Abstract
V134, a marine isolate of the Vibrio genus, was found to produce a new beta-agarase of the GH16 family. The relevant agarase gene agaV was cloned from V134 and conditionally expressed in Escherichia coli. Enzyme activity analysis revealed that the optimum temperature and pH for the purified recombinant agarase were around 40°C and 7.0. AgaV was demonstrated to be useful in two aspects: first, as an agarolytic enzyme, the purified recombinant AgaV could be employed in the recovery of DNA from agarose gels; second, as a secretion protein, AgaV was explored at the genetic level and used as a reporter in the construction of a secretion signal trap which proved to be a simple and efficient molecular tool for the selection of genes encoding secretion proteins from both gram-positive and gram-negative bacteria.
Agarose is a type of polysaccharide produced by some species of red-purple marine algae and functions as a component of the cell wall. Agar has been extensively used as a common gelling substance in microbiological culturing media and as an ingredient stabilizer in the food industry. In addition to these classical applications, recent research has discovered various new biological and therapeutic properties in many agar derivatives and hence the potential for their application in the medicinal and pharmaceutical industries (32, 33). Chemically, agarose is made up of subunits of galactose that form a polymer through alternating 1,3-linked β-d-galactopyranose and 1,4-linked 3,6-anhydro-α-l-galactopyranose; these linkages can be specifically cleaved by certain agarolytic enzymes known as agarases, which are a group of glycoside hydrolases (GH) that catalyze the degradation of agarose polysaccharide into neoagarooligosaccharides. Based on the mechanisms of their actions, agarases are classified as alpha-agarases, which cleave the α-1,3-galactosidic linkages in agarose, and beta-agarases, which cleave the β-1,4 linkages. On the basis of primary sequences, a number of families have been set up under the category of GH, and except perhaps in one case (3), families GH16, GH50, and GH86 encompass all the beta-agarases that have been characterized at the sequence level. Of these three families of beta-agarases, GH16 is the largest and most heterogeneous group, with members differing from one another in substrate specificity, which has served as the basis for the redivision of the GH16 beta-agarases into several subfamilies (1, 7, 8).
Agarases are the natural products of certain agar-degrading organisms found mostly in marine habitats, which is consistent with the fact that agar, being a product of marine algae, is available to and utilized by some marine organisms as a convenient carbon and energy source. Except in a few cases, most of these agarolytic organisms are bacteria. To date, agarase-producing bacteria of many different genera have been found, including species of Alteromonas (14, 16, 21, 31), Asticcacaulis (9), Bacillus (13, 28), Pseudomonas (6, 11), Pseudoalteromonas (24), Streptomyces (12), and Vibrio (27). However, despite the discovery of a large and increasing number of agarases, few of these enzymes have been successfully applied in industry or scientific research.
In this report, we describe the isolation and identification of an agar-digesting bacterium from marine environments, the cloning and expression of the agarase gene in Escherichia coli, and the biochemical analysis of the purified recombinant agarase. We also explored the secretion and enzymatic features of this agarase and constructed an efficient molecular tool for the trapping of secretion signal sequences from both gram-negative and gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains DH5α and BL21(DE3) were grown on Luria-Bertani (LB) medium (23) at 37°C with appropriate antibiotics. Vibrio sp. strain V134 was cultured at 28°C in LB medium supplemented with 2.5% NaCl. Vibrio harveyi strain T4 and Streptococcus iniae strain G26 were cultured at 28°C in TSAYE medium (peptone, 1.7%; polypeptone, 0.3%; yeast extract, 0.6%; NaCl, 1.5%; K2HPO4, 0.327%; glucose, 0.25%).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid(s) | Relevant characteristic(s)a | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Aps Kns; host strain for general cloning | Clontech (United States) |
| BL21(DE3) | Aps Kns; host strain for expression of recombinant agaV | Novagen (United States) |
| Vibrio spp. | ||
| V134 | Marine isolate, carries agaV | This study |
| T4 | Fish pathogen | This study |
| S. iniae | ||
| G26 | Fish pathogen | This study |
| Plasmids | ||
| pACYC177 | Apr Knr; cloning vector | New England Biolabs (China) |
| pBR322 | Apr; cloning vector | TaKaRa (China) |
| pBK | Knr; pBR322 derivative | This study |
| pBS-T | Apr; TA cloning vector | Tiangen, China |
| pBU | Knr; pBR322 derivative, carries signal sequence-lacking agaV | This study |
| pBUS series | pBU derivatives, carry secretion signal sequences derived from T4 | This study |
| pBUG series | pBU derivatives, carry secretion signal sequences derived from G26 | This study |
| pET25b | Apr; general expression vector | Novagen (United States) |
| pET28a | Knr; general expression vector | Novagen (United States) |
| pET258 | Knr; pET28 derivative | This study |
| pETUA | pET258 carrying agaV structural gene | This study |
| pUALS | Apr; carries signal sequence-lacking agaV | This study |
| pUA series | pBR322 derivatives, carry agaV | This study |
Ap, ampicillin; Kn, kanamycin.
