Abstract
We developed a 12-h Salmonella detection method, based on 8 h of preenrichment, followed by automated DNA extraction and a sensitive real-time PCR. The method was optimized to obtain the highest possible yield of cells and DNA. The growth of different Salmonella strains in various preenrichment media and the effects of adding growth-promoting and selective reagents were explored, taking into account their PCR compatibility. The effects of (i) analyzing larger volumes (1 to 5 ml) from preenriched samples and introducing wash steps prior to DNA extraction, (ii) regulating the amount of paramagnetic particles (increasing it from 60 to 90 μl) in the DNA extraction, (iii) eluting the DNA in reduced volumes (25 or 50 μl rather than 100 μl), and (iv) increasing the PCR template volume (from 5 to 20 μl) were investigated. After 8 h of preenrichment, buffered peptone water yielded the highest number of salmonellae. When analyzing minced meat samples, positive effects of increasing the initial sampling volume from 1 to 5 ml and increasing the amount of paramagnetic particles to 90 μl were observed. However, washing the pellet and eluting the DNA in reduced volumes (25 and 50 μl) had no positive effects and resulted in decreased reproducibility. Increasing the amount of PCR template DNA from 5 to 20 μl improved the threshold cycle value by approximately 2. The improved 12-h PCR method was successfully compared to a reference culture method with 100 minced meat and poultry samples, with a relative accuracy of 99%, a relative sensitivity of 98%, and a relative specificity of 100%.
Bacteriological detection of Salmonella in foods and environmental samples is costly, laborious, and time-consuming, requiring up to 5 days to obtain a confirmed result. Thus, rapid and cost-effective detection of Salmonella is of major interest to the food industry and the public. Real-time PCR technology offers several advantages compared to classical bacteriology in terms of speed, detection limit, potential for automation, and cost (17, 24). However, it is essential that new PCR methods be reliable and robust. They have to comply with legislation on microbiological criteria for foodstuffs and be able to detect as few as one Salmonella bacterium per 25-g sample. They should be validated against reference culture methods, and last, but not least, they should be sufficiently robust to be transferred from the expert laboratory to end users.
Several PCR-based methods for the detection of Salmonella in foodstuff have been published. Most of these methods operate with preenrichment periods of 16 to 24 h, followed by DNA purification and gel-based or real-time PCR (8, 11, 13, 16, 18, 34). Only a few of them report reduced preenrichment times of 6 to 12 h (1, 12, 27). Both Ellingson et al. (12) and Agarwal et al. (1) reported a preenrichment period of only 6 h for the detection of Salmonella by PCR. In a study by Ellingson et al. (12), the results obtained by real-time PCR correlated 100% with a reference culture method. However, for both studies, the samples analyzed were inoculated with at least 1 CFU/g (not per 25 g) food sample; thus, it is questionable if these methods can meet the legislative demand of detection of 1 CFU/25-g sample. In a recent study by Myint et al. (27) five Salmonella-positive chicken samples were subjected to PCR after 2 to 18 h of preenrichment, with a sampling interval of 2 h. Even though two out of the five samples were detectable by PCR after 8 h of preenrichment, it required 18 h of preenrichment for all five samples to be detected.
The present study describes the development and optimization of a 12-h Salmonella analysis for the meat industry, enabling a faster release of Salmonella-free fresh meat and meat products. The method is based on a shortened preenrichment period combined with increased detection sensitivity in a real-time PCR.
Because of low levels of Salmonella in meat from subclinically infected herds, a preenrichment step is usually included prior to PCR. The preenrichment was followed by a TaqMan PCR assay including an internal amplification control (IAC) (25). However, the shortened preenrichment period of only 8 h did not produce Salmonella counts that could be detected consistently by this PCR. For this reason, critical steps throughout the method were optimized in order to obtain the highest possible yield of cells and DNA recovery after automated DNA extraction. In addition, the detection limit of the PCR method was optimized by implementing the locked nucleic acid (LNA) technology (30). LNA probes have a higher melting temperature than TaqMan probes because the LNA modifications provide stronger hybridization between double-stranded DNAs and are reported to be more sensitive (28).
Besides evaluating alternative preenrichment broths, optimization of pre-PCR treatment was attempted by increasing the sampling volume and introducing washing steps prior to DNA extraction. The DNA extraction protocol used in this study has been shown to be a promising method for extracting bacterial DNA from other matrices (23). The DNA loss in this method was evaluated, and the method was optimized with regard to the amounts of paramagnetic particles and elution buffer. Finally, the effect of increasing the volume of template DNA was studied.
The improved 12-h PCR method was compared to the reference culture method for Salmonella from the Nordic Committee on Food Analysis (NMKL no. 71; reference 3) with 50 artificially inoculated samples of minced pork meat and 50 artificially inoculated poultry samples.
MATERIALS AND METHODS
Optimization of growth conditions.
In the experiments described below, all of the samples and media were preheated to 37°C prior to preenrichment.
The first step was to confirm if shaking during preenrichment had any beneficial effect on the number of salmonellae present after 8 h (29). Overnight cultures of four of the Salmonella strains most frequently isolated from pork were prepared in buffered peptone water (BPW; Oxoid, Basingstoke, United Kingdom). The number of CFU per milliliter was determined by plating 10-fold dilution series on blood agar (Statens Serum Institute, Copenhagen, Denmark) in duplicate.
Salmonella enterica serovar Typhimurium CCUG 31939, S. enterica serovar Enteritidis CCUG 32352, S. enterica serovar Dublin, and S. enterica serovar Infantis (in-house collection) were inoculated at a level of 1 to 10 CFU into 100 ml of BPW in quadruplicate and incubated at 37°C. Half of the preenrichments were shaken (60 rpm, Certomat U; B. Braun Biotech International, Göttingen, Germany), and half were incubated without shaking. The numbers of salmonellae in the preenrichments were determined by plating on blood agar after 6, 8, and 24 h of incubation.
The second step was to examine whether alternative nutrient media would be superior to BPW in supporting the growth of Salmonella and if these were PCR compatible, i.e., not inhibiting the PCR. One-hundred-milliliter volumes of brain heart infusion (BHI) broth (Oxoid) and tryptone soya broth (TSB; Becton Dickinson, Franklin Lakes, NJ) were inoculated in duplicate with 1 to 10 CFU of each aforementioned Salmonella strain and incubated at 37°C. The number of salmonellae in the preenrichments was determined by plating on blood agar after 6, 8, and 24 h of incubation. To test the PCR inhibition of the media, an overnight culture containing 1.9 × 109 CFU/ml was diluted in BPW, BHI, and TSB to 10−5 to 10−8 and the DNA was extracted and analyzed in duplicate in the PCR assay.
Growth-promoting reagents.
The third step in the optimization process was to study the effect of adding sodium pyruvate to the preenrichment media (26). One-hundred-milliliter volumes of BPW containing 0, 0.2, and 0.4 g/liter sodium pyruvate (Sigma) were inoculated in duplicate with 1 to 10 CFU of each Salmonella strain and incubated at 37°C. The numbers of salmonellae in the BPW were determined by plating on blood agar after 6, 8, and 24 h of incubation.
The effect of adding egg yolk to the preenrichment media was investigated. S. enterica serovar Typhimurium CCUG 31939 and S. enterica serovar Infantis (in-house collection) were inoculated at a level of 1 to 10 CFU into 100 ml BPW containing 0.5, 1.0, and 5.0% egg yolk (Oxoid) and incubated at 37°C. The number of salmonellae in the BPW was determined by plating on blood agar after 8 h of incubation.
Selective reagents.
In order to suppress competitive flora and thereby improve the growth conditions for Salmonella, the effect of adding a range of different selective reagents to the BPW was investigated. To BPW were added novobiocin (20, 50, and 100 mg/liter; Fluka, Buchs, Switzerland), brilliant green (10, 20, and 50 mg/liter; Fluka), malachite green oxalate salt (50, 100, and 250 mg/liter, Fluka), tergitol 4 (1, 2, and 4 ml/liter; Fluka), sodium deoxycholate (2.5, 5, and 7.5 g/liter; Fluka), and finally sulfamandelate supplement (1, 2, and 3 vials/liter; Oxoid). Samples of minced pork meat (10 g), frozen at −18°C and thawed, were transferred to 90 ml of BPW with the selective reagents added and inoculated with 1 to 10 and 10 to 100 CFU/g sample by using freeze-stressed S. enterica serovar Typhimurium CCUG 31939 and S. enterica serovar Infantis (in-house collection). Stressed cells were prepared from a BHI culture grown at 37°C for 20 to 24 h and frozen at −18°C. Before use, both minced meat samples and freeze-stressed cells were thawed at 4°C. The samples were incubated at 37°C for 20 h. Aliquots for PCR were drawn after 6, 8, and 20 h. DNA extraction was performed prior to analysis by the PCR assay (see below).
Automated DNA extraction.
One-, 2-, and 5-ml aliquots were drawn from the preenrichments for DNA extraction. The aliquots were centrifuged at 3,000 × g for 5 min at 4°C, and DNA extraction was performed with a KingFisher (Thermo Labsystems, Helsinki, Finland) and a DNA isolation kit for blood, stool, cells, and tissue (Magnesil KF, Genomic System; Promega) as specified by the manufacturer. Briefly, the sample pellet was resuspended in lysis buffer and transferred to a 96-well plate (Thermo Labsystems) containing paramagnetic particles, washing buffers, and elution buffer. The DNA extraction program consisted of two salt buffer washing steps and two alcohol buffer washing steps, followed by a final elution step (for a detailed protocol, see InnovationsPCR at www.foodpcr.com). Five to 20 μl of the extracted DNA was used as the template in the PCR.
TaqMan PCR.
A TaqMan real-time PCR method, targeting a region within the ttrRSBCA locus required for tetrathionate respiration, for the specific detection of Salmonella was set up (adopted from reference 25, with the following modifications). The PCR was performed on an Mx3005P (Stratagene, La Jolla, CA) in a total reaction volume of 25 μl, consisting of 1.5 U of Tth DNA polymerase (Roche Applied Science, Mannheim, Germany), 2.5 μl of 10× PCR buffer for Tth DNA polymerase (Roche Applied Science), 500 μM deoxynucleoside triphosphate blend with dUTP (Applied Biosystems, Foster City, CA), 4.0 mM MgCl2 (Roche Applied Science), 8% pure glycerol (Merck, Darmstadt, Germany), 1 g/liter bovine serum albumin (Roche Applied Science), 2% dimethyl sulfoxide (Sigma, Steinheim, Germany), 240 nM both LNA target probe (6-FAM [6-carboxyfluorescein]-CG+ACGGCG+AG+ACCG-BHQ1; Sigma-Proligo, Paris, France) and an IAC probe (JOE-CACACGGCGACGCGAACGCTTT-BHQ1; MWG Biotech, Ebersberg, Germany), and 5 μl of purified DNA. The cycle temperature profile was initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 65°C for 60 s, and 72°C for 30 s. Fluorescence measurements were obtained online and analyzed with the MxPro-Mx3005P software (version 3.00). The threshold was assigned by using the software option background-based threshold; i.e., the standard deviation of all amplifications was determined from cycle 5 to cycle 9, and this value was multiplied by a background sigma multiplier of 10. Each PCR run included three positive DNA controls (S. enterica serovar Typhimurium 51K61; Institute for Reference Material and Measurements [IRMM], Geel, Belgium) at final concentrations of 0.5, 0.05, and 0.005 ng/PCR tube, a nontemplate control (only the master mix and PCR grade water), and a negative DNA control (Escherichia coli O157; IRMM) at a concentration of 5 ng/PCR tube.
Optimization of sample preparation.
The effects of analyzing larger volumes from BPW and introducing washing of the pellet prior to DNA extraction were investigated. A 25-g sample of minced pork meat was inoculated with 1 to 10 CFU of S. enterica serovar Livingstone (in-house collection), transferred to 225 ml of BPW, and incubated at 37°C for 8 h. One-, 2-, and 5-ml aliquots were drawn (eight replicates). DNA was extracted from half of the replicates directly, and pellets from the remaining replicates were washed before DNA extraction; i.e., twice they were centrifuged at 3,000 × g for 5 min and the pellet was resuspended in 1 ml of physiological saline. After DNA extraction, replicates were analyzed in the PCR.
Loss of DNA during extraction.
The amount of DNA lost in the extraction procedure was evaluated by comparing CT (threshold cycle) values of samples containing a known amount of reference DNA before and after automated DNA extraction. As shown in Table 1, the experiment was designed so that theoretically equivalent amounts of DNA were analyzed in the PCR. Four samples were prepared from reference DNA (S. enterica serovar Typhimurium 51K61; IRMM) and 1× Tris-EDTA buffer to final concentrations of 0.1, 0.05, 0.01, and 0.005 ng/μl. The samples were analyzed in duplicate in the PCR before and after automated DNA extraction.
TABLE 1.
Loss of DNA in the extraction procedure
| Treatment and DNA concn (ng/μl) in PCR mixture | Amt (ng) of DNA entering KingFisher | Elution vol (μl), KingFisher |
CT valuea
|
|
|---|---|---|---|---|
| Salmonella/FAM | IAC/HEX | |||
| Direct PCR | ||||
| 0.1 | 20.5 | 30.6 | ||
| 0.1 | 20.3 | 31.4 | ||
| DNA extraction | ||||
| 0.1b | 8 | 80 | 25.6 | 30.1 |
| 0.1b | 8 | 80 | 25.5 | 30.3 |
| Direct PCR | ||||
| 0.05 | 20.6 | 29.8 | ||
| 0.05 | 20.0 | 30.2 | ||
| DNA extraction | ||||
| 0.05b | 4 | 80 | 25.0 | 30.1 |
| 0.05b | 4 | 80 | 25.5 | 30.1 |
| Direct PCR | ||||
| 0.01 | 23.8 | 30.3 | ||
| 0.01 | 23.0 | 31.0 | ||
| DNA extraction | ||||
| 0.01b | 0.8 | 80 | 28.7 | 29.8 |
| 0.01b | 0.8 | 80 | 27.6 | 29.7 |
| Direct PCR | ||||
| 0.005 | 22.7 | 30.3 | ||
| 0.005 | 22.3 | 30.4 | ||
| DNA extraction | ||||
| 0.005b | 0.4 | 80 | 26.9 | 29.9 |
| 0.005b | 0.4 | 80 | 27.0 | 30.2 |
CT values obtained in PCR from samples with similar DNA concentrations subjected directly to PCR and following DNA extraction.
Estimated DNA concentration if the extraction procedure was 100% efficient.
In the same experimental setup, the effect of regulating the amount of paramagnetic particles was investigated. The DNA was extracted in triplicate from samples containing the aforementioned concentrations of DNA by using 60, 75, and 90 μl of paramagnetic particles, respectively. The samples were subsequently analyzed in replicate in the PCR.
Increasing the concentration of DNA.
The effect of reducing the volume of elution buffer to increase the DNA concentration was investigated. A 25-g sample of minced pork meat was inoculated with 1 to 10 and 10 to 100 CFU of S. enterica serovar Typhimurium CCUG 31939, transferred to 225 ml of BPW, and incubated at 37°C for 8 h. One-milliliter aliquots were drawn (nine replicates from each preenrichment), and the DNA was extracted from the replicates with 100, 50, and 25 μl of elution buffer, respectively. The replicates were analyzed in the PCR.
Increasing the PCR template DNA volume.
The effect of increasing the PCR template volume was investigated. A 25-g sample of minced pork meat was inoculated with 1 to 10 or 10 to 100 CFU of S. enterica serovar Typhimurium CCUG 31939, transferred to 225 ml of BPW, and incubated at 37°C for 8 h. One-milliliter aliquots were drawn (three replicates from each preenrichment). DNA was extracted and subsequently analyzed in the PCR with 5 and 10 μl of template DNA in a total volume of 25 μl of master mix and 20 μl of template DNA in a total volume of 50 μl of master mix.
Validation against a reference culture method.
The final 12-h PCR method was compared to the reference culture method for Salmonella from the Nordic Committee on Food Analysis (NMKL no. 71; reference no. 3) with 100 artificially inoculated minced meat and poultry neck skin samples. As the prevalence of Salmonella-positive pork meat samples is 1 to 2% and that of Salmonella-positive broiler meat samples is 1.7% at the moment (5), a statistically valid study would require a very large number of samples. For this reason, the comparison was performed with samples artificially inoculated with Salmonella in the exponential growth phase. This alternative to naturally contaminated samples is in compliance with international guidelines (6, 7).
Twenty-five grams of Salmonella-free fresh minced pork meat was transferred to 225 ml of BPW (37°C). Nine samples were inoculated with 1 to 10 CFU of S. enterica serovar Typhimurium, 10 were inoculated with 1 to 10 CFU of S. enterica serovar Livingstone, and 14 were inoculated with 10 to 100 CFU of S. enterica serovar Livingstone. The remaining 17 samples were left uninoculated.
Twenty-five grams of poultry neck skin was cut into small pieces and transferred to 225 ml of BPW (37°C). Fifteen samples were inoculated with 1 to 10 CFU of S. enterica serovar Enteritidis, and 14 were inoculated with 1 to 10 CFU of S. enterica serovar Typhimurium. The remaining samples were left uninoculated.
All of the samples were preheated to 37°C and homogenized by hand for 20 s. After 8 h of preenrichment at 37°C, 5-ml aliquots were drawn for DNA extraction with 75 μl of paramagnetic beads, followed by a PCR with 10 μl of the extracted DNA as the template.
The enrichment was thereafter continued for up to 24 h according to NMKL no. 71 (3). The next day, 100 μl was transferred to 10 ml of Rappaport-Vassiliadis soy peptone (Oxoid) broth preheated to 37°C. The Rappaport-Vassiliadis soy peptone broth was incubated at 41.5°C for 24 h and inoculated onto the surface of the selective plating media xylose lysine deoxycholate (Oxoid) and Rambach (Merck). The plates were incubated at 37°C for 24 h, and presumptive colonies were transferred to 5% blood agar plates (Statens Serum Institute) and confirmed by API 20E (BioMérieux, Marcy l'Étoile, France) and by serotyping.
Statistical analysis.
A paired-sample t test was performed according to Campbell (10) on the data obtained by comparing preenrichments that were shaken and those that were not shaken. The data obtained by comparing the three preenrichment broths (BPW, BHI, and TSB) and adding growth-promoting reagents to the preenrichments were analyzed in a single-factor one-way analysis of variance according to Campbell (10), with an α of 0.05.
When comparing the improved 12-h PCR method to the reference culture method, the test characteristics relative accuracy (AC), sensitivity, and specificity were evaluated (7). AC is defined as the degree of correspondence between the responses obtained by the PCR method and the reference culture method with identical samples, as follows: (PA + NA) × 100/(PA + NA + PD + ND), where PA refers to positive agreement, NA is negative agreement, PD is positive deviation, and ND is negative deviation. Sensitivity is defined as the ability of the PCR method to detect the target compared to the reference culture method, as follows: PA × 100/(PA + FN), where FN refers to false negatives. Specificity is defined as the ability of the PCR method not to detect the target when it is not detected by the reference culture method, as follows: (NA × 100)/(NA + FP), where FP refers to false positives.
RESULTS
Optimization of growth conditions.
Shaking the preenrichment culture had no beneficial effect on the growth of any of the Salmonella strains after 6 and 8 h. After 6 h of incubation, the average number of salmonellae was 9.6 × 103 CFU/ml in the samples not shaken and 2.6 × 103 CFU/ml in the samples shaken (P = 0.2). The average number of salmonellae after 8 h was 2.3 × 105 CFU/ml in the samples not shaken and 9.7 × 104 CFU/ml in the samples shaken (P = 0.4). However, after incubation for 24 h, shaking increased the number of CFU per milliliter by 1 log unit (P = 0.0003), as the average number reached 6.5 × 109 CFU/ml, compared to 5.8 × 108 CFU/ml for those not shaken.
Analysis of the results obtained by BHI, TSB, and BPW showed no significant difference in the abilities of the three different preenrichment media to support growth of Salmonella after 6 and 8 h. After 6 h, the average number of salmonellae reached 9.6 × 103 CFU/ml in BPW, 1.2 × 103 CFU/ml in TSB, and 1.4 × 103 CFU/ml in BHI (P = 0.15). After 8 h, these values were 2.3 × 105 CFU/ml in BPW, 6.4 × 104 CFU/ml in TSB, and 4.3 × 104 CFU/ml in BHI (P = 0.21). After incubation for 24 h, both of the nutrient-rich media, BHI and TSB, resulted in increased growth of Salmonella (BPW, ∼5.8 × 108 CFU/ml; TSB, ∼6.1 × 109 CFU/ml; BHI, ∼5.3 × 109 CFU/ml [averages]; P = 0.005). These average results were also reflected on the strain level. The PCR results obtained with the different preenrichment media indicated no difference in the inhibition of PCR among the three media. After 8 h, the average CT values obtained were 20 for BPW, 21.5 for TSB, and 21.1 for BHI. After 24 h of preenrichment, these values were 15.3 for BPW, 16.7 for TSB, and 15.2 for BHI.
No difference in growth was observed for any of the strains tested when sodium pyruvate or egg yolk was added, regardless of the concentration (data not shown).
Addition of tergitol 4 and sulfamandelate to preenrichment of minced pork meat had no effect on the CT values, regardless of the concentrations applied. Malachite green oxalate salt and deoxycholate resulted in slightly higher CT values after 20 h of preenrichment, while no differences in CT values were observed after 6 and 8 h. Addition of novobiocin (20 and 50 mg/liter) and brilliant green (10 mg/liter) had a tendency to improve the PCR results obtained after 8 h of preenrichment.
Optimization of sample preparation.
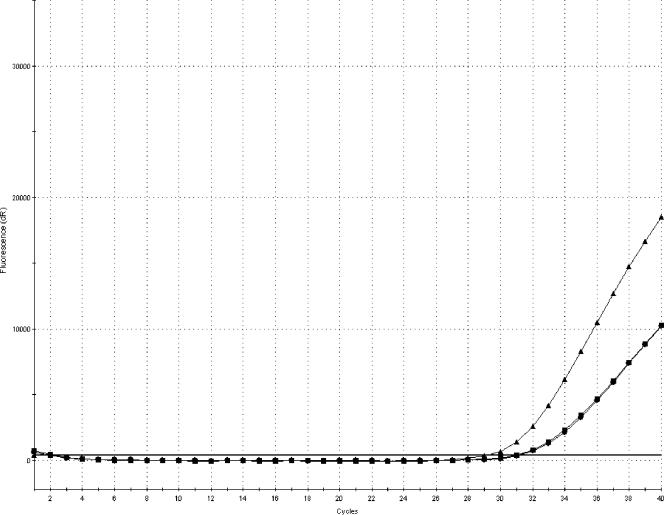
Increasing the sampling volume of BPW to 5 ml resulted in an improved detection limit and steeper amplification curves, as shown in Fig. 1. The average CT values were 31.8 for a 1-ml volume, 31.6 for a 2-ml volume, and 29.7 for a 5-ml volume. Washing of pellets produced higher CT values and flatter PCR amplification curves. Some samples were not even PCR positive following the two washing steps.
FIG. 1.
Amplification plot (FAM) showing the difference in amplification curves obtained by sampling 1 ml (•), 2 ml (▪), and 5 ml (▴) of minced pork meat inoculated with 1 to 10 CFU of S. enterica serovar Livingstone and enriched for 8 h at 37°C. Each amplification curve represents an average of four replicates.
Loss of DNA during extraction.
The amount of DNA lost in the extraction procedure when extracting purified DNA resulted in an average increase in CT values of 4.8 (Table 1). The DNA losses were more pronounced with the higher concentrations of DNA.
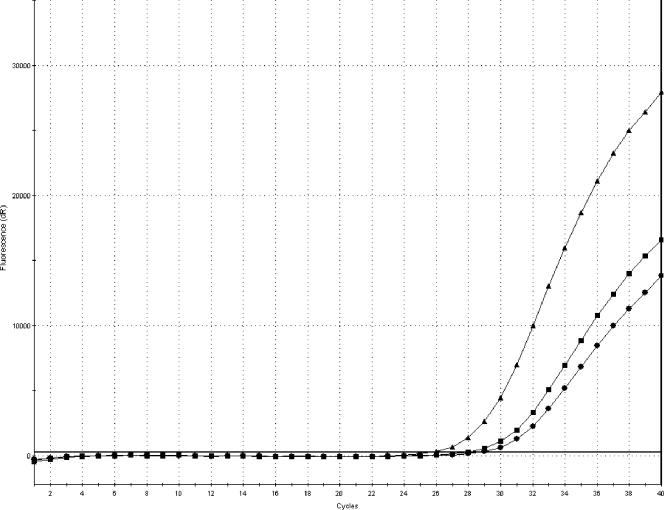
Table 2 shows the reverse correlation between the amount of paramagnetic particles and the CT values. The lowest CT values were obtained with 90 μl of paramagnetic particles. The effect of increasing the amount of paramagnetic particles was not pronounced, and the CT value was lowered approximately 1, on average, when using 90 μl of paramagnetic particles compared to 60 μl. Steeper amplification curves were, however, obtained with 90 μl of paramagnetic particles, as shown in Fig. 2.
TABLE 2.
CT values obtained in PCR comparing duplicate samples with similar DNA concentrations extracted with 60, 75, or 90 μl of paramagnetic particles per sample
| DNA concn (ng/μl) | Vol of paramagnetic particles (μl/sample) |
CT value
|
|||
|---|---|---|---|---|---|
| Salmonella/FAM | IAC/HEX | ||||
| 0.1 | 60 | 22.1 | 22.2 | 27.0 | 27.0 |
| 0.1 | 60 | 22.2 | 22.3 | 26.9 | 26.8 |
| 0.1 | 60 | 22.7 | 22.8 | 27.1 | 26.6 |
| 0.1 | 75 | 21.8 | 21.9 | 26.9 | 26.8 |
| 0.1 | 75 | 22.7 | 22.5 | 26.6 | 26.5 |
| 0.1 | 75 | 21.9 | 21.9 | 26.9 | 26.6 |
| 0.1 | 90 | 21.9 | 21.6 | 26.9 | 26.7 |
| 0.1 | 90 | 21.8 | 21.9 | 26.6 | 26.3 |
| 0.1 | 90 | 21.8 | 21.8 | 26.5 | 26.4 |
| 0.05 | 60 | 24.5 | 24.6 | 26.6 | 27.1 |
| 0.05 | 60 | 23.8 | 23.7 | 26.8 | 27.1 |
| 0.05 | 60 | 24.1 | 24.3 | 26.5 | 26.9 |
| 0.05 | 75 | 23.6 | 23.5 | 27.3 | 26.6 |
| 0.05 | 75 | 23.1 | 23.1 | 26.7 | 26.9 |
| 0.05 | 75 | 24.5 | 24.4 | 26.8 | 26.5 |
| 0.05 | 90 | 23.9 | 23.9 | 26.7 | 26.8 |
| 0.05 | 90 | 23.3 | 23.4 | 26.8 | 27.2 |
| 0.05 | 90 | 23.2 | 23.4 | 27.2 | 26.5 |
| 0.01 | 60 | 26.9 | 26.9 | 27.1 | 26.8 |
| 0.01 | 60 | 26.7 | 26.3 | 26.4 | 26.6 |
| 0.01 | 60 | 26.9 | 27.6 | 27.0 | 27.4 |
| 0.01 | 75 | 24.8 | 24.9 | 26.8 | 26.7 |
| 0.01 | 75 | 26.3 | 26.8 | 26.8 | 27.2 |
| 0.01 | 75 | 25.0 | 24.9 | 26.8 | 27.0 |
| 0.01 | 90 | 24.8 | 25.2 | 26.5 | 26.7 |
| 0.01 | 90 | 25.7 | 25.7 | 27.1 | 26.5 |
| 0.01 | 90 | 24.7 | 24.9 | 26.9 | 27.2 |
| 0.005 | 60 | 26.7 | 26.6 | 26.7 | 26.7 |
| 0.005 | 60 | 27.2 | 27.4 | 27.0 | 26.8 |
| 0.005 | 60 | 27.3 | 27.3 | 26.9 | 27.1 |
| 0.005 | 75 | 26.6 | 26.6 | 27.0 | 26.5 |
| 0.005 | 75 | 26.0 | 26.3 | 26.7 | 26.8 |
| 0.005 | 75 | 27.8 | 27.6 | 27.1 | 27.2 |
| 0.005 | 90 | 26.2 | 26.2 | 27.4 | 26.8 |
| 0.005 | 90 | 25.8 | 25.6 | 26.9 | 26.8 |
| 0.005 | 90 | 26.3 | 26.3 | 27.0 | 26.8 |
FIG. 2.
Amplification plot (FAM) showing the difference in amplification curves obtained from a sample containing 0.01 ng/μl DNA extracted with 60 (•), 75 (▪), or 90 (▴) μl of paramagnetic particles, respectively. The amplification curves represent average values of duplicate analyses.
Increasing the concentration of DNA.
Ambiguous results were obtained when attempting to increase the concentration of DNA by eluting in reduced volumes. Elution of DNA in a 25-μl volume resulted in very high and nonreproducible CT values. Reducing the elution volume from 100 to 50 μl did not improve the results.
Increasing the PCR template DNA.
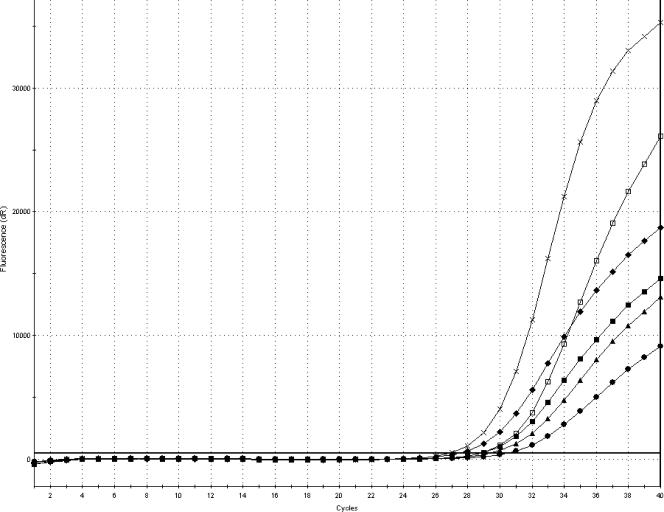
As shown in Fig. 3, increasing the amount of template DNA markedly reduced the CT values and resulted in steeper amplification curves. There was a reverse correlation between the amount of template DNA and the CT value, and the lowest CT values were obtained by addition of 20 μl of template DNA. Addition of 10 μl, compared to 5 μl, of template DNA also reduced the CT values. The average CT value was reduced from 30.2 for 5 μl of template DNA to 29.1 for 10 μl of template DNA and to 28.6 for 20 μl of template DNA for the samples inoculated with 1 to 10 CFU and from 28.6 for 5 μl of template DNA to 27.1 for 10 μl of template DNA and to 26.6 for 20 μl of template DNA. However, the most significant effect was the steepness and reproducibility of the curves.
FIG. 3.
Amplification plot (FAM) showing the difference in amplification curves obtained by analysis of 5, 10, and 20 μl of template DNA in the PCR from samples of minced pork meat inoculated with 1 to 10 or 10 to 100 CFU of S. enterica serovar Typhimurium CCUG 31939 and enriched for 8 h at 37°C. The amplification curves represent average values from triplicate analyses. •, 1 to 10 CFU and 5 μl of template DNA; ▪, 10 to 100 CFU and 5 μl of template DNA; ▴, 1 to 10 CFU and 10 μl of template DNA; ⧫, 10 to 100 CFU and 10 μl of template DNA; □, 1 to 10 CFU and 20 μl of template DNA; ×, 10 to 100 CFU and 20 μl of template DNA.
Validation against a reference culture method.
The reference culture method identified 33 Salmonella-positive and 17 Salmonella-negative minced pork meat samples. For the 12-h PCR method, these values were 32 and 18, respectively. The CT values of the PCR-positive samples inoculated with 1 to 10 CFU were in the range of 23.3 to 37.8, with an average of 29.6 (Table 3). Samples inoculated with 10 to 100 CFU gave CT values in the range of 21.1 to 26.7, with an average of 24.6 (Table 3). The relative AC of the PCR method was 98%, its relative sensitivity was 97%, and its relative specificity was 100%.
TABLE 3.
Results obtained by analyzing artificially inoculated minced pork meat and poultry samples for the presence of Salmonella by the PCR methoda
| Sample | No. of CFU inoculated (organism)b | PCR (CT value)
|
|
|---|---|---|---|
| FAMc | HEXd | ||
| Minced pork meat | |||
| 1 | 1-10 (st) | 28.7 | 33.3 |
| 2 | 1-10 (st) | 37.7 | 32.9 |
| 3 | 1-10 (st) | NAe | 32.6 |
| 4 | 1-10 (st) | 36.4 | 32.4 |
| 5 | 1-10 (st) | 33.7 | 32.1 |
| 6 | 1-10 (st) | 33.6 | 32.3 |
| 7 | 1-10 (st) | 37.8 | 32.4 |
| 8 | 1-10 (st) | 33.7 | 32.5 |
| 9 | 1-10 (st) | 35.2 | 32.1 |
| 10 | 1-10 (sl) | 24.8 | 34.9 |
| 11 | 1-10 (sl) | 23.7 | 34.8 |
| 12 | 1-10 (sl) | 23.3 | 34.5 |
| 13 | 1-10 (sl) | 25.5 | 33.5 |
| 14 | 1-10 (sl) | 25.8 | 25.7 |
| 15 | 1-10 (sl) | 24.5 | 34.1 |
| 16 | 1-10 (sl) | 27.1 | 34.7 |
| 17 | 1-10 (sl) | 29.9 | 33.9 |
| 18 | 1-10 (sl) | 27.2 | 34.6 |
| 19 | 1-10 (sl) | 24.7 | 34.9 |
| 20 | 10-100 (sl) | 24.0 | 31.4 |
| 21 | 10-100 (sl) | 24.2 | 32.1 |
| 22 | 10-100 (sl) | 21.1 | 32.6 |
| 23 | 10-100 (sl) | 23.6 | 31.5 |
| 24 | 10-100 (sl) | 23.9 | 31.0 |
| 25 | 10-100 (sl) | 25.6 | 31.2 |
| 26 | 10-100 (sl) | 24.2 | 31.1 |
| 27 | 10-100 (sl) | 25.1 | 31.5 |
| 28 | 10-100 (sl) | 26.6 | 31.6 |
| 29 | 10-100 (sl) | 22.1 | 32.0 |
| 30 | 10-100 (sl) | 26.2 | 31.8 |
| 31 | 10-100 (sl) | 25.5 | 31.3 |
| 32 | 10-100 (sl) | 26.7 | NA |
| 33 | 10-100 (sl) | 24.9 | 32.5 |
| Poultry skin | |||
| 34 | 1-10 (se) | 28.0 | 28.2 |
| 35 | 1-10 (se) | 29.2 | 28.7 |
| 36 | 1-10 (se) | 31.5 | 27.3 |
| 37 | 1-10 (se) | 29.6 | 27.6 |
| 38 | 1-10 (se) | 33.2 | 27.5 |
| 39 | 1-10 (se) | 28.4 | 28.0 |
| 40 | 1-10 (se) | 30.6 | 27.2 |
| 41 | 1-10 (se) | 29.0 | 28.5 |
| 42 | 1-10 (se) | 29.3 | 28.4 |
| 43 | 1-10 (se) | 29.4 | 28.9 |
| 44 | 1-10 (se) | 28.3 | 28.1 |
| 45 | 1-10 (se) | 29.7 | 28.6 |
| 46 | 1-10 (se) | 28.2 | 27.3 |
| 47 | 1-10 (se) | 28.8 | 28.1 |
| 48 | 1-10 (se) | 29.8 | 27.5 |
| 49 | 1-10 (st) | 24.8 | 27.3 |
| 50 | 1-10 (st) | 26.7 | 27.5 |
| 51 | 1-10 (st) | 26.9 | 27.7 |
| 52 | 1-10 (st) | 26.3 | 27.4 |
| 53 | 1-10 (st) | 27.5 | 28.2 |
| 54 | 1-10 (st) | 25.2 | 27.3 |
| 55 | 1-10 (st) | 26.6 | 28.3 |
| 56 | 1-10 (st) | 25.1 | 27.4 |
| 57 | 1-10 (st) | 24.9 | 27.5 |
| 58 | 1-10 (st) | 25.5 | 27.3 |
| 59 | 1-10 (st) | 25.6 | 27.7 |
| 60 | 1-10 (st) | 26.9 | 25.8 |
| 61 | 1-10 (st) | 23.8 | 25.9 |
| 62 | 1-10 (st) | 25.7 | 25.9 |
All samples were found positive by the reference culture method.
st, S. enterica serovar Typhimurium; sl, S. enterica serovar Livingstone; se, S. enterica serovar Enteritidis.
Salmonella target.
Internal amplification control.
NA, no amplification.
The reference culture method and the PCR method identified 29 Salmonella-positive and 21 Salmonella-negative poultry samples, resulting in a relative AC, sensitivity, and specificity of 100%. The CT values of the PCR-positive samples were in the range of 23.8 to 33.2, with an average of 27.7 (Table 3).
For details of the PCR results obtained by addition of selective reagents to BPW or reducing the elution, volume, see the supplemental material.
DISCUSSION
The 12-h PCR described in the present study is intended as a diagnostic tool for routine use in the meat industry, and therefore a high degree of robustness and reproducibility is imperative. As a tool for ensuring food safety and thereby public health, the method has to be reliable and consistent, day after day, in the hands of different personnel and on different sample matrices. Preliminary studies revealed that the limited preenrichment period of 8 h in BPW was unable to produce sufficient cell counts to meet these demands; thus, crucial steps of the method were optimized to improve sensitivity and robustness.
The concept of proving the presence or absence of Salmonella with only 8 h of preenrichment entails the need for optimization of this growth phase to yield the greatest possible amount of cells. The majority of the existing culture methods recommend BPW for resuscitation and preenrichment of Salmonella (2). The findings of the present study gave no reason to alter this. This point was further emphasized by the fact that shaking of the preenrichment cultures, and thereby improving the accessibility of the nutrients, did not increase the number of salmonellae during short-term incubation. This could be due to overgrowth of competitive flora, as found in other studies (33). Fricker (15) found BPW to be superior to lactose broth for preenrichment of Salmonella from sewage sludge. Other studies have shown no significant difference in the diagnostic sensitivity of BPW and universal preenrichment broth in an examination of fecal samples from swine (19). Similar results were found for BPW, TSB, and glucose mineral salt medium used for preenrichment of frozen or fresh samples of minced meat (35).
Adding growth-promoting reagents to BPW did not seem to provide Salmonella with a competitive edge, so as a last resort, addition of selective reagents directly to BPW was attempted. According to reference culture methods, a selective advantage is introduced when transferring preenriched cells to the next enrichment step (2, 4). However, because of the time limitations of the present method, this was not possible. In the present study, the experiments with selective reagents were performed with freeze-stressed Salmonella, taking into account the potential presence of damaged cells in the samples. During slaughter of pigs in Denmark, wind chilling is applied, rendering part of the bacterial flora freeze damaged. The lack of a beneficial effect of addition of most of the selective reagents could be due to the restriction of resuscitation and growth of the stressed Salmonella cells caused by the selective pressure in the preenrichment media. Delayed addition of selective reagents could be an alternative way to overcome this problem. Joosten et al. (21) showed increased recovery of Salmonella in infant formula containing high levels of probiotic microorganisms when malachite green was added to the preenrichment medium at a concentration of 100 mg/liter. However, the recovery rate improved when nonfat dry milk powder was added to the preenrichments to reduce the toxicity of malachite green toward Salmonella, which was also shown previously by van Schothorst and Renaud (36). In a study by Blivet et al. (9), malachite green (20 mg/liter) and brilliant green (5 mg/liter) were found to inhibit the growth of various Salmonella strains, while novobiocin (up to 40 mg/liter) enhanced their growth. Novobiocin addition (22 mg/liter) has likewise been reported to increase the recovery of Salmonella from fecal samples (20). Even though addition of novobiocin and brilliant green slightly improved the PCR results obtained after 8 h in the present study, it was decided not to add them to the BPW. The reference culture method does not include novobiocin or brilliant green, and adding these to the preenrichment would necessitate two preenrichment protocols of parallel samples, thus compromising the validation.
Another subject of importance is the PCR compatibility of the media used. In a study by Stone et al. (32), Rappaport-Vassiliadis and tetrathionate broths were found to be inhibitory to PCR, whereas BHI—as shown in this study—and selenite cystine broth were not. Eyigor et al. (13), on the other hand, found that a PCR performed directly on tetrathionate enrichment broth extracted by boiling was more sensitive than the reference culture method. Knutsson et al. (22) showed that both BPW and BHI inhibited the PCR. They also found that the rTth PCR mixture, with the same DNA polymerase as in the present study, was less influenced by the presence of BPW than the AmpliTaq Gold PCR mixture. In other words, the DNA polymerase type plays a central role.
For the minced meat examined in the present study, a 5-ml volume resulted in markedly improved PCR results compared to a 1-ml volume. A high number of target organisms in a given sample can, however, influence the outcome of a larger sampling volume, and the possibility of overloading the PCR with target DNA should always be considered. Increasing the volume taken from preenrichment will, of course, result in higher numbers of target cells but, inconveniently, also increase the amount of other bacteria and PCR inhibitors. Accordingly, it is essential to find a balance where the advantage of the larger volume is not obscured by more inhibitors.
The automated DNA extraction procedure applied in this study was selected because the 12-h PCR method was developed for use in a routine laboratory with a high throughput and the need for a high degree of quality control. However, the DNA loss during extraction was shown to be high, as an average of 4.8 CT units was lost by running pure DNA through the magnetic DNA extraction system. As a rule of thumb, 3.3 CT units corresponds to 1 log unit. For example, for viral DNA in various clinical samples, Schuurman et al. (31) showed a DNA recovery of approximately 50% from magnetic DNA extraction. Ferreira-Gonzalez et al. (14) showed close to 100% recovery of viral DNA from plasma with a silicone-based kit. Previous in-house comparisons have shown automated DNA extraction with a KingFisher to be similar to, or better than, various manual and kit-based extraction methods (data not shown).
Increasing the amount of initial DNA in a PCR may result in rapid accumulation of high numbers of PCR products and in lower CT values. However, when setting up a routine PCR test, the financial aspect should also be considered. Running a PCR with 20 μl of template DNA in a total volume of 50 μl of master mix would double the cost of PCR analysis compared to using 10 μl of template DNA in a total of 25 μl of master mix.
In conclusion, it was successfully demonstrated that the optimized 12-h PCR method for Salmonella detection produced results comparable to those of the reference culture method with artificially inoculated pork meat and poultry samples. Further studies with naturally contaminated samples are needed. The main advantage of the method developed is the reduced time of analysis, enabling faster release of Salmonella-free fresh meat. Moreover, the sample price and workload are markedly reduced compared to those of the reference culture method.
The strategies described in the present study are, in most cases, not unique to the detection of Salmonella but could be used to improve the sensitivity and/or shorten the preenrichment time of other real-time PCR-based methods.
Supplementary Material
Acknowledgments
This work was supported in part by Danish Directorate for Food, Fisheries and Agri-Business (DFFE) grant 3414-04-01032, in part by grant F040301 from the CampyFood project of the Nordic Innovation Centre (NICE), and in part by the European Union project BIOTRACER (FOOD-2006-CT-036272).
We thank Kirsten Michaelis and Julia Christensen for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Agarwal, A., A. Makker, and S. K. Goel. 2002. Application of the PCR technique for a rapid, specific and sensitive detection of Salmonella spp. in foods. Mol. Cell. Probes 16:243-250. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Food-borne pathogens. Monograph number 1, Salmonella. Oxoid Limited, Hampshire, England.
- 3.Anonymous. 1999. Salmonella. Detection in foods, 5th ed. NMKL no. 71. Nordic Committee on Food Analysis, Esbo, Finland.
- 4.Anonymous. 2002. Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp. ISO 6579, 4th edition. International Organization for Standardization, Geneva, Switzerland.
- 5.Anonymous. 2004. Annual report on zoonosis in Denmark 2003. Ministry of Food, Agriculture, and Fisheries, Copenhagen, Denmark.
- 6.Anonymous. 2004. Microbiology of food and animal feeding stuffs—protocol for the validation of alternative methods, ISO 16140, 1st ed. International Organization for Standardization, Geneva, Switzerland.
- 7.Anonymous. 2005. Protocol for the validation of alternative microbiological methods. NordVal-doc-2005-01-01. Danish Institute for Food and Veterinary Research, Søborg, Denmark.
- 8.Bennett, A. R., D. Greenwood, C. Tennant, J. G. Banks, and R. P. Betts. 1998. Rapid and definitive detection of Salmonella in foods by PCR. Lett. Appl. Microbiol. 26:437-441. [DOI] [PubMed] [Google Scholar]
- 9.Blivet, D., G. Salvat, F. Humbert, and P. Colin. 1998. Development of a new culture medium for the rapid detection of Salmonella by indirect conductance measurements. J. Appl. Microbiol. 84:399-403. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, R. C. 1989. Statistics for biologists, 3rd ed. Cambridge University Press, Cambridge, Great Britain.
- 11.Chen, S., A. Yee, M. Griffiths, C. Larkin, C. T. Yamashiro, R. Behari, C. Paszko-Kolva, K. Rahn, and S. A. De Grandis. 1997. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. J. Food Microbiol. 35:239-250. [DOI] [PubMed] [Google Scholar]
- 12.Ellingson, J. L. E., J. L. Anderson, S. A. Carlson, and V. K. Sharma. 2004. Twelve hour real-time PCR technique for the sensitive and specific detection of Salmonella in raw and ready-to-eat meat products. Mol. Cell. Probes 18:51-57. [DOI] [PubMed] [Google Scholar]
- 13.Eyigor A., K. T. Carli, and C. B. Unal. 2002. Implementation of real-time PCR to tetrathionate broth enrichment step of Salmonella detection in poultry. Lett. Appl. Microbiol. 34:37-41. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira-Gonzalez, A., S. Yanovich, M. R. Langley, L. A. Weymouth, D. S. Wilkinson, and C. T. Garrett. 2000. Enhanced analytical sensitivity of a quantitative PCR for CMV using a modified nucleic-acid extraction procedure. J. Clin. Lab. Anal. 14:32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fricker C. R. 1984. A comparison of two methods for the isolation of salmonellae from sewage sludge. Zentbl. Bakteriol. Mikrobiol. Hyg. B 179:170-178. [PubMed] [Google Scholar]
- 16.Gadó, I., P. Major, M. Kiraly, and M. G. Plaveczky. 2000. Rapid combined assay for Salmonella detection in food samples. Acta Microbiol. Immunol. Hung. 47:445-456. [PubMed] [Google Scholar]
- 17.Hanai, K., M. Satake, and T. J. White. 1997. Comparison of commercially available kits for detection of Salmonella strains in foods. Appl. Environ. Microbiol. 63:775-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hein, I., G. Flekna, M. Krassnig, and M. Wagner. 2006. Real-time PCR for the detection of Salmonella spp. in food: an alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 66:538-547. [DOI] [PubMed] [Google Scholar]
- 19.Hoorfar, J, and A. V. Mortensen. 2000. Improved culture methods for isolation of Salmonella organisms from swine feces. Am. J. Vet. Res. 61:1426-1429. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, A. N., G. Sørensen, D. L. Baggesen, R. Bødker, and J. Hoorfar. 2003. Addition of novobiocin in preenrichment step can improve Salmonella culture protocol of modified semisolid Rappaport-Vassiliadis. J. Microbiol. Methods 55:249-255. [DOI] [PubMed] [Google Scholar]
- 21.Joosten, H., E. Bidlas, and N. Garofalo. 2006. Salmonella detection in probiotic products. Int. J. Food Microbiol. 110:104-107. [DOI] [PubMed] [Google Scholar]
- 22.Knutsson, R., C. Löfström, H. Grage, J. Hoorfar, and P. Rådström. 2002. Modeling of 5′ nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica. J. Clin. Microbiol. 40:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause, M., M. H. Josefsen, M. Lund, N. R. Jacobsen, L. Brorsen, M. Moos, A. Stockmarr, and J. Hoorfar. 2006. Comparative, collaborative and on-site validation of a TaqMan PCR method as a tool for certified production of fresh, Campylobacter-free chickens. Appl. Environ. Microbiol. 72:5463-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lübeck, P. S., and J. Hoorfar. 2003. PCR technology and applications to zoonotic food-borne pathogens. Methods Mol. Biol. 216:65-84. [DOI] [PubMed] [Google Scholar]
- 25.Malorny, B., E. Paccasonni, P. Fach, C. Bunge, A. Martin, and R. Helmuth. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70:7046-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, A., and S. E. Katz. 1991. A resuscitation/selection system for rapid determination of Salmonella in foods. J. Assoc. Off. Anal. Chem. 74:522-525. [PubMed] [Google Scholar]
- 27.Myint, M. S., Y. J. Johnson, N. L. Tablante, and R. A. Heckert. 2006. The effect of preenrichment protocol on the sensitivity and specificity of PCR for detection of naturally contaminated Salmonella in raw poultry compared to conventional culture. Food Microbiol. 23:599-604. [DOI] [PubMed] [Google Scholar]
- 28.Proudnikov, D., V. Yuferov, Y. Zhou, K. S. LaForge, A. Ho, and M. J. Kreek. 2003. Optimizing primer-probe design for fluorescent PCR. J. Neurosci. Methods 123:31-45. [DOI] [PubMed] [Google Scholar]
- 29.Reissbrodt, R., E. Vielitz, E. Kormann, W. Rabsch, and H. Kuhn. 1996. Ferrioxamine E-supplemented preenrichment and enrichment media improve various isolation methods for Salmonella. Int. J. Food Microbiol. 29:81-91. [DOI] [PubMed] [Google Scholar]
- 30.Reynisson, E., M. H. Josefsen, M. Krause, and J. Hoorfar. 2006. Evaluation of probe chemistries and platforms to improve the detection limit of real-time PCR. J. Microbiol. Methods 66:206-216. [DOI] [PubMed] [Google Scholar]
- 31.Schuurman, T., A. van Breda, R. de Boer, M. Kooistra-Smid, M. Beld, P. Savelkoul, and R. Boom. 2005. Reduced PCR sensitivity due to impaired DNA recovery with the MagNA Pure LC total nucleic acid isolation kit. J. Clin. Microbiol. 43:4616-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone, G. G., R. D. Oberst, M. P. Hays, S. McVey, and M. M. Chengappa. 1994. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J. Clin. Microbiol. 32:1742-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomason, B. M., D. J. Dodd, and W. B. Cherry. 1977. Increased recovery of salmonellae from environmental samples enriched with buffered peptone water. Appl. Environ. Microbiol. 34:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uyttendaele, M., K. Vanwildemeersch, and J. Debevere. 2003. Evaluation of real-time PCR versus automated ELISA and a conventional culture method using a semi-solid medium for detection of Salmonella. Lett. Appl. Microbiol. 37:386-391. [DOI] [PubMed] [Google Scholar]
- 35.van Schothorst, M., R. J. Gilbert, R. W. Harvey, O. Pietzsch, and E. H. Kampelmacher. 1978. Comparative studies on the isolation of Salmonella from minced meat. Zentbl. Bakteriol. B 167:138-145. [PubMed] [Google Scholar]
- 36.van Schothorst, M., and A. M. Renaud. 1985. Malachite green preenrichment medium for improved salmonella isolation from heavily contaminated samples. J. Appl. Bacteriol. 59:223-230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.