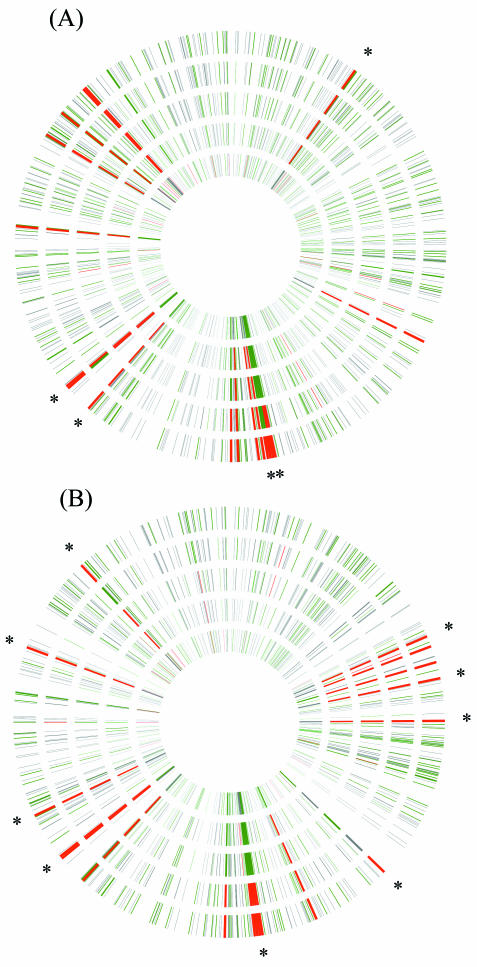
FIG. 2.
Physical map of the C. acetobutylicum genome color-coded with the degree of fragment enrichment as determined by DNA microarray analysis for (A) biological replicate 1 and (B) biological replicate 2 of stationary-phase transfer challenges (protocol II). Plasmid library insert DNA isolated from the initial challenge inoculum was PCR amplified, Cy3 labeled, and hybridized against Cy5-labeled, amplified library insert DNA from the 4th, 7th, 10th, and 13th challenge transfers. After subtraction of background and nonspecific hybridization signals and averaging of replicate spots, gene signal intensities were ranked within a given challenge transfer. Concentric circles represent (starting with the innermost circle) the ranks of individual genes from the initial inoculum and 4th, 7th, 10th, and 13th transfers, respectively. Genes were color-coded according to their percentile rank in a given transfer, such that the top 5% are red, 5 to 33% are green, and 33 to 100% are gray. Genes that did not generate a signal-to-noise ratio greater than 3 on a given array are white.
High-resolution DNA microarray-based visualization of library enrichment.
To visualize the dynamics of library enrichment with high genetic resolution, DNA microarrays were hybridized with Cy5-labeled PCR products from the 4th, 7th, 10th, and 13th transfers of each biological replicate challenge against PCR-amplified, Cy3-labeled insert DNA isolated from the test tube used to inoculate the replicate challenge series. The resulting signal intensities (after background and nonspecific-signal subtraction) of all genes in the challenged DNA channel were then sorted and grouped by intensity rank for each microarray. The first group consisted of genes in the top 5% of intensity rank at a given transfer, the second group consisted of genes in the 5 to 33% of intensity rank, and the third group consisted of genes in the bottom two-thirds of intensity rank. The enrichment process at the genomic level is shown in Fig. 2. The C. acetobutylicum genome (chromosome and megaplasmid) is represented as a closed circle with the ordinal gene position increasing in a clockwise direction and megaplasmid genes falling roughly between 11 and 12 o'clock. Individual concentric circles show the relative gene ranks of a single transfer starting from the initial inoculum (innermost circle) and working outwards to the 13th transfer (outermost circle). Enrichment of contiguous genes and operons is apparent (Fig. 2). Enrichment of any particular gene is discernible by moving from the inner circle outward, with a change from green to red indicative of increasing enrichment. An additional illustration of the dynamic nature of gene enrichment is shown in Fig. S3 in the supplemental material, where the signal intensity ranks for the top-10-ranking genes as of the 13th transfer are tracked over the course of butanol challenge transfers. Several genes (e.g., CAC0742, CAC3289, and CAC3005) quickly increase in signal intensity rank with increasing transfers, while other genes (e.g., CAC1868 to CAC1870) are more gradually enriched during the selection process. For each biological replicate challenge, a tabular list of the top-70-ranking genes as of the 13th transfer is shown in Table S3 in the supplemental material.
Further selection and likely role of enriched genes.
The pIMP1 shuttle vector does not contain a constitutive promoter to transcribe library inserts separated from their natural promoter during library generation. Therefore, solvent tolerance as a result of protein overexpression would be from a natural promoter. C. acetobutylicum preferentially took up plasmids with smaller library inserts (Fig. 1), with an average insert size of ∼600 bp. Based on these facts, an enriched gene would most likely be the first gene of a transcriptional unit (TU). Sixteen genes between the two biological replicate experiments are marked with asterisks in Fig. 2 (14 asterisks because only one asterisk is used to mark the contiguous genes CAC1868 to CAC1870). These 16 genes not only were ranked in the top 5% as of the 13th transfer but are also the start of a predicted TU (38). These 16 genes are the most likely genes to function directly (i.e., by overexpression of a relevant protein from an intact natural promoter) for imparting butanol tolerance, thus resulting in preferential enrichment.
Of these 16 genes that both were significantly enriched and are the first gene of a predicted operon, 4 are annotated as transcriptional regulators (CAC0977, CAC1463, CAC1869, and CAC2495) (35). Such genes have the ability to directly or indirectly regulate a large set of other genes, thus possibly affecting an array of solvent tolerance mechanisms. In particular, CAC0977 is annotated as an Lrp (leucine-responsive regulatory) protein (35) for which homologs have been found in 45% of available bacterial genomes (47). Lrp has been shown to control a regulon of upwards of 10% of the E. coli genome, including genes for sugar transport as well as amino acid biosynthesis and degradation (12, 34, 45). It should be clarified that, at least for CAC1869 and CAC2495, neighboring genes (CAC1868 and CAC1870, and CAC2494, respectively) are also highly enriched genes, and each contiguous set of genes may exist on a single genomic fragment.
Also among the 16 identified enriched genes is CAC0854, annotated as a GTPase subunit with a role in antibiotic resistance (35). CAC0395 is annotated as kdgK, coding for 2-keto-3-deoxygluconate kinase, a carbohydrate metabolism enzyme that may be growth rate limiting under butanol stress, and CAC2413 is a predicted membrane protein (35). CAC0742, CAC1870, CAC1874, and CAC3435 are an uncharacterized protein and three hypothetical proteins, respectively (35). CAC1868 and CAC2633 are similarly uncharacterized, although they do have homologs (also uncharacterized) in B. subtilis (35). Finally, CAC3143 is the DNA-dependent RNA polymerase (35).
Individual plasmid inserts from both biological replicate experiments were sequenced, and sequence information confirmed the microarray data. For instance, 30 inserts (11 from replicate 1 and 19 from replicate 2) were found to be identical and comprised the genomic region spanning CAC1868 to CAC1870 (86% of CAC1868, all of CAC1869, and 61% of CAC1870). Similarly, nine sequences from the second biological replicate experiment were themselves identical, beginning at the intergenic region between CAC2493 and CAC2494 and ending just inside the CAC2495 ORF. This indicates that CAC2494 (annotated as a predicted membrane protein) is a complete gene within this genomic fragment. CAC2413 and CAC3359 were also identified in at least one sequenced insert.
Strain 824(pCAC1869) shows increased tolerance to butanol and prolonged metabolic activity.
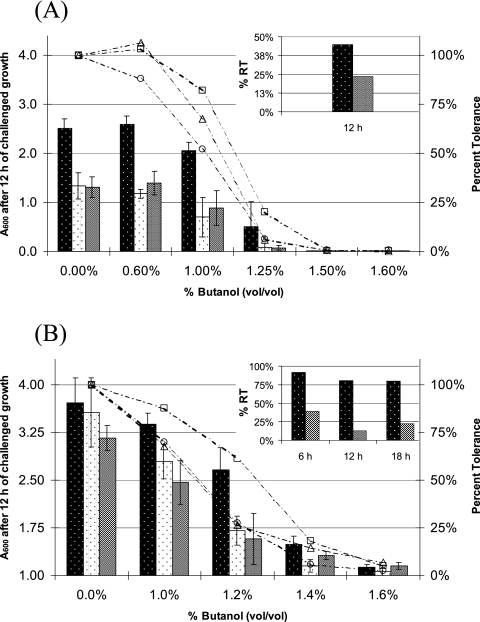
Sequenced plasmids carrying intact CAC0003 and CAC1869 ORFs (hereafter referred to as pCAC0003 and pCAC1869, respectively) were electroporated back into wild-type C. acetobutylicum to examine whether enriched gene fragments confer solvent tolerance in the absence of serial enrichment. First, the MIC of butanol in newly inoculated test tube cultures was measured, and results for 824(pCAC1869), 824(pCAC0003), and 824(pIMP1) are shown in Fig. 3A. At all challenge levels, strains 824(pCAC1869) and 824(pCAC0003) displayed greater percent tolerance (relative to their respective uninhibited cultures) than the plasmid control strain 824(pIMP1). The improvement in tolerance (inset graph in Fig. 3A) was 45% and 24% for 824(pCAC1869) and 824(pCAC0003), respectively, compared to 824(pIMP1). Two of the four 824(pCAC1869) biological replicates grew in medium containing 1.25% butanol, while none of the four test tubes showed growth at 1.25% butanol for strain 824(pCAC0003) or 824(pIMP1). However, the MIC of butanol in newly inoculated cultures was not affected with statistical significance, even after several attempts with different inoculum levels, incubation times, and butanol test tube concentrations (data not shown).
FIG. 3.
MIC assays of strains 824(pCAC1869) (
), 824(pIMP1) ( ), and 824(pCAC0003) (□). Growth measurements were used to calculate the percent tolerance at each challenge level for 824(pCAC1869) (□), 824(pIMP1) (○), and 824(pCAC0003) (▵). (A) Inoculation of a dilution series of butanol in CGM. (B) Butanol challenge of actively growing cultures (A600 of ∼1.0). Growth data for 12 h postchallenge are displayed. Inset graphs show the percent relative tolerance (% RT) to butanol for library strains relative to the pIMP1 control strain. See the text for details.
), and 824(pCAC0003) (□). Growth measurements were used to calculate the percent tolerance at each challenge level for 824(pCAC1869) (□), 824(pIMP1) (○), and 824(pCAC0003) (▵). (A) Inoculation of a dilution series of butanol in CGM. (B) Butanol challenge of actively growing cultures (A600 of ∼1.0). Growth data for 12 h postchallenge are displayed. Inset graphs show the percent relative tolerance (% RT) to butanol for library strains relative to the pIMP1 control strain. See the text for details.
The MIC of butanol on actively growing cultures was measured next, and data from 12 h postchallenge are shown in Fig. 3B. Strain 824(pCAC1869) again showed dramatically improved butanol tolerance at all challenge levels relative to the 824(pIMP1) control strain, with 91%, 81%, and 80% increases in percent relative tolerance at 6, 12, and 18 h postchallenge, respectively (shown in the inset graph of Fig. 3B). Relative to 824(pIMP1), strain 824(pCAC0003) showed 39%, 13%, and 22% increases in percent relative tolerance after 6, 12, and 18 h of butanol challenge, respectively.
Biological replicate static-flask batch cultures of 824(pCAC1869) and 824(pIMP1) were grown in the absence of external butanol stress and sampled periodically for A600 and metabolite analysis (see Fig. S4A to C in the supplemental material). Strain 824(pCAC1869) showed increased metabolic activity through a prolonged transitional phase, consuming more glucose and producing more butanol than 824(pIMP1). In doing so, 824(pCAC1869) overcame documented host-plasmid interactions that result in a biphasic-growth pattern (50).
DISCUSSION
Two selection protocols were used to screen a C. acetobutylicum genomic library for preferential growth in the presence of butanol, with protocol II providing superior outcomes. Monitoring the progression of enrichment over serial challenges generated greater genetic detail of the enrichment process.
Among the many identified candidate genes imparting solvent tolerance, we examined the impact of two genes: CAC0003 and CAC1869. The improvement in solvent tolerance was markedly better for 824(pCAC1869) than for 824(pCAC0003). When the MIC of actively growing cultures was measured (Fig. 3B), both 824(pCAC1869) and 824(pCAC0003) continued to grow in medium containing as much as 1.4% (vol/vol) butanol, while the 824(pIMP1) control strain was not similarly capable. For comparison, the 80 to 90% improvement in solvent tolerance for strain 824(pCAC1869) is equivalent to the 85% improvement in solvent tolerance found for strain 824(pGROE1) (50).
The transcriptional patterns of the 16 enriched genes that are also the start of a predicted TU were examined using data from two DNA microarray transcriptional analyses from our laboratory (2, 3). One study detailed the transcriptional pattern of exponential- and transitional-phase events (3), while another detailed the transcriptional response of exponential-phase C. acetobutylicum cultures to a single bolus of 0.46% (vol/vol) butanol (2). Of the 16 enriched genomic inserts, CAC1869, CAC3435, and CAC3359 were maximally transcribed at the onset of the transitional phase. Conversely, CAC1463 and CAC1874 were transcribed predominantly in the exponential phase, while CAC0003, CAC3143, CAC0854, and CAC0395 were all downregulated at the onset of the transitional phase (3). Interestingly, transcription of these 16 enriched genes was not significantly affected by butanol stress (2). We conclude that a stress response is not necessarily a good method for identifying genes imparting tolerance to the stressor.
CAC1869 is annotated as a singleton transcriptional regulator, and for several reasons it is reasonable to assign a role in the regulation of transitional-phase events. CAC1869 is maximally transcribed just prior to the induction of the solventogenic genes aad, ctfA, and ctfB and remains actively transcribed through the transitional phase (3). Also, protocol II favored inserts that delay transitional-phase phenomena, making cells that harbor them better able to continue vegetative growth upon inoculation into the next challenge flask. Consistent with that, 824(pCAC1869) exhibited prolonged metabolic activity through the transitional phase compared to that of the 824(pIMP1) control strain.
Transcriptional regulators are important for tolerant phenotypes (1, 5, 30). CAC1869 has homology to the xenobiotic-responsive element (XRE) family of regulatory proteins (35). Twenty-four other XRE-like regulatory proteins reside in the C. acetobutylicum genome (35). CAC1869 shows the closest homology to the Bacillus cereus NVH 391-98 gene bcer98draft_2450 (34% identity), but no similar homolog is present in the other functionally annotated clostridia (Clostridium perfringens and Clostridium tetani). XRE-like proteins have been linked to a wide variety of cellular functions (9, 17) and tolerant phenotypes (6, 26, 29). The protein product of la867 is critical for acid tolerance in Lactobacillus acidophilus (6) and has homology to gadR, a positive regulator of the Lactococcus lactis acid resistance operon gadBC (42). One of the primary genes for regulating the recovery of Deinococcus radiodurans from ionizing radiation is the XRE-like regulator dr2574. Finally, SinR is an XRE-like regulator that forms a tetramer that binds promoter regions and represses the transcription of genes (e.g., spo0A) that are necessary for the initiation of the sporulation cascade (17, 37). In the case of CAC1869, further study is necessary in order to determine which genes are directly regulated as well as the precise regulatory mechanism for tolerance imparted by CAC1869 overexpression.
With both library selection protocols, mixed-cell populations exhibited growth in media containing 1.56% butanol. However, neither 824(pIMP1) nor 824(pCAC0003) nor 824(pCAC1869) was able to grow when inoculated in medium containing more than 1.3% butanol (Fig. 3A), and actively growing cultures were completely inhibited at concentrations exceeding 1.6% butanol (Fig. 3B). This suggests that library-bearing cultures with a mixture of inserts reproducibly tolerated higher butanol levels than cultures with only a single insert or the pIMP1 control. Two explanations are that this is either an inherent trait of the mixed population or the result of unidentified chromosomal events leading to improved solvent tolerance. The 824(pIMP1) control strain, when subjected to stationary-phase challenges with increasing butanol concentrations, apparently did not display a similar chromosomal event, surviving at only up to 1.3% butanol (transfer 10) (see Table S2 in the supplemental material) and indicating either a low frequency of occurrence or a role for library inserts in producing these beneficial chromosomal modifications.
The identification of additional solvent tolerance elements is possible for several reasons. First, the pLib1 library provided coverage for approximately 73% of the C. acetobutylicum genome. Second, the lack of a constitutive promoter on the pIMP1 vector precluded the expression of library inserts separated from their natural promoter. Finally, the preferential uptake of plasmids containing smaller library inserts limited the sizes of genes and operons screened in protocols I and II. This likely explains why previously identified tolerance elements, such as the groESL operon (∼2.2 kb) (50), were not identified in this study.
Despite these shortcomings of the pLib1 genomic library, the method and results presented here should prove useful for future efforts to define the genetic basis for complex phenotypes such as tolerance to various toxic chemicals. Visualizing the dynamic process of library insert enrichment with DNA microarrays provides greater genetic detail than previous efforts that aimed to characterize the result of the enrichment process (18, 24). Enriched genes identified in this study require further characterization of their role in solvent tolerance through a combination of constitutive gene overexpression and DNA microarray transcriptional profiling. In addition, co-overexpression of multiple enriched genes in a single culture may provide additional tolerance above that imparted singly.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant BES-0331402, Department of Energy grant DE-FG36-03GO13160, and an NIH/NIGMS biotechnology training grant (T32-GM08449) fellowship to Jacob R. Borden.
We thank the Northwestern University Biotechnology Core Laboratory for assistance with Agilent 2100 Bioanalyzer analysis and sequencing of plasmid library inserts. We also thank Muhammad A. Ali, John S. Park, and Carlos J. Paredes for assistance with subcloning, tolerance assays, and illustration of microarray enrichment data, respectively.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alper, H., J. Moxley, E. Nevoigt, G. R. Fink, and G. Stephanopoulos. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565-1568. [DOI] [PubMed] [Google Scholar]
- 2.Alsaker, K. V. 2006. Microarray-based transcriptional analyses of stationary phase phenomena and stress responses in Clostridium acetobutylicum. Ph.D. thesis. Northwestern University, Evanston, IL.
- 3.Alsaker, K. V., and E. T. Papoutsakis. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 187:7103-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsaker, K. V., C. J. Paredes, and E. T. Papoutsakis. 2005. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng. 10:432-443. [Google Scholar]
- 5.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azcarate-Peril, M. A., E. Altermann, R. L. Hoover-Fitzula, R. J. Cano, and T. R. Klaenhammer. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70:5315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baer, S. H., H. P. Blaschek, and T. L. Smith. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baer, S. H., D. L. Bryant, and H. P. Blaschek. 1989. Electron-spin resonance analysis of the effect of butanol on the membrane fluidity of intact cells of Clostridium acetobutylicum. Appl. Environ. Microbiol. 55:2729-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barragan, M. J., B. Blazquez, M. T. Zamarro, J. M. Mancheno, J. L. Garcia, E. Diaz, and M. Carmona. 2005. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280:10683-10694. [DOI] [PubMed] [Google Scholar]
- 10.Bowles, L. K., and W. L. Ellefson. 1985. Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 50:1165-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunka, D. H., and P. G. Stockley. 2006. Aptamers come of age—at last. Nat. Rev. Microbiol. 4:588-596. [DOI] [PubMed] [Google Scholar]
- 12.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, S. W., G. N. Bennett, and F. B. Rudolph. 1989. Isolation and characterization of mutants of Clostridium acetobutylicum ATCC 824 deficient in acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase (EC 2.8.3.9) and in other solvent pathway enzymes. Appl. Environ. Microbiol. 55:970-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ezeji, T., N. Qureshi, and H. Blaschek. 2005. Industrially relevant fermentations, p. 903. In P. Durre (ed.), Handbook on clostridia. Taylor & Francis, Boca Raton, FL.
- 16.Fieseler, L., A. Quaiser, C. Schleper, and U. Hentschel. 2006. Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ. Microbiol. 8:612-624. [DOI] [PubMed] [Google Scholar]
- 17.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill, R. T., S. Wildt, Y. T. Yang, S. Ziesman, and G. Stephanopoulos. 2002. Genome-wide screening for trait conferring genes using DNA microarrays. Proc. Natl. Acad. Sci. USA 99:7033-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, L. M., R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1-11. [PubMed] [Google Scholar]
- 20.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingram, L. O. 1976. Adaptation of membrane lipids to alcohols. J. Bacteriol. 125:670-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain, M. K., J. Gleeson, A. Upreti, and G. C. Upreti. 1978. Intrinsic perturbing ability of alkanols in lipid bilayers. Biochim. Biophys. Acta 509:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Jain, M. K., and N. M. Wu. 1977. Effect of small molecules on dipalmitoyl lecithin liposomal bilayer. 3. Phase-transition in lipid bilayer. J. Membr. Biol. 34:157-201. [Google Scholar]
- 24.Jin, Y. S., H. Alper, Y. T. Yang, and G. Stephanopoulos. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilman, S. 2006. DuPont-BP venture will make competing product to ethanol. Wall Street J. 21 June, p. A2.
- 26.Labie, C., F. Bouche, and J. P. Bouche. 1989. Isolation and mapping of Escherichia coli mutations conferring resistance to division inhibition protein DicB. J. Bacteriol. 171:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. Y. 1991. Construction of Escherichia coli-Clostridium acetobutylicum shuttle vectors and transformation and characterization of Clostridium acetobutylicum strains using these vectors. Ph.D. thesis. Northwestern University, Evanston, IL.
- 28.Lepage, C., F. Fayolle, M. Hermann, and J. P. Vandecasteele. 1987. Changes in membrane lipid-composition of Clostridium acetobutylicum during acetone butanol fermentation—effects of solvents, growth temperature and pH. J. Gen. Microbiol. 133:103-110. [Google Scholar]
- 29.Liu, Y., J. Zhou, M. V. Omelchenko, A. S. Beliaev, A. Venkateswaran, J. Stair, L. Wu, D. K. Thompson, D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucau-Danila, A., G. Lelandais, Z. Kozovska, V. Tanty, T. Delaveau, F. Devaux, and C. Jacq. 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 25:1860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mermelstein, L. D., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermelstein, L. D., N. E. Welker, G. N. Bennett, and E. T. Papoutsakis. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 10:190-195. [DOI] [PubMed] [Google Scholar]
- 33.Nair, R. V., E. M. Green, D. E. Watson, G. N. Bennett, and E. T. Papoutsakis. 1999. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J. Bacteriol. 181:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman, E. B., and R. T. Lin. 1995. Leucine-responsive regulatory protein—a global regulator of gene-expression in Escherichia coli. Annu. Rev. Microbiol. 49:747-775. [DOI] [PubMed] [Google Scholar]
- 35.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ounine, K., H. Petitdemange, G. Raval, and R. Gay. 1985. Regulation and butanol inhibition of d-xylose and d-glucose uptake in Clostridium acetobutylicum. Appl. Environ. Microbiol. 49:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paredes, C. J., K. V. Alsaker, and E. T. Papoutsakis. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969-978. [DOI] [PubMed] [Google Scholar]
- 38.Paredes, C. J., I. Rigoutsos, and E. T. Papoutsakis. 2004. Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32:1973-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reardon, C. 21 August 2003, posting date. Indirect genomic labeling. FAS Center for Systems Biology 1.0. http://www.sysbio.harvard.edu/csb/resources/downloads/Bauer_Core_Genomic_DNA_Labeling_Protocol.doc
- 40.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 43.Sardessai, Y., and S. Bhosle. 2002. Tolerance of bacteria to organic solvents. Res. Microbiol. 153:263-268. [DOI] [PubMed] [Google Scholar]
- 44.Sinensky, M. 1974. Homeoviscous adaptation—homeostatic process that regulates viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terracciano, J. S., and E. R. Kashket. 1986. Intracellular conditions required for initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thaw, P., S. E. Sedelnikova, T. Muranova, S. Wiese, S. Ayora, J. C. Alonso, A. B. Brinkman, J. Akerboom, J. van der Oost, and J. B. Rafferty. 2006. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 34:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thormann, K., and P. Durre. 2001. Orf5/SolR: a transcriptional repressor of the sol operon of Clostridium acetobutylicum? J. Ind. Microbiol. Biotechnol. 27:307-313. [DOI] [PubMed] [Google Scholar]
- 49.Thormann, K., L. Feustel, K. Lorenz, S. Nakotte, and P. Durre. 2002. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J. Bacteriol. 184:1966-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomas, C. A., N. E. Welker, and E. T. Papoutsakis. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and large changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Westhuizen, A., D. T. Jones, and D. R. Woods. 1982. Autolytic activity and butanol tolerance of Clostridium acetobutylicum. Appl. Environ. Microbiol. 44:1277-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vollherbst-Schneck, K., J. A. Sands, and B. S. Montenecourt. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47:193-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber, F. J., and J. A. de Bont. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225-245. [DOI] [PubMed] [Google Scholar]
- 54.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods, D. R. 1993. The clostridia and biotechnology. Butterworth-Heinemann, Boston, MA.
- 57.Woolley, R. C., and J. G. Morris. 1990. Stability of solvent production by Clostridium acetobutylicum in continuous culture—strain differences. J. Appl. Bacteriol. 69:718-728. [Google Scholar]
- 58.Zhao, Y., C. A. Tomas, F. B. Rudolph, E. T. Papoutsakis, and G. N. Bennett. 2005. Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl. Environ. Microbiol. 71:530-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.