Abstract
The degradation pathway of 2,4,6-trichlorophenol (2,4,6-TCP), a hazardous pollutant, in the aerobic bacterium Cupriavidus necator JMP134(pJP4) (formerly Ralstonia eutropha JMP134) is encoded by the tcp genes. These genes are located in a genetic context, tcpRXABCYD, which resembles a putative catabolic operon. In this work, these gene sequences were individually disrupted and mutant strains were evaluated for their ability to grow on or degrade 2,4,6-TCP. The tcpX and tcpA mutants completely failed to degrade this compound. Although the tcpC mutant was also unable to grow on 2,4,6-TCP, it still transformed this chlorophenol to 6-chlorohydroquinol. In contrast, the tcpD mutant grew on 2,4,6-TCP, suggesting the presence of additional maleylacetate reductase-encoding genes. Five other open reading frames encoding maleylacetate reductases, in addition to the tcpD gene, were found in the genome of C. necator, and two of them provide this function in the tcpD mutant. The tcpR gene, encoding a putative LysR-type transcriptional regulator, was disrupted, and this mutant strain completely failed to grow on 2,4,6-TCP. Transcriptional fusion studies demonstrated that TcpR activates the expression of the tcp genes, responding specifically to 2,4,6-TCP. The transcriptional start of the tcp operon was mapped, and a putative σ70-type promoter was identified.
2,4,6-Trichlorophenol (2,4,6-TCP), widely used as a biocide and preservative, is considered a priority environmental pollutant worldwide (14, 32). Aerobic bacteria degrade this pollutant and, in several cases, grow on it as the sole carbon source (3, 5, 15, 20). A catabolic pathway for 2,4,6-TCP (Fig. 1A) has been proposed (21, 26, 33, 37, 39). The pathway is initiated by the conversion of 2,4,6-TCP to 2,6-dichloro-p-hydroquinone (2,6-DCHQ) and then to 6-chlorohydroxyquinol (6-CHQ); both steps are catalyzed by flavin adenine dinucleotide (FAD)-dependent 2,4,6-TCP monooxygenase (TCP-MO). 6-CHQ is transformed to 2-chloromaleylacetate (2-CMA) by 6-chlorohydroxyquinol 1,2-dioxygenase (HQDO), and 2-CMA is converted to β-ketoadipate by NADH-dependent maleylacetate reductase (MAR). Recently, the tcpABC genes from Cupriavidus necator JMP134(pJP4), a well-known chloroaromatic compound-degrading strain (5, 8, 29), were shown to encode enzymes that convert 2,4,6-TCP to 2-CMA (21). However, no evidence about a gene encoding MAR activity was provided. The tcpABC genes in C. necator are adjacent to four other open reading frames (ORFs) (tcpY, tcpD, tcpR, and tcpX), forming a putative catabolic operon (Fig. 1B) (23). The tcpR gene has significant identity to the pcpR gene, which encodes a LysR-type regulator involved in the degradation of pentachlorophenol in Sphingobium chlorophenolicum (4). The TcpX protein is believed to provide FADH2 to TcpA because it has high identity to the TftC protein, which putatively performs this function in the degradation of 2,4,5-TCP in Burkholderia cepacia AC1100 (10, 13). This flavin reductase activity would also be encoded by the tcpB gene, which shows sequence similarity to genes coding for nitroreductases. However, a tcpB mutant still degrades 2,4,6-TCP (21). Whereas the tcpY gene does not possess homology to any other gene sequence, the tcpD gene shows high sequence identity to maleylacetate reductase genes. In this work, we investigated the function of each of the tcp gene sequences in the tcpRXABCYD cluster from C. necator JMP134(pJP4). We report that this gene cluster encodes all of the functions required for transformation of 2,4,6-TCP to β-ketoadipate and that tcpR is the regulatory gene controlling tcp gene expression.
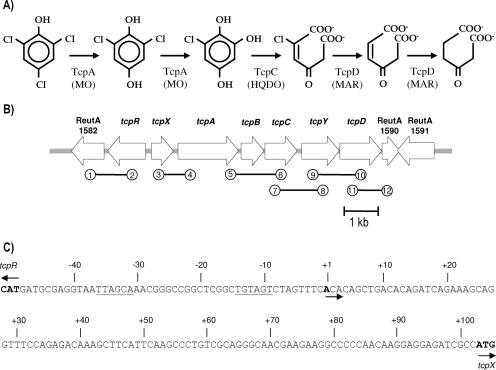
FIG. 1.
(A) Degradative pathway proposed for 2,4,6-TCP in C. necator JMP134. 2,4,6-TCP-MO, HQDO, and MAR catalyze the conversion of 2,4,6-trichlorophenol to 2,6-dichlorohydroquinone, 6-chlorohydroxyquinol, 2-chloromaleylacetate, maleylacetate, and β-ketoadipate, respectively. (B) Genetic context of the tcpRXABCYD gene cluster. The bar represents 1 kb. The numbers in circles correspond to the primers described in Table 2: 1, FAD2; 2, tcpR2; 3, tcpX1; 4, tcpA3; 5, tcpAend; 6, tcpC2; 7, tcpC1; 8, tcpY2; 9, tcpY1; 10, tcpD2; 11, tcpD1; and 12, TCPout. (C) Schematic representation of the organization of the tcp promoter region. The arrows indicate the starts of transcription and translation. The transcriptional initiation nucleotide (+1) and the putative −35 and −10 motifs are underlined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. C. necator JMP134(pJP4) and its 2,4,6-TCP-mineralizing derivatives were grown at 30°C in minimal medium (16) with 0.4 to 2 mM 2,4,6-TCP as the sole carbon source. To determine growth at higher 2,4,6-TCP levels, C. necator JMP134 and its derivatives were initially grown on 0.4 mM 2,4,6-TCP in a 50-ml Erlenmeyer flask, and every 6 days of incubation, the culture was transferred to fresh medium containing a higher concentration (0.2 mM increases), up to 2 mM. Growth was determined by measuring the optical density at 600 nm (OD600). Determination of OD600 in cell-free supernatants of these cultures discarded any effect of 2,4,6-TCP or its catabolic intermediates on the OD600 values. C. necator derivatives not able to proliferate on 2,4,6-TCP were grown on 1 mM phenol plus the appropriate antibiotic (Table 1). Escherichia coli strains were maintained on Luria-Bertani (LB) agar plates containing 50 μg of kanamycin ml−1 or 50 μg of trimethoprim ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| C. necator | ||
| JMP134 | 2,4,6-TCP+ 2,4-D+; pJP4 | DSMZb |
| JMP134 tcpR | 2,4,6-TCP− Apr Kmr | This study |
| JMP134 tcpX | 2,4,6-TCP− Apr Kmr | This study |
| JMP134 tcpA | 2,4,6-TCP− Apr Kmr | This study |
| JMP134 tcpC | 2,4,6-TCP− Apr Kmr | This study |
| JMP134 tcpY | 2,4,6-TCP+ Apr Kmr | This study |
| JMP134 tcpD | 2,4,6-TCP+ Apr Kmr | This study |
| JMP134 ReutA1582 | 2,4,6-TCP+ Apr Kmr | This study |
| JMP134 tfdFI | 2,4,6-TCP+ Kmr | This study |
| JMP134 tfdFII | 2,4,6-TCP+ Kmr | This study |
| JMP134 ReutB4129 | 2,4,6-TCP+ Apr Kmr | This study |
| JMP134 ReutB4694 | 2,4,6-TCP+ Apr Kmr | This study |
| JMP134 ReutC5982 | 2,4,6-TCP+ Apr Kmr | This study |
| E. coli BW25113 | pKD46; red recombinase | 6 |
| Plasmids | ||
| pRK600 | Cmr | 12 |
| pCR2.1 TOPO-TA | Apr Kmr | Invitrogen |
| pMLS7 | PS7 promoter; Tpr | 19 |
| pS7X | tcpX; pMLS7 derivative | This study |
| pS7XA | tcpX-tcpA; pMLS7 derivative | This study |
| pS7A | tcpA; pMLS7 derivative | This study |
| pS7AB | tcpA-tcpB; pMLS7 derivative | This study |
| pS7C | tcpC; pMLS7 derivative | This study |
| pS7D | tcpD; pMLS7 derivative | This study |
| pCM132 | lacZ promoter-probe vector; Kmr | 22 |
| pPRTCP | tcpR-PtcpX-tcpX′; pCM132 derivative | This study |
| pPTCP | PtcpX-tcpX′; pCM132 derivative | This study |
| pJP4-tfdFI mutant | tfdFI Kmr | This study |
| pJP4-tfdFII mutant | tfdFII Kmr | This study |
2,4,6-TCP+ and 2,4-D+, able to grow in 2,4,6-TCP or 2,4-D, respectively; tcp, catabolic genes involved in 2,4,6-TCP degradation; FAD, FAD synthase; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Tpr, trimethoprim resistance; Kmr, kanamycin resistance.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany.
Analytical methods.
The presence of 2,4,6-TCP and its derivatives was determined by UV spectroscopy or by high-performance liquid chromatography (HPLC) using cell-free supernatants from suspensions of cells grown in 8 mM succinate and induced for 3 h with 0.5 mM 2,4,6-TCP. Cells were centrifuged at 6,000 × g for 5 min, washed twice, and resuspended in minimal medium to an OD600 of 1.0. 2,4,6-TCP (0.3 mM) was added, and the cells were incubated at 30°C in a shaker. Samples of cell-free supernatants were analyzed by UV spectroscopy in a diode array HP8452-A UV-visible spectrophotometer. Samples (20 μl) from cell-free supernatants were taken at different times and injected into a 126/166 System Gold Beckman liquid chromatograph equipped with a Waters Symmetry C18 4.6-μm-diameter column (Beckman Instruments, Fullerton, CA). A methanol-H2O (80:20) mixture containing 0.1% (vol/vol) phosphoric acid was used as the solvent at a flow rate of 1 ml min−1. The column effluent was monitored at 210 nm.
DNA manipulation.
Restriction, ligation, and dephosphorylation reactions, purification, and electroporation of DNA were performed by standard procedures (1). Derivatives of the broad-host-range plasmid vector pMLS7 (19) and of pCM132 (22) were mobilized from E. coli DH5α to derivatives of C. necator JMP134 by triparental mating with the helper strain E. coli HB101(pRK600), as previously described (27). Transconjugants were selected on minimal medium agar plates supplemented with 1 mM phenol plus kanamycin or trimethoprim.
Chromosomal disruption of gene sequences in C. necator JMP134.
An internal fragment (250 to 500 bp) of each gene sequence was PCR amplified from DNA of strain JMP134 using primer pairs listed in Table 2. The PCR products were cloned into pCR2.1-TOPO (Invitrogen Life Technologies, Carlsbad, CA), and each plasmid was electroporated into cells from strain JMP134 to obtain a one-recombination-event chromosomal disruption of each target gene. The electrocompetent cells were obtained as follows: cells from a 50-ml culture (OD600 of 0.4 to 0.5) were grown in LB medium and collected by centrifugation at 6,000 × g for 5 min at 4°C. The cell pellet was washed three times with cold distilled water, and the cells were collected by centrifugation at 6,000 × g and resuspended in 50 μl of distilled water. Recombinant strains were selected on LB agar containing kanamycin. Chromosomal disruptions were checked by the presence/absence of the expected PCR products in the wild-type and the mutant strains, using primer pairs described in Table 2.
TABLE 2.
PCR primers used in this work
| Primer and function(s) | Sequence (5′→3′) | Primer | Sequence (5′→3′) |
|---|---|---|---|
| Chromosomal disruption, inactivation of tfdFI and tfdFII genes,a and overlapping RT-PCR | |||
| tcpR1 | TCAGCTTCTTCCTGCTCGAT | tcpR2 | GGATACCAGCACATGCTTCTC |
| tcpX1 | TCACGTGCTCGGCTGTTTGT | tcpX2 | ACCATGGCGGACTTGCTGTA |
| tcpA1 | CCAAGAAGCACGACCTGA | tcpA2 | CTTGTTGGTGCCGATATG |
| tcpC1 | AGGAGTTCATCCTGCTCAG | tcpC2 | CGCACGCCGAACACGGCATC |
| tcpY1 | CGTGGTACCGATCGTGTCT | tcpY2 | GATATCGACCTTGGGAGTCG |
| tcpD1 | GGCTCGGAGATGACTACGAT | tcpD2 | CATGGGCGTACAAACCTTCT |
| ReutB4129 1 | ATCTACGATCCCGCACTGAC | ReutB4129 2 | ATGGCACAGCTTGTGATGAA |
| ReutB4694 1 | GGTCTACGATCCCGAACTGA | ReutB4694 2 | GTATGGCACAGCTTGTGGTG |
| ReutC5982 1 | ACCTGCACCAACAGCAGAC | ReutC5982 2 | TCTAACACGGCGAAAAATCG |
| FAD1 | TCGATGGTGTACATTGCGGG | FAD2 | AAGTCATCCGCACTGGCTTC |
| MUTF1FW | GACCCTTCATGAAGAAGTTCACGCTTGACTACCTGAGCCCGTGTAGGCTGGAGCTGCTTC | MUTF1RE | GCGGAGTTGCAGGTCACATTATTTGAAATCCGGTCTTCGCCATTCCGGGGATCCGTCGACC |
| MUTF2FW | CCGGCGATCTGAATGAATTCGTTGCGCACTTCTGGCCGGTGTGTAGGCTGGAGCTGCTTC | MUTF2RE | AGAGGTCCATGGGATGTCCGGTTCACGCCGGCATTTCTCCATTCCGGGGATCCGTCGACC |
| FORF1 | ATGAAGAAGTTCACGCTTGACTACCTGAGC | REVF1 | ACCGTACTAAACGCGGAGTG |
| FORF2 | GCACTAGTAGTGACCGGCGAT | REVF2 | CTTATCGATAGGTCGGGTCG |
| TcpA3 | GTGCAGGTCGTAGAAGTCG | tcpAend | GAGGGCCACGACAGCGAATAC |
| TCPout | AACCTTCCACATTTTGTGCC | ||
| RT-PCR MARs | |||
| RTtfdFI1 | ACGCGAGTTAGCGAAGGATA | RTtfdFI2 | GAGATAGCAAGCGGCAAATC |
| RTtfdFII1 | GAATTCGTTGCGCACTTCTG | RTtfdFII2 | GGCAAGGAGGTCAGGTGAT |
| RTtcpD1 | TCGCGCACGCAGCAGAAGGTTTGT | RTtcpD2 | ACGCGGGTTCGGGTACTGGTTCTG |
| RT ReutB4129 1 | ATCTACGATCCCGCACTGAC | RT ReutB4129 2 | ATGGCACAGCTTGTGATGAA |
| RT ReutB4694 1 | GGTCTACGATCCCGAACTGA | RT ReutB4694 2 | GTATGGCACAGCTTGTGGTG |
| RT ReutC5982 1 | ACCTGCACCAACAGCAGAC | RT ReutC5982 2 | TCTAACACGGCGAAAAATCG |
| RT358F | CCTACGGGAGGCAGCAG | RT907R | CCGTCAATTCTTTTRAGTTT |
| Transcription start mapping (RACE) | |||
| X1 | CGCTCACGGAACAAACAGC | X2 | GTGGCGATGACGGTCACG |
| X3 | ACTGCACGCGACAAGGCG | AUAP | GGCCACGCGTCGACTAGTAC |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | ||
| Constitutive expression of tcp gene sequencesb | |||
| tcpRlacZ | ATCGAAGGTCAGCAATACGG | tcpB-XbaI | ATGCTGTCTAGACTCTTGCATCACTGGACTCC |
| tcpR-EcoRI | GCTAATGAATTCGCATCATGGACACTATTCCC | tcpR-HindIII | CCGCGGAAGCTTATCGAAGGTCAGCAATACGG |
| tcpX-EcoRI | GGAGGAGAATTCCATGTCGTCCGCAGTCTTC | tcpX-XbaI | CGCGGGTCTAGACGCTTCAAGTCGCGCTAGG |
| tcpA-EcoRI | AGGAGAGAATTCGATGATTCGCACTGGCAAGC | tcpA-XbaI | CTGCGTTCTAGACGGAAGATCTTGTCAAGCAG |
| tcpC-EcoRI | CGGGAGGAATTCGATGCAAGAGTATGACCAGC | tcpC-HindIII | GACCTTAAGCTTATCTGTCGAACCCATTTGCC |
| tcpD-EcoRI | TTTGACGAATTCCCCTACGATGAAAGCATTCC | tcpD-HindIII | GTATTGAAGCTTAACCTTCCACATTTTGTGCC |
Primers to inactivate either tfdFI or tfdFII genes (MUTF1FW, MUTF1RE, MUTF2FW, and MUTF2RE) contain nearly 40-bp homology extensions of the tfdFI or tfdFII gene sequence (in boldface) and nearly 20-bp priming sequences for pKD4 (6).
Primers include a restriction site which is underlined.
Inactivation of tfdFI and tfdFII genes.
The tfdFI and tfdFII genes were independently inactivated in E. coli BW25113(pJP4) cells by the method of Datsenko and Wanner (6). PCR primers MUTF1FW and MUTF1RE (for the tfdFI gene) and MUTF2FW and MUTF2RE (for the tfdFII gene), which contain 40-bp homology extensions of the tfdFI or tfdFII gene sequence (Table 2, in boldface) and 20-bp priming sequences for pKD4 (6), were synthesized. These primer pairs were used to amplify the kanamycin resistance gene flanked by 40 bp of the tfdFI or tfdFII gene sequence, with pKD4 as a template. The PCR program was 95°C for 5 min; 28 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 90 s; and then 72°C for 10 min. PCR products were used to inactivate the tfdFI or tfdFII gene in E. coli strain BW25113(pJP4) harboring RecBCD recombinase by a procedure described previously (28). pJP4 derivatives containing the inactivated tfdFI or tfdFII gene were then transferred to strain JMP134 by biparental conjugation as described previously (27), and the transconjugants were selected on minimal medium agar plates supplemented with 1 mM phenol plus kanamycin. Each transconjugant was transferred five times to the same liquid minimal medium until the pJP4 plasmid was completely removed and only the corresponding pJP4 tfdFI(II) mutant was present in C. necator JMP134. Primer pairs FORF1/REVF1 and FORF2/REVF2 were used to verify correct recombinational insertion of the kanamycin resistance cassette in place of each tfdF gene. This was then confirmed by direct sequencing of the insertion region using these primers.
Constitutive expression of tcp gene sequences.
To obtain constitutive expression of each tcp gene, the corresponding PCR product was cloned into the pMLS7 expression vector (19), using the primers listed in Table 2. The corresponding translation start is included in the primer sequence, and it is located immediately downstream of the EcoRI site. Each PCR product was cloned into pCR2.1 TOPO-TA (Invitrogen Life Technologies, Carlsbad, CA), and the resulting plasmids were digested with EcoRI and HindIII or XbaI to obtain a fragment which was inserted into the pMLS7 plasmid. The identity of the DNA fragments cloned in pMLS7 was confirmed by sequencing.
Detection of transcripts by RT-PCR.
Cells from a 50-ml minimal medium culture grown on 8 mM succinate to a OD600 of 1.0 were centrifuged at 6,000 × g for 5 min, washed twice, and resuspended in the same volume of minimal medium containing 0.5 mM 2,4,6-TCP. When over 50% of chlorophenol degradation had taken place, total RNA was obtained by the Trizol reagent method (Invitrogen Life Technologies, Carlsbad, CA). In brief, 10 ml of culture was centrifuged at 10,000 × g for 5 min and the pellet was resuspended in 1 ml of Trizol and incubated for 5 min at room temperature. Two hundred microliters of chloroform was added, and the mixture was agitated vigorously for 15 s. The samples were left for 3 min at room temperature and then centrifuged at 12,000 × g for 15 min at 4°C. The liquid phase was transferred to a clean tube, and 500 μl of isopropanol was added. After 10 min of incubation at room temperature, the samples were centrifuged at 12,000 × g for 10 min at 4°C, and the pellet was washed out with 1 ml of cold 70% ethanol. The samples were centrifuged at 7,500 × g for 5 min at 4°C and dried for 10 min. The pellets were resuspended in 20 μl of distilled water and 10 μl of 10× MOPS (1× MOPS buffer is 20 mM morpholinepropanesulfonic acid [pH 7.0], 8 mM sodium acetate, and 1 mM EDTA). To remove any DNA contamination, the RNA was treated and cleaned with TURBO DNase kit (Ambion The RNA Company, Austin, TX). The reverse transcription-PCR (RT-PCR [20 μl of reaction mixture]) was carried out using the ImProm-II reverse transcription system (Promega Corporation, Madison, WI) with 1 μg of total RNA. PCRs (25 ul) were performed using specific primer pairs (Table 2) with mixtures that contained 1 μl of total cDNA, 50 pmol of each primer, 50 μM deoxynucleoside triphosphates (dNTPs), 1 mM MgCl2, and 5 U of Taq DNA polymerase, prepared in the reaction buffer supplied by the manufacturer. After RT, the mixtures were treated to 95°C for 2 min and subjected to 30 cycles of 30 s at 95°C, 30 s at 54°C, and 1 min at 72°C. Negative control reactions were performed in the same way, except that the RT addition was omitted.
Construction of tcpR-tcpX′-lacZ fusions.
The vector pCM132 containing the promoterless lacZ-Kmr cassette was used (22). A DNA fragment including the complete tcpR gene sequence and the first 403 bp of the tcpX gene sequence was amplified by PCR with primer pairs tcpRlacZ and tcpX2 (Table 2), using Pfx Platinum DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA). The PCR product was cloned into pTOPO-BLUNT (Invitrogen Life Technologies, Carlsbad, CA), and the resulting plasmid was digested with EcoRI to release a 1.7-kb fragment that was then cloned into pCM132. This procedure created a tcpR-PtcpX-tcpX′-lacZ gene fusion in pCM132 named pPRTCP. Using the same procedure, a PtcpX-tcpX′-lacZ gene fusion in pCM132 was created, named pPTCP. The identity of the DNA fragments cloned in pCM132 was confirmed by sequencing. The expression of lacZ was determined by β-galactosidase assays performed as follows. Cells were grown in 10 ml of minimal medium containing 8 mM succinate and 50 μg of kanamycin ml−1 until an OD600 of 0.5. At this point, a 0.1 mM concentration of the putative inducer compound was added and the incubation was prolonged by 3 h. Cells were lysed with chloroform and sodium dodecyl sulfate, and β-galactosidase activities were determined as described previously (24).
Transcription start mapping.
The transcription start of the tcpX gene was mapped using a protocol for rapid amplification of cDNA ends (RACE). Total RNA (2 μg in a 20-μl reaction) was submitted to RT, as described above, using the primer X1 (Table 2), which anneals 183 bp downstream of the tcpX translational start. Clean up of the resulting cDNA fragments was done using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). A homopolymeric tail was added to the 3′ end of the cDNA (corresponding to the 5′ end of mRNA that was reverse transcribed) using terminal deoxytransferase (Invitrogen Life Technologies, Carlsbad, CA) and dCTPs and incubated at 37°C for 10 min. The dC-tailed cDNA was amplified by PCR using the abridged anchor primer and primer X2 (Table 2), which anneals 127 bp downstream of the tcpX translational start. To eliminate any nonspecific PCR products from the first reaction, this PCR product was then PCR amplified with primer X3 (Table 2), which anneals 103 bp downstream of the tcpX translational start, and the abridged universal amplification primer. The amplified products were purified using a Qiaquick PCR clean up column (QIAGEN, Hilden, Germany) and were cloned into the pCR 2.1 vector (Invitrogen Life Technologies, Carlsbad, CA).
DNA sequencing and sequence analysis methods.
Nucleotide sequences of both DNA strands were determined with an ABI PRISM 377 DNA sequencer (Perkin-Elmer, Foster City, CA). PCR primers were designed with the aid of the Lasergene software package (DNAStar, Inc., Madison, WI).
RESULTS AND DISCUSSION
The tcpRXABCYD gene cluster encodes key functions involved in 2,4,6-TCP degradation.
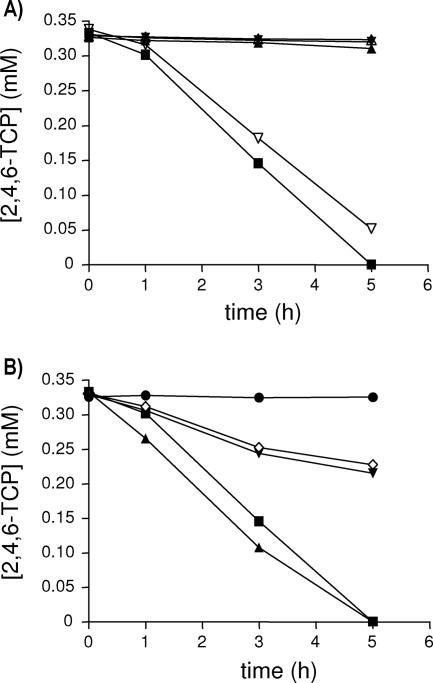
A 2,4,6-TCP degradation pathway (Fig. 1A) has been determined in several bacteria, including C. necator JMP134 (21, 26, 33, 39). The genetic context related to 2,4,6-TCP degradation has been identified in the C. necator JMP134 genome (23), and the functions of the tcpA, tcpB, and tcpC genes have been described (21). To assess the role in 2,4,6-TCP degradation of all the genes from the tcpRXABCYD cluster of C. necator JMP134, insertional inactivation of each putative gene was carried out. The tcpA mutant did not grow on 2,4,6-TCP (not shown). Degradation tests performed with resting cells of this mutant indicated that inactivation completely prevented removal of the chlorophenol (Fig. 2A). The tcpA mutant harboring the tcpA gene cloned in pS7A degraded 2,4,6-TCP to an extent similar to that of the wild-type strain (Fig. 2A), but was still not able to grow on this compound. A reddish orange metabolite, possibly derived from 6-CHQ, was observed in the culture of the tcpA (pS7A) mutant, suggesting a polar effect on the tcpC gene encoding HQDO. Such a polar effect is caused by the insertion of a complete copy of the plasmid containing the sequence of the inactivated gene.
FIG. 2.
Degradation of 2,4,6-TCP by tcpX and tcpA mutant strains of C. necator JMP134 and some of its derivatives. (A) Removal of 2,4,6-TCP was detected by high-performance liquid chromatography using samples of supernatants after incubation of preinduced cell suspensions (OD600 of 1.0). ▪, strain JMP134; ▴, JMP134 tcpX; ▾, JMP134 tcpX (S7X) constitutively expressing the tcpX gene; ▵, JMP134 tcpA; ▿, JMP134 tcpA (S7A) constitutively expressing the tcpA gene. (B) Removal of 2,4,6-TCP by strains constitutively expressing the tcpX, tcpA, or tcpB genes. ▪, strain JMP134; ▴, JMP134 tcpR (S7XA) constitutively expressing the tcpXA genes; ▾, JMP134 tcpR (S7A) constitutively expressing the tcpA gene; ⋄, JMP134 tcpR (S7AB) constitutively expressing the tcpAB genes; •, JMP134 tcpR. The values are averages from three or four replicates, with standard deviations of less than 5%.
As with the tcpA mutant, the tcpX mutant was also unable to grow on or degrade 2,4,6-TCP (Fig. 2A). The pS7X plasmid, containing the tcpX gene, was introduced into the tcpX mutant. The resulting strain was still unable to grow on or degrade 2,4,6-TCP (Fig. 2A). A polar effect on the tcpA gene may prevent recovery of the wild-type phenotype. This inactivation approach does not allow us to know if the tcpX gene is essential or not. The tcpX gene may code for the FAD reductase activity required by TcpA originally ascribed to the tcpB gene, but without experimental support (21). The tcpX gene putatively encodes a protein with a 54% amino acid identity to the TftC protein from the 2,4,5-TCP degradation pathway in B. cepacia AC1100 (10, 13). Partner flavin reductases are usually located in the same operon or physically linked (2, 9, 13, 36), a condition that is fulfilled by both tcpB and tcpX genes. To elucidate which is the cognate flavin reductase of the TcpA protein (protein TcpX or TcpB), we compared the degradation rates of 2,4,6-TCP among different derivatives of the tcpR mutant, which is completely unable to grow on or degrade 2,4,6-TCP (see below). This background avoids the effect of any flavin reductase function associated with tcp functions. To provide to the tcpR mutant the functional monooxygenase and its putative reductase pair, the tcpX-tcpA or the tcpA-tcpB sequences were cloned into pMLS7, generating the plasmids pS7XA and pS7AB, respectively. The pS7A plasmid was also tested in the tcpR mutant. tcpR mutants containing both pS7A and pS7AB were able to transform 0.3 mM of 2,4,6-TCP, but at low rate (Fig. 2B). This observation is in agreement with a previous report (21) and indicates that TcpB is either not required for 2,4,6-TCP degradation or is replaced by other reductases in C. necator. In contrast, the tcpR (pS7XA) mutant transformed this compound at a high rate (Fig. 2B), strongly indicating the interdependence between the TcpX and TcpA proteins as a functional pair: monooxygenase-reductase.
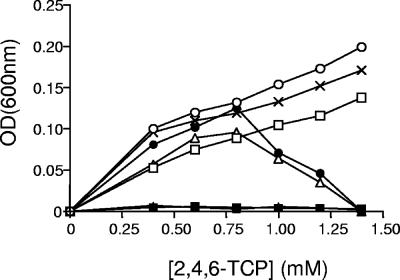
6-CHQ is the central intermediate of the 2,4,6-TCP degradation pathway (11, 18, 21, 26), and the TcpC protein catalyzes the ring cleavage that produces 2-CMA. The tcpC mutant failed to grow on 2,4,6-TCP (Fig. 3) and only transformed this chlorophenol to an oxidized product of 6-CHQ, as previously reported (21). The tcpC mutant complemented with the tcpC gene cloned in pS7C grew on 2,4,6-TCP (Fig. 3). This result also indicates that the tcpY and tcpD genes, located downstream to the tcpC gene, are not affected by a potential polar effect of the tcpC gene inactivation or are not essential for growth on 2,4,6-TCP. Both, tcpY and tcpD mutants were able to degrade and to grow on 2,4,6-TCP. The tcpY mutant was able to grow at the same concentrations of 2,4,6-TCP as the wild-type strain, and no differences in the removal rate were detected (data not shown). The tcpY gene does not present homology to any known gene, and its product contains the cluster of orthologous groups (COG) 4313. This COG has been found in proteins from different bacterial strains, but mainly β- and γ-proteobacteria. The function of this COG has been related to regulation of phenolic compounds degradation in Azoarcus sp. strain EbN1 (30) or the meta-pathway of phenol degradation in Acinetobacter calcoaceticus (35), but not experimentally proved. In contrast to the tcpY mutant, the tcpD mutant grew on 2,4,6-TCP but at lower concentrations (up to 1.2 mM) than the wild-type strain (Fig. 3). Complementation of this mutant with the pS7D plasmid restored the wild-type phenotype (Fig. 3).
FIG. 3.
Growth yields obtained after growing C. necator JMP134 and some of its derivatives on different 2,4,6-TCP concentrations: C. necator JMP134 (X), JMP134 tcpR (▪), JMP134 tcpR (pPRTCP) (□), JMP134 tcpC (▴), JMP134 tcpC (pS7C) constitutively expressing tcpC (▵) JMP134 tcpD (•), and JMP134 tcpD (pS7D) constitutively expressing the tcpD gene (○). Values shown are means of three or four independent experiments, with standard deviations of less than 5%. An OD600 of 1.0 corresponds to about 5 × 108 CFU per ml.
tfdFI and ReutB4129 genes, two other maleylacetate reductase genes carried in C. necator JMP134, contribute to degradation of higher levels of 2,4,6-TCP.
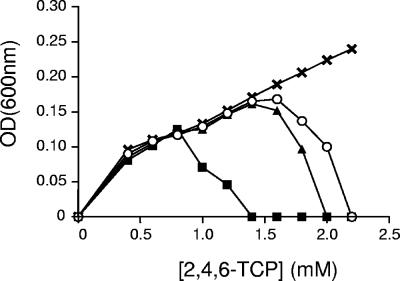
The catabolic phenotype of the tcpD mutant suggests that other MAR-encoding genes in C. necator may provide such function. In addition to the tcpD gene, which has 63% amino acid identity to the MAR-encoding gene macA from C. necator 335, strain JMP134 harbors five other ORFs encoding MAR in its genome. The tfdFI and tfdFII genes located in the catabolic plasmid pJP4 are involved in the turnover of chlorocatechols (28). Three other ORFs, ReutB4129, ReutB4694, and ReutC5982, are scattered on the chromosomes of C. necator JMP134 (unpublished data), showing an amino acid identity of 69% to the pnpD gene of Ralstonia sp. strain SJ98, 62% identity to the tftE gene of B. cepacia AC1100, and 29% identity to the tfdFI gene of C. necator JMP134(pJP4), respectively. Several microorganisms have been reported to have two or more functional MAR activities (17, 31). Interestingly, one of these cases is observed in the polychlorophenol-degrading bacterium Sphingobium chlorophenolicum (4). The corresponding mutants in C. necator JMP134 were obtained by insertional inactivation and were evaluated for growth on 2,4,6-TCP. All mutant strains were able to grow on 2,4,6-TCP; tfdFII, ReutB4694, and ReutC5982 mutants did not show any difference with respect to the wild-type strain (data not shown). In contrast, at higher concentrations of 2,4,6-TCP, the ReutB4129 and tfdFI mutants reached lower yields than the wild-type strain (Fig. 4). The RT-PCR analysis of the RNA purified from 2,4,6-TCP-grown C. necator JMP134 cells showed that only tfdFI, tcpD, and ReutB4129 sequences were induced (data not shown). This indicates that these genes can contribute to the MAR function during growth on 2,4,6-TCP. The role of the tfdFI gene in degradation of 2,4,6-TCP is further supported by the lower growth yields in 2,4,6-TCP observed in strain JMP222 (5), a pJP4-cured derivative of strain JMP134 lacking the two plasmid-encoded MAR (26).
FIG. 4.
Involvement of MARs in 2,4,6-TCP degradation. (A) Growth yields of C. necator JMP134 derivatives at different concentrations of 2,4,6-TCP. Strain JMP134 and mutant strains (JMP134 tfdFII, JMP134 ReutB4694, and JMP134 ReutC5982) were without difference with respect to the wild type (X), JMP134 tfdFI (▴), JMP134 tcpD (▪), and JMP134 ReutB4129 (○). Values shown are means of three or four independent experiments, with standard deviations of less than 5%. An OD600 of 1.0 corresponds to about 5 × 108 CFU ml−1.
tcpR gene encodes a putative LysR-type transcriptional activator that controls tcp gene expression using 2,4,6-TCP as an inducer.
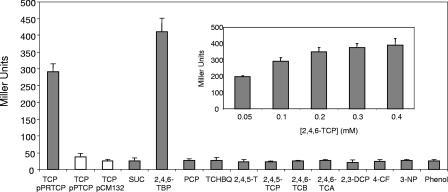
The tcpR mutant did not grow on 2,4,6-TCP at any concentration tested (Fig. 3), and resting cells of the tcpR mutant were also unable to transform 2,4,6-TCP. Furthermore, complete lack of transcription of any of the tcpXABCYD genes in the tcpR mutant was determined by RT-PCR (data not shown). The tcpR mutant harboring the tcpR gene cloned in pPRTCP grew on 2,4,6-TCP, supporting the role of the TcpR protein in expression of the tcp genes (Fig. 3). To look for putative inducers required by the TcpR protein, the levels of expression of the tcpR-PtcpX-tcpX′-lacZ fusion in cells exposed to several chlorophenols or related compounds were determined. Cells of the tcpR (pPRTCP) mutant were grown until an OD600 of 0.5, exposed to a 0.1 mM concentration of each compound for 3 h, and assayed for β-galactosidase expression. A significant induction of the tcpX expression was only observed with 2,4,6-TCP and 2,4,6-tribromophenol (Fig. 5). 2,4,6-TCP was able to promote tcpX expression in a range of concentrations (Fig. 5). The levels of expression of the PtcpX-tcpX′-lacZ construct in the pPTCP plasmid into C. necator JMP134 tcpR were not significant (Fig. 5). 2,4,5-TCP and pentachlorophenol, two compounds that produce catabolic intermediates similar to those produced in the degradation of 2,4,6-TCP, did not act as inducers for the tcp system. Curiously, 2,4,6-tribromophenol is a better inducer of the tcp genes than 2,4,6-TCP. It is possible to speculate that the tcp genes would be recruited initially as a pathway for degradation of bromophenols, which are widespread in soils and oceans (34). It is also possible that bromophenols simply bind to promoting elements more readily than chlorophenols.
FIG. 5.
β-Galactosidase activities from tcpR-tcpX′-lacZ fusions with different aromatic compounds. C. necator JMP134 tcpR (pPRTCP) was grown until the OD600 was 0.5 and exposed to 0.1 mM aromatic compounds for 3 h. Abbreviations: SUC, succinate; 2,4,6-TBP, 2,4,6-tribromophenol; PCP, pentachlorophenol; TCHBQ, tetrachlorohydroxibenzoquinone; 2,4,5-T, 2,4,5-trichlorophenoxyacetic acid; 2,4,5-TCP, 2,4,5-trichlorophenol; 2,4,6-TCB, 2,4,6-trichlorobenzoate; 2,4,6-TCA, 2,4,6-trichloroanisole; 2,3-DCP, 2,3-dichlorophenol; 4-CP, 4-chlorophenol; and 3-NP, 3-nitrophenol. The insert shows activity of the tcpR-tcpX′-lacZ fusion at different concentrations of 2,4,6-TCP. The values are averages for three independent tests conducted in duplicate.
tcpRXABCYD genes constitute a catabolic operon in C. necator.
Based on sequence analysis (23), it has been previously proposed that the tcp genes in C. necator JMP134 are expressed as a single operon. To test if the six ORFs located divergently from tcpR were transcribed in only one transcript, an RT-PCR analysis of RNA purified from strain JMP134 cells grown on 2,4,6-TCP was conducted, using primer pairs from overlapping tcp gene sequences (Fig. 1B). The expected sizes and DNA sequences for all of the PCR products were obtained with these primer pairs (data not shown), indicating that tcp genes are transcribed as one transcript. Since TcpA requires FAD for catalysis, it is worth noting that ReutA1582, encoding a putative FAD synthetase (amino acid identity of 39% to SCO5711 of Streptomyces coelicolor A3) (3), is located downstream of the tcpR gene (Fig. 1B). Use of the overlapping RT-PCR primer-pair approach showed that this ORF is not cotranscribed with tcpR. Consistently, the ReutA1582 mutant grew on 2,4,6-TCP like the wild-type strain (not shown). In addition, no PCR product was obtained with primer pairs 11 and 12 (Fig. 1B), targeting ReutA1590 putatively encoding a formiminoglutamase function (amino acid identity of 66% with Rmet_5045 of Ralstonia metallidurans CH34), further supporting that the tcpD gene is the last gene of the operon.
To determine the transcriptional initiation site of the tcp operon, putatively located in the tcpR-tcpX intergenic region, 5′ RACE PCR experiments were carried out with RNA extracted from cells of C. necator JMP134 grown on 2,4,6-TCP. The nucleotide sequence of the 5′ RACE PCR product showed that transcription starts at the adenine located 103 bases upstream of the tcpX gene translational start site (Fig. 1C). A putative −10 sequence, TGTAGT, separated by 16 bp from a putative −35 sequence, TTAGCA, is located upstream of this transcriptional start site (Fig. 1C). These sequences are good candidates for the tcp gene promoter, since they resemble the −10 and −35 promoter consensus sequences, TATAAT and TTGACA, respectively (7). More than 100 bp between the +1 site and the ATG site are also found for the tftC gene in B. cepacia AC1100 (13). However, the −10 and −35 consensus sequences do not show significant similarities.
The organization of the tcp genes has similarities to and differences from those of other gene clusters involved in chlorophenol degradation. The tcpXA genes are homologous to and are located in the same order as the corresponding tftCD genes of the 2,4,5-TCP pathway encoded in B. cepacia AC1100 (10). However, these genes in strain AC1100 are in two identical copies and in different replicons (13). Furthermore, the tftEFGH genes are located in an unrelated genetic context compared with the other tft functions (13, 38). Ralstonia pickettii DTP0602 also harbors homologues to the tcp genes. In this case, the hadABC genes have the same genetic organization of the corresponding tcpABC genes (15). Unfortunately, no information on the hadABC gene flanking sequences is available. In contrast, the 4-chlorophenol pathway in Arthrobacter chlorophenolicus (25), encoded by genes localized in two clusters [cphF(I)C(I)SRBA(I)X and cphC(II)F(II)A(II)], and the PCP pathway in S. chlorophenolicum (4), encoded by the transcriptional units pcpE, pcpMA, pcpC, and pcpBDR, have an organization completely different from that of tcp genes. These observations suggest that the genetic abilities to degrade chlorophenols in bacteria have different evolutionary origins.
Acknowledgments
This work was financed by FONDECYT 1030493 and Millennium Nucleus Microbial Ecology and Environmental Microbiology and Biotechnology grant P/04-007-F. M.A.S. is a CONICYT and DIPUC Ph.D. fellow.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology, 2nd ed. Greene Publishing Associates, New York, NY.
- 2.Bohuslavek, J., J. W. Payne, Y. Liu, H. Bolton, Jr., and L. Xun. 2001. Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1. Appl. Environ. Microbiol. 67:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briglia, M., F. A. Rainey, E. Stackebrandt, G. Schraa, and M. S. Salkinoja-Salonen. 1996. Rhodococcus percolatus sp. nov., a bacterium degrading 2,4,6-trichlorophenol. Int. J. Syst. Bacteriol. 46:23-30. [DOI] [PubMed] [Google Scholar]
- 4.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.deHaseth, P. L., M. L. Zupancic, and M. T. Record, Jr. 1998. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 180:3019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 10.Gisi, M. R., and L. Xun. 2003. Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:flavin adenine dinucleotide oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Bacteriol. 185:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatta, T., O. Nakano, N. Imai, N. Takizawa, and H. Kiyohara. 1999. Cloning and sequence analysis of hydroxyquinol 1,2-dioxygenase gene in 2,4,6-trichlorophenol-degrading Ralstonia pickettii DTP0602 and characterization of its product. J. Biosci. Bioeng. 87:267-272. [DOI] [PubMed] [Google Scholar]
- 12.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hübner, A., C. E. Danganan, L. Xun, A. M. Chakrabarty, and W. Hendrickson. 1998. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and location of the tft operons on multiple replicons. Appl. Environ. Microbiol. 64:2086-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan, M., M. A. Sánchez, L. Padilla, R. Céspedes, M. Osses, and B. González. 2002. Kraft mill residues effects on Monterey pine growth and soil microbial activity. J. Environ. Qual. 31:1004-1009. [DOI] [PubMed] [Google Scholar]
- 15.Kiyohara, H., T. Hatta, Y. Ogawa, T. Kakuda, H. Yokoyama, and N. Takizawa. 1992. Isolation of Pseudomonas pickettii strains that degrade 2,4,6-trichlorophenol and their dechlorination of chlorophenols. Appl. Environ. Microbiol. 58:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kröckel, L., and D. D. Focht. 1987. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl. Environ. Microbiol. 53:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, C. M., J. H. J. Leveau, A. J. B. Zehnder, and J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182:4165-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latus, M., H.-J. Seitz, J. Eberspächer, and F. Lingens. 1995. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 61:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, D.-Y., J. Eberspächer, B. Wagner, J. Kuntzer, and F. Lingens. 1991. Degradation of 2,4,6-trichlorophenol by Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 57:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie, T. M., C. M. Webster, and L. Xun. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J. Bacteriol. 184:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marx, C. J., and M. E. Lidström. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 23.Matus, V., M. A. Sánchez, M. Martínez, and B. González. 2003. Efficient degradation of 2,4,6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 69:7108-7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Nordin, K., M. Unell, and J. K. Jansson. 2005. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71:6538-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padilla, L., V. Matus, P. Zenteno, and B. González. 2000. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J. Basic Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Pantoja, D., L. Guzmán, M. Manzano, D. H. Pieper, and B. González. 2000. Role of tfdCIDIEIFI and tfDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Pantoja, D., T. Ledger, D. H. Pieper, and B. González. 2003. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134(pJP4) in 3-chlorobenzoic acid. J. Bacteriol. 185:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieper, D. H., W. Reineke, K. H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 30.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 31.Seibert, V., E. M. Kourbatova, L. A. Golovleva, and M. Schlömann. 1998. Characterization of the maleylacetate reductase MacA of Rhodococcus opacus 1CP and evidence for the presence of an isofunctional enzyme. J. Bacteriol. 180:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sittig, M. 1981. Handbook of toxic and hazardous chemicals. Noyes Publications, Park Ridge, NJ.
- 33.Wieser, M., B. Wagner, J. Eberspächer, and F. Lingens. 1997. Purification and characterization of 2,4,6-trichlorophenol-4-monooxygenase, a dehalogenating enzyme from Azotobacter sp. strain GP1. J. Bacteriol. 179:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 2005. 2,4,6-Tribromophenol and other simple brominated phenols. Concise international chemical assessment document 66. World Health Organization, Geneva, Switzerland.
- 35.Xu, Y., M. Chen, W. Zhang, and M. Lin. 2003. Genetic organization of genes encoding phenol hydroxylase, benzoate 1,2-dioxygenase alpha subunit and its regulatory proteins in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol. 46:235-240. [DOI] [PubMed] [Google Scholar]
- 36.Xu, Y., M. W. Mortimer, T. S. Fisher, M. L. Kahn, F. J. Brockman, and L. Xun. 1997. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J. Bacteriol. 179:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xun, L., and C. M. Webster. 2004. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Biol. Chem. 279:6696-6700. [DOI] [PubMed] [Google Scholar]
- 38.Zaborina, O., D. L. Daubaras, A. Zago, L. Xun, K. Saido, T. Klem, D. Nikolic, and A. M. Chakrabarty. 1998. Novel pathway for conversion of chlorohydroxyquinol to maleylacetate in Burkholderia cepacia AC1100. J. Bacteriol. 180:4667-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaborina, O., M. Latus, J. Eberspächer, L. A. Golovleva, and F. Lingens. 1995. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J. Bacteriol. 177:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]