Plasmids pET258 and pETUA were generated as follows. pET25b was digested with BglII-NdeI, and the 104-bp DNA fragment was ligated to the compatible sites of pET28a, yielding plasmid pET258. By using V134 genomic DNA as the template, the agaV structural gene was PCR amplified with primers UAF3 and UAR3 (Table 2), and the PCR products were ligated as an NdeI-XhoI fragment to the 5,126-bp DNA fragment derived from pET258 digested with the same enzymes, resulting in plasmid pETUA.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| KnF2 | ATGGATCCACGGTTGATGAGAGC(BamHI) |
| KnR3 | GCCGTCGACCAATTAACCAATTC(HincII) |
| UAF5 | ATGGATCCGAAGACTGGCGAGAAATC(BamHI) |
| UAR3 | CACTCGAGTTGTATAAATTTCAATCGC(XhoI) |
| UAF3 | GAGCATATGAAATCCATAATTAAAAC(NdeI) |
| ApF2 | CAACAGCTGACGAAAGGGCCTCGT |
| ApR1 | TACCAGCTGACAGTTACCAATGC |
| 8F | AGAGTTTGATCCTGGCTCAG |
| 1492R | GGTTACCTTGTTACGACTT |
Underlined nucleotides are restriction sites of the enzymes indicated in parentheses at the ends of the sequences.
The construction of plasmids pBK, pUALS, and pBU was as follows. The kanamycin resistance (Knr) gene was obtained by PCR with primers KnF2 and KnR3 (Table 2) by using plasmid pACYC177 as the template. The PCR products were digested with BamHI and HincII and ligated to the 3.47-kb DNA fragment of pBR322 digested with BamHI and ScaI, resulting in plasmid pBK. The DNA fragment encoding the mature AgaV protein was PCR amplified from V134 genomic DNA by using primers UAF5 and UAR3 (Table 2), and the PCR products were ligated into the TA cloning vector pBS-T, generating plasmid pUALS. The plasmid pBK was digested with BamHI-BsaBI, and the 3.2-kb DNA fragment was ligated to the 1.3-kb DNA fragment derived from pUALS digested with BamHI-SmaI, yielding plasmid pBU.
DNA techniques.
Plasmid preparation, the extraction of DNA fragments from agarose gels, and the purification of PCR products were performed using the respective kits from Omega Bio-Tek (GA) according to the manufacturer's instructions. PCR amplifications were carried out in a Biometra Tpersonal thermocycler. When the TA cloning strategy was employed, the PCR products were directly ligated to the TA cloning plasmid pBS-T (Tiangen, Beijing, China). Restriction endonucleases, alkaline phosphatase, T4 polynucleotide kinase, and T4 DNA ligases were purchased from New England Biolabs and used in accordance with the manufacturer's specifications. Genome walking was performed with the BD GenomeWalker universal kit (Clontech). The 16S rRNA gene of V134 was PCR amplified with primers 8F and 1492R according to the method of Lane et al. (15).
Chemicals.
Unless otherwise stated, all chemicals used in this study were purchased from Sangon (Shanghai, People's Republic of China).
Cloning of the beta-agarase gene.
The chromosomal DNA of V134 was extracted by the method described by Syn and Swarup (29). The genomic DNA was partially digested with Sau3A1 and separated on a 0.8% agarose gel, and the fragments of between 4 and 8 kb were recovered, purified, and ligated into BamHI-linearized pBR322. E. coli DH5α was transformed with the ligation mixture, and the transformants were selected on LB plates supplemented with 100 μg of ampicillin/ml. Colonies with craters formed around them were picked and further tested for agarolytic activity by pouring I2 vapor onto the plates and examining for the formation of clear zones around the colonies. I2 vapor was generated by placing a few grains of iodine (purchased from Sangon, Shanghai, People's Republic of China) onto the inner side of the lid of a petri dish and heating over a flame or in an oven until the iodine sublimated into vapor.
Expression and purification of the recombinant agarase.
E. coli BL21(DE3) containing the plasmid pETUA was grown to mid-log phase, and the expression of the recombinant agaV was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1uM. After growth at 10°C for an additional 4 h, the cells were harvested by centrifugation and the His-tagged agarase was purified with nickel-nitrilotriacetic acid agarose under native conditions according to the recommendations of the manufacturer (QIAGEN, Valencia, CA).
Enzyme assay.
The enzyme activity of the purified recombinant AgaV was assayed under the standard conditions as follows: in a small glass test tube, 10 μl of diluted enzyme solution was mixed with 390 μl of 50 mM sodium phosphate buffer (pH 7.0) containing 0.2% low-melting-point agarose. After incubation at 40°C for 30 min, the sample was mixed with 400 μl of dinitrosalicylic acid reagent solution (6.5 g of dinitrosalicylic acid, 325 ml of 2 M NaOH, and 45 ml of glycerol in 1,000 ml) and heated in boiling water for 5 to 10 min until the color developed. The reaction was stopped by cooling the test tube in a cold-water bath. The absorbance at 540 nm was then recorded. The amounts of reducing sugars generated were determined using d-galactose as a standard.
TLC.
Thin-layer chromatography (TLC) analysis of the agarose hydrolysis products of AgaV was performed as described by Ohta et al. (19). The hydrolysis reactions were carried out at 40°C in 50 mM sodium phosphate buffer (pH 7.0) containing the purified agarase and 0.25% substrate. Samples were taken at different time intervals and, together with the neoagarooligosaccharide standards (kindly provided by W. Yu of Ocean University of China and described in reference 17), applied to Silica Gel 60 TLC plates (Merck, Darmstadt, Germany), and the plates were developed twice with n-butanol-acetic acid-water (2:1:1, vol/vol/vol). The oligosaccharides were detected by rapidly immersing the plates in 10% H2SO4 (vol/vol, in ethanol), followed by heating the plates on top of a burning oven until the spots became visible.
Effects of temperature, pH, and metal ions on the activity of the recombinant beta-agarase.
The effects of temperature and metal ions on the activity of the purified agarase were determined under the standard assay conditions. The effect of pH was determined by the methods of Suzuki et al. (28) and Wang et al. (31) in which a combination of three different buffering systems was used: 50 mM citric acid-sodium phosphate (pH 3 to 5.3), 50 mM KH2PO4-NaOH buffer (pH 5 to 9), and 50 mM glycine-NaOH (pH 8.8 to 11).
Screening for prokaryotic genes encoding secretion proteins.
The genomic DNA of strains T4 and G26 was prepared by the method of Syn and Swarup (29) with the following modification: for G26, the final concentration of lysozyme was increased to 10 mg/ml and, after incubation at 37°C for 0.5 to 1 h, a 0.12 volume of 10% sodium dodecyl sulfate was added to the sample mixture, followed by incubation at 37°C for 10 min. Genomic DNA of T4 and G26 was partially digested with Sau3A1 and ligated into BamHI-linearized pBU, and E. coli DH5α was transformed with the resulting plasmid. After 24 to 48 h of growth at 37°C on LB plates, transformants were examined for the pit-forming phenotype.
Database search and analysis of nucleotide and amino acid sequences.
The searches for nucleotide and amino sequence similarities and conserved domains were conducted with the BLAST programs from the National Center for Biotechnology Information (NCBI). Signal peptide searches and predictions were performed with SignalP 3.0 and LipoP 1.0. Subcellular localization predictions were performed with the CELLO II subcellular localization predictor (34) and PSORTb version 2.0.4.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA genes of V134, agaV, bus1, bus3, bus4, bus5, bug1, bug2, bug3, and bug4 have been deposited in the GenBank database under accession numbers EF100710, EF100711, EF157302, EF157305, EF157303, EF432376, EF166068, EF432378, EF157304, and EF432377, respectively.
RESULTS
Screening and isolation of agarolytic bacteria.
Seawater collected from a fish farm in Shandong Province, China, was concentrated 10 to 20 times by filtration through a 0.45-μm-pore-size filter membrane and plated onto ZoBell 2216E medium supplemented with 1.2% agar. After incubation at 25°C for 2 to 3 days, colonies were examined for the agarolytic phenotype. Of the ∼600 colonies that appeared, one had a clear hollow formed around it which grew deeper and larger as the time of incubation increased. This isolate was named V134. To confirm its production of agarase, V134 was streaked onto ZoBell 2216E agar plates for the formation of single colonies. The colonies were then subjected to the I2 test, which gave rise to clear zones surrounding the colonies and growth lawns, suggesting that V134 produced an active agarase under the experimental conditions. To examine whether glucose had any effect on its agar-degrading capacity, V134 was plated onto TSAYE medium supplemented with 1% glucose, and no detectable pits developed after prolonged incubation, suggesting that the production of agarase by V134 was subjected to catabolite repression.
Genetic identification of V134.
Since V134 grew well on the Vibrio-selective medium TCBS (thiosulfate-citrate-bile salts-sucrose), it was tentatively placed in the genus Vibrio. To genetically verify this classification and further position the isolate within the Vibrio genus, the 16S rRNA genes of V134 were amplified by PCR and the PCR products were purified and submitted directly for sequencing. Comparison with the known 16S rRNA gene sequence data indicated that the best matches for the16S rRNA gene sequence of V134 were those of Vibrio brasiliensis, Vibrio sp. strain O2, Vibrio sp. strain CAIM 695, and V. halioticoli, which showed, respectively, 97.5, 97.4, 97.3, and 97.2% identity to the sequence of V134. Within the genus Vibrio, V. alginolyticus and V. parahaemolyticus exhibited the lowest level of homology to V134, both having 16S rRNA gene sequences with only 95.5% identity to the sequence of V134. These results indicated that V134 is a member of the Vibrio genus and, considering that its 16S rRNA gene sequence showed only 95.5 to 97.5% identity to those of the known Vibrio species, probably represents a new Vibrio species that is relatively closely related to V. brasiliensis and V. halioticoli and distantly related to V. alginolyticus and V. parahaemolyticus.
Cloning, sequencing, and characterization of the agaV gene.
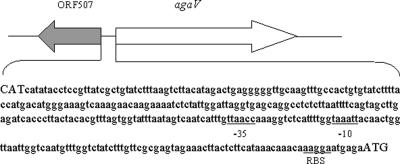
Six clones with apparent agarolytic activity from the V134 genomic DNA library were screened, and the recombinant plasmids in these clones were designated pUA1 to pUA6, respectively. Restriction enzyme analyses revealed that pUA1 harbored the smallest insert, which was approximately 2 kb. The nucleotide sequence of this 2-kb insert was determined and found to contain an open reading frame (ORF) of 1,359 bp (orf1359), preceded by a putative ribosome binding site. The 600-bp DNA sequence immediately upstream of orf1359 was obtained by sequencing the plasmid pUA4 with primers based on the 5′ DNA sequence of orf1359. The combined 2.6-kb DNA sequence possessed two putative operons that overlapped with and diverged from each other: one was that containing orf1359, and the other had a predicted ORF of 507 bp encoding a hypothetical protein of unknown function (Fig. 1). A putative σ70-dependent promoter, with TTAACC as the −35 element and TAAATT as the −10 element, and a ribosome binding site (AAGGAA) were found to be present 111 and 6 bp, respectively, upstream from the translation initiation codon of orf1359.
FIG. 1.
Schematic representation of the agaV region in V134 and the nucleotide sequence between orf507 and agaV. The translation start codons of orf507 and agaV are shown in uppercase letters. The putative −35 and −10 promoter elements and the ribosome binding site (RBS) of agaV are underlined.
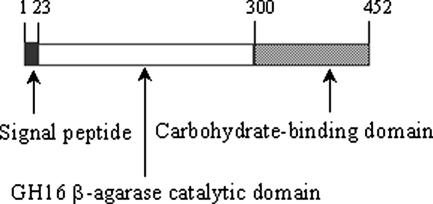
orf1359 encoded a protein of 452 amino acids with an estimated molecular mass of 51.7 kDa and a pI of 4.52. By using the SignalP 3.0 server, a putative signal peptide at the N terminus was identified, with the most likely cleavage site situated between the amino acids Ala and Glu at positions 22 and 23, respectively. Immediately downstream from the signal peptide sequence was a beta-agarase domain corresponding to the GH16 family, which was followed by a ricin-type carbohydrate binding module (Fig. 2), both identified with the NCBI conserved-domain database search program. An amino acid sequence BLAST search indicated that the only matches for the translated product of orf1359 (named AgaV) were the beta-agarase AgaB from Pseudoalteromonas sp. strain CY24 (GenBank accession no. AY150179) and the beta-agarase AgaD from Vibrio sp. strain PO-303 (GenBank accession no. AB221476), which showed 74 and 72% overall identity to AgaV, respectively. The identities between AgaV and these two enzymes were located mainly in the catalytic domains, which, corresponding to amino acids Asp24 to Gly300 in AgaV, were 84% identical between AgaV and AgaB and 83% identical between AgaV and AgaD. Though similar to these two enzymes, AgaV differed significantly from other known agarases in the catalytic domain and more so in other regions and showed only 37 and 33% overall identity to the amino acid sequences of the agarases of Pseudomonas sp. strains ND137 and BK1, respectively, which were, next to AgaB and AgaD, the closest homologues to AgaV that could be found. From these data, we concluded that orf1359 encoded a beta-agarase that belonged to the GH16 group of glycosyl hydrolases. The gene of this agarase was subsequently named agaV.
FIG. 2.
Schematic representation of the domain organization in AgaV. The numbers refer to the positions of the amino acids of the predicted protein.
Expression and purification of the recombinant beta-agarase.
The agaV structural gene was introduced into the Novagen pET expression system and conditionally expressed in the E. coli strain BL21(DE3) as a C-terminally His-tagged recombinant protein. After induction, agarase activity and the recombinant protein were detected in both the culturing supernatant and the pellet, but mostly in the former. To retain its bioactivity, the recombinant protein was purified from the supernatant under native conditions. The purity and homogeneity of the purified protein were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which displayed a single band corresponding in size to the expected recombinant agarase, with an apparent molecular mass approximating the estimated molecular mass of the recombinant AgaV as calculated based on the amino acid sequence (50.35 kDa including the six-His tag).
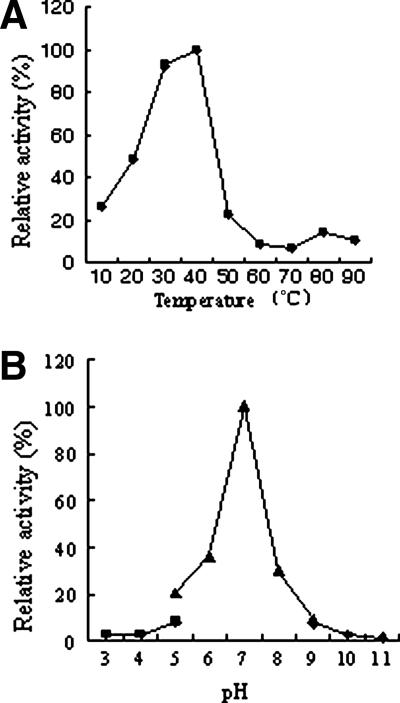
Effects of temperature and pH on the activity of the purified agarase.
Both temperature and pH were found to have a strong impact on the activity of the recombinant agarase. As shown in Fig. 3, within the range of the tested temperatures, the activity of the enzyme increased with the temperature until the latter reached 40°C, at which the enzyme exhibited the maximum activity. Once the temperature rose above 40°C, the activity of the enzyme decreased precipitously and was barely detectable at 50°C. To determine the effect of pH on the activity of the recombinant enzyme, a combination of three different buffering systems was used, and the results indicated that the activity of the purified agarase was highly pH dependent and measurable only within a narrow window centered around pH 7.0, which was the apparent optimum pH; deviations from this optimum led to drastic diminutions in enzyme activity.
FIG. 3.
Effects of temperature (A) and pH (B) on the activity of the purified recombinant AgaV. The effect of temperature on the enzyme activity was determined under standard assay conditions at temperatures ranging between 10 and 90°C. The effect of pH on the enzyme activity was determined under standard assay conditions but in three different buffers: 50 mM citric acid-sodium phosphate (▪), 50 mM KH2PO4-NaOH (▴), and 50 mM glycine-NaOH (⧫).
Effects of cations on the activity of the purified agarase.
To investigate the effects of various metal ions on the activity of the purified agarase, standard enzyme assays were performed at 40°C in 50 mM sodium phosphate buffer (pH 7.0) supplemented with the tested cation at three different concentrations (1, 10, and 100 mM). The results (Table 3) showed that in contrast to several of the divalent ions tested (Mn2+, Co2+, and Fe2+), which exhibited an apparent negative effect on the enzyme activity, Na+, K+, and to certain extent, Mg2+ had a clear positive effect on the enzyme activity. The effect of Ca2+, however, was undetectable at concentrations lower than 10 mM and negative at higher concentrations. To examine the composite effects of different cations, the enzyme activities were measured in the presence of different combinations of the three cations (Na+, K+, and Mg2+) that had demonstrated a clear stimulatory effect on the enzyme activity. The results showed that, rather than having a synergistic or redoubling effect, the presence of K+ and Mg2+ appeared to reduce the stimulatory effect of Na+.
TABLE 3.
Effects of different cations on the activity of the purified recombinant AgaVa
| Cation | Relative activity (%) of AgaV at cation concn of:
|
||
|---|---|---|---|
| 1 mM | 10 mM | 100 mM | |
| Na+ | 170 | 206 | 340 |
| K+ | 151 | 205 | 252 |
| Ca2+ | 108 | 107 | 1.2 |
| Co2+ | 104 | 0 | 0 |
| Fe2+ | 62 | 0 | 0 |
| Mg2+ | 241 | 137 | 45 |
| Mn2+ | 0.64 | 0 | 0 |
The activities are expressed as percentages of the enzyme activity measured in 50 mM sodium phosphate buffer (pH 7.0) alone.
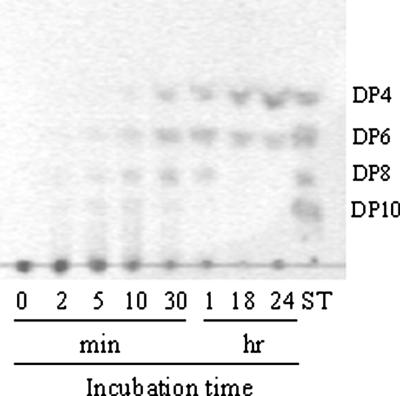
Analysis of the hydrolysis pattern and products of the purified AgaV.
To find out whether AgaV was an endo- or exo-glycoside hydrolase, as well as to determine its end hydrolysis products, a time course hydrolysis analysis was carried out with the purified recombinant enzyme. As shown in Fig. 4, the agarose hydrolysis products detectable in the first 10 min of incubation were of various degrees of polymerization (DP), with the DP10 and DP8 products appearing first, followed by DP6 (neoagarohexaose) and DP4 (neoagarotetraose). This hydrolysis pattern indicated that AgaV was an endoagarase. With the increase of the incubation time, the proportions of larger oligosaccharides (DP10 and DP8) gradually decreased while those of smaller products (neoagarohexaose and neoagarotetraose) increased. After 24 to 48 h of incubation, neoagarotetraose and neoagarohexaose became the sole hydrolysis products, with the former being the predominant one. Consistent with these results, when neoagarohexaose and neoagarotetraose were used as substrates for AgaV, no hydrolysis products were observed, suggesting that AgaV could not cleave DP6 and shorter oligomers.
FIG. 4.
TLC analysis of the products of agarose hydrolysis by AgaV. Hydrolysis reactions were conducted at 40°C in 50 mM sodium phosphate buffer (pH 7.0) containing 0.25% substrate. Samples were taken at the indicated incubation times and analyzed by TLC as described in Materials and Methods. ST, neoagarooligosaccharide standards.
Application of the recombinant agarase in extracting DNA from agarose gels.
One of the most important applications of agarase is in the recovery of DNA or other biological molecules embedded in agarose gels. To test the applicability of the purified AgaV in this respect, a 1,068-bp DNA fragment was obtained by PCR from the plasmid pBR322 with primers ApF2 and ApR1 (Table 2). PCR products were separated on both 1% regular and 1% low-melting-point agarose gels. After electrophoresis, the 1,068-bp DNA fragment was excised from each gel, mixed with 200 μl of 50 mM sodium phosphate buffer (pH 7.0) containing the purified agarase, and incubated at 40°C for 2 h. After incubation, the undegraded agarose was removed by centrifugation and the DNA was precipitated from the solution by adding a 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 volumes of ethanol, followed by incubation at −20°C for 1 h. The DNA was recovered by centrifugation at 11,000 × g for 5 min, and the pellet was dissolved in H2O and subjected to electrophoresis. The results indicated that the recombinant AgaV could be used for the extraction of DNA from both regular and low-melting-point agarose gels, although the recovery from the former (∼35%) was almost threefold lower than that from the latter (∼90%). The purity and integrity of the recovered DNA were examined subsequently by ligation and transformation tests, which showed that the quality of the DNA extracted with the purified AgaV was comparable to that of DNA extracted with the commercial kit (Omega Bio-Tek, GA) (data not shown).
Application of AgaV in the screening of secretion signal sequences from bacteria.
Given that AgaV could be efficiently processed and transported in E. coli and that the secretion of the enzyme was easily detectable, we thought it feasible to use the signal sequence-lacking agaV as a reporter for the screening of DNA sequences encoding secretion signals. To explore this idea, we constructed a secretion signal selection trap in the form of plasmid pBU, which was basically a pBR322 derivative carrying a copy of the agaV structural gene devoid of the first 66 nucleotides that constitute the putative secretion signal. Heterologous DNA inserts could be introduced into pBU at the BamHI site located immediately upstream of the agaV gene. The selection rationale was as follows: if the introduced DNA fragment harbored a partial transcriptional unit that was truncated at the coding region but retained all the promoter elements and hence the capacity to be transcribed and translated and if the truncated heterologous ORF formed an in-frame fusion with agaV, then agaV would be transcribed as part of a chimeric operon and subsequently translated into the C-terminal part of a fusion protein; if the introduced ORF happened to encode a secretion protein with characteristics that are recognizable by some E. coli secretion apparatuses, then with certain chances, depending on the biological nature of the carrier protein and the precise position where the fusion of the two proteins took place, AgaV could be transported as a passenger protein out of the cytoplasm and into the extracellular space. To test the workability of this scenario, we applied it to the screening of secretion sequences from the genomic DNA libraries of T4, a V. harveyi strain, and G26, an S. iniae strain. Of the agarolytic transformants that appeared, we randomly chose six colonies from the T4 genomic library and five from the G26 genomic library for detailed analysis. The results showed that, although two transformants originating from the T4 genomic library turned out to be duplicates, each of the other nine agarolytic transformants harbored a different recombinant plasmid with a unique DNA insert carrying a truncated gene which, as we had expected, formed an in-frame fusion with the agaV gene. The five truncated genes captured from the G26 genomic library were designated bug1 to bug5, respectively, and the five from the T4 genomic library were designated bus1 to bus5, respectively. The complete coding regions of bus1, bus3, bus4, bus5, bug1, bug3, and bug4 were subsequently obtained by genome walking (25). Amino acid sequence analysis using the SignalP and LipoP tools revealed that a putative signal peptide sequence existed in all but one (BUG2) of the captured carrier proteins (Fig. 5). Though the above-mentioned predictive methods failed to identify a signal peptide in BUG2, this protein was predicted by CELLO II to be extracellular or/and membrane localized. Similarly, the other nine captured proteins, consistent with the identification of a signal peptide in each of them, were predicted to have subcellular locations in the extracellular space, the outer membrane, and for the BUS series from the gram-negative strain T4, the periplasm (Table 4). From these data we concluded that, as we had speculated, AgaV could indeed serve as a reporter for the screening and selection of transcription units encoding secretion proteins from both gram-positive and gram-negative bacteria.
FIG. 5.
Sequences of the N-terminal 50 amino acids of the captured secretion proteins. Predicted signal peptides are underlined.
TABLE 4.
Summary of the secretion proteins captured with pBU
| Source strain | Protein | Signal peptidea | Predicted subcellular location(s)b | Reliability score(s) | Putative function |
|---|---|---|---|---|---|
| G26 | BUG1 | + | Membrane | 3.602 | Unknown |
| BUG2 | − | Extracellular space/membrane | 2.263/1.844 | Unknown | |
| BUG3 | + | Cytoplasm/membrane | 2.239/1.720 | Lipoprotein | |
| BUG4 | + | Extracellular space/membrane | 2.404/2.230 | Oligopeptide permease | |
| BUG5 | + | Extracellular space | 2.408/2.295 | N-Acetylmuramidase | |
| T4 | BUS1 | + | Periplasm | 2.337 | Glucose dehydrogenase-B |
| BUS2 | + | Periplasm | 2.890 | Soluble lytic murein transglycosylase | |
| BUS3 | + | Outer membrane/extracellular space | 1.875/1.772 | Unknown | |
| BUS4 | + | Periplasm | 4.045 | ABC transporter | |
| BUS5 | + | Periplasm | 3.749 | Metal binding protein |
Plus and minus signs indicate the presence (+) or absence (−) of the predicted signal peptide.
Protein subcellular localizations were predicted by using the CELLO II program.
DISCUSSION
In this report we described the cloning, expression, and characterization of a new beta-agarase gene from a marine isolate of the Vibrio genus. To date, at least five different beta-agarases in Vibrio species have been discovered (2, 3, 4, 26, 27). Of these, four have been analyzed at the primary-structure level, resulting in the classification of AgaA and AgaB (from Vibrio sp. strain JT0107) into the GH50 group and AgaD (from Vibrio sp. strain PO-303) into the GH16 family, while AgaC, which is also from Vibrio sp. strain PO-303, is a unique agarase unrelated to the presently established GH families and hence has been proposed to represent a new type of glycoside hydrolase (3). The AgaV presented in this report revealed no significant homology to the above-mentioned Vibrio AgaA, AgaB, and AgaC but was significantly similar to AgaD and the beta-agarase AgaB of the Pseudoalteromonas sp. strain CY24 (17), especially in the catalytic domain (83 to 84% identity). The AgaB from the Pseudoalteromonas sp. strain CY24 (hereafter referred to simply as AgaB, not to be confused with the AgaB from Vibrio sp. strain JT0107) and AgaD, though differing in sources of origin, are highly homologous in both amino acid and DNA sequences (sharing 98% overall identity in each case). In comparing AgaV with these two enzymes, we found that, like AgaB, AgaV was unable to degrade neoagarohexaose and smaller neoagarooligosaccharides, in contrast to AgaD, which can utilize DP6 as a substrate. However, similar to those of AgaD, the end hydrolysis products of AgaV were DP4 and DP6, while those of AgaB were DP8 and DP10 (4, 17).
Like most of the GH16 beta-agarases that have been characterized, which generally have a temperature optimum around 40°C and a preferential pH that is neutral to mildly alkaline, the recombinant AgaV demonstrated maximum activity at 40°C and pH 7.0. As observed with other beta-agarases, we found that NaCl, KCl, and to certain extent, MgCl2 acted positively on the activity of the recombinant AgaV, which was consistent with the fact that V134, the source strain of AgaV, was a marine inhabitant. CaCl2, however, exhibited no measurable effect on the activity of the enzyme at concentrations of up to 10 mM and exerted an apparently inhibitory effect at higher concentrations, in contrast to the observations with the GH86 beta-agarase AgaO and the AgaB of the Pseudoalteromonas sp. reported by Ohta et al. (18) and Ma et al. (17), who found that CaCl2 stabilized the activity of AgaO and enhanced the activity of AgaB.
It has been recognized that agarases have potentially wide applications in many areas of industry and scientific research, such as generating simple neo-oligosaccharides from complex polysaccharides, liberating DNA and other embedded molecules from agarose, and extracting bioactive or medicinal compounds from algae and seaweed, etc. All these applications rely on the natural functionality of purified agarases and require, as a prerequisite, the large-scale production and purification of high-quality enzymes. To our knowledge, no study has been reported concerning the exploration at the genetic level of agarases as secretion proteins. With the understanding that in E. coli the agaV gene could be expressed at relatively high levels without apparent detrimental effects to the host (W. Zhang and L. Sun, unpublished data) and that the extracellular presence of the enzyme could be easily detected, we speculated that agaV might be used as a reporter in the engineering of a molecular trap for the selection of secretion signal sequences from bacteria in general. The notion of a secretion signal trap was first proposed by Tashiro et al. in 1993 (30); since then, similar ideas in various forms have been contrived and tested by other investigators with a variety of different proteins with extracellular and membrane destinations. Of the successful cases, those that are comparable to our study include experiments using the Staphylococcus aureus nuclease and the Bacillus subtilis alpha-amylase (5, 10, 20, 22) as the trapping agents. Similar to some of these researchers who made observations regarding different trapping systems, we found that most of the genes identified with our pBU system encoded putative proteins belonging to the category of the classical type I transmembrane proteins, as marked by a secretion signal typical of that recognized and cleaved by signal peptidase I. This limitation is probably due to the facts that AgaV itself is a type I transmembrane protein, that the general Sec translocon-dependent translocation system is the most conserved, and that secretion proteins that utilize this pathway are abundant in both gram-negative and gram-positive bacteria, which may also account for our observation that the pBU trap worked equally well, or without differentiation, with V. harveyi and S. iniae. The success of the pBU system led us to believe that similar explorations should, in principle, be applicable to agarases other than AgaV, as the design of the system is based not on any features unique to AgaV but on the biochemical and enzymatic characteristics that are common to all agarases.
Since extracellular and membrane-targeted proteins are usually endowed with vital biological functions, such as participating in the sensing and transduction of environmental signals, cell-cell communications, and host-pathogen interactions, etc., these proteins have become targets for the study and development of therapeutic drugs and vaccines. Considering these facts, we believe that pBU will be a useful genetic tool for the selection of secretion proteins from bacteria in general and particularly from those with unresolved genomic sequences, as well as for the functional testing of putative signal peptide sequences. The secretion proteins obtained herein could be used for various purposes, such as identifying possible vaccine candidate proteins encoded by pathogenic bacteria.
Acknowledgments
We thank R. Z. Jiang of Nankai University, People's Republic of China, for useful discussions.
This work was supported by National Natural Science Foundation of China (NSFC) grant 40576071 and 973 Project of China grant 2006CB101807.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Allouch, J., M. Jam, W. Helbert, T. Barbeyron, B. Kloareg, B. Henrissat, and M. Czjzek. 2003. The three-dimensional structures of two beta-agarases. J. Biol. Chem. 278:47171-47180. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, T., T. Araki, and M. Kitamikado. 1990. Purification and characterization of β-agarase from Vibrio sp. AP-2. Eur. J. Biochem. 187:461-465. [DOI] [PubMed] [Google Scholar]
- 3.Dong, J., S. Hashikawa, T. Konishi, Y. Tamaru, and T. Araki. 2006. Cloning of the novel gene encoding β-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl. Environ. Microbiol. 72:6399-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, J., Y. Tamaru, and T. Araki. 2007. Molecular cloning, expression, and characterization of a beta-agarase gene, agaD, from a marine bacterium, Vibrio sp. strain PO-303. Biosci. Biotechnol. Biochem. 71:38-46. [DOI] [PubMed] [Google Scholar]
- 5.Downing, K. J., R. A. McAdam, and V. Mizrahi. 1999. Staphylococcus aureus nuclease is a useful secretion reporter for mycobacteria. Gene 239:293-299. [DOI] [PubMed] [Google Scholar]
- 6.Ha, J. C., G. T. Kim, S. K. Kim, T. K. Oh, J. H. Yu, and I. S. Kong. 1997. β-Agarase from Pseudomonas sp. W7: purification of the recombinant enzyme from Escherichia coli and the effects of salt on its activity. Biotechnol. Appl. Biochem. 26:1-6. [PubMed] [Google Scholar]
- 7.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosoda, A., and M. Sakai. 2006. Isolation of Asticcacaulis sp. SA7, a novel agar-degrading Alphaproteobacterium. Biosci. Biotechnol. Biochem. 70:722-725. [DOI] [PubMed] [Google Scholar]
- 10.Jeong, D. W., Y. C. Choi, J. M. Lee, J. M. Seo, J. H. Kim, J. H. Lee, K. H. Kim, and H. J. Lee. 2004. Screening and characterization of secretion signals from Lactococcus lactis ssp cremoris LM0230. J. Microbiol. Biotechnol. 14:1052-1056. [Google Scholar]
- 11.Kang, N., Y. Choi, Y. Cho, B. Kim, B. Jeon, J. Cha, C. Kim, and Y. Lee. 2003. Cloning, expression and characterization of a β-agarase gene from a marine bacterium, Pseudomonas sp. SK38. Biotechnol. Lett. 25:1165-1170. [DOI] [PubMed] [Google Scholar]
- 12.Kendall, K., and J. Cullum. 1984. Cloning and expression of an extracellular agarase from Streptomyces coelicolor A3(2) in Streptomyces lividans 66. Gene 29:315-321. [DOI] [PubMed] [Google Scholar]
- 13.Kim, B. J., H. J. Kim, S. D. Ha, S. H. Hwang, D. S. Byun, T. H. Lee, and J. Y. Kong. 1999. Purification and characterization of β-agarase from marine bacterium Bacillus cereus ASK202. Biotechnol. Lett. 21:1011-1015. [Google Scholar]
- 14.Kirimura, K., N. Masuda, Y. Iwasaki, H. Nakagawa, R. Kobayashi, and S. Usami. 1999. Purification and characterization of a novel beta-agarase from an alkalophilic bacterium, Alteromonas sp. E-1. J. Biosci. Bioeng. 87:436-441. [DOI] [PubMed] [Google Scholar]
- 15.Lane, D., B. Pace, G. Olsen, D. Stahl, M. Sogin, and N. Pace. 1985. Rapid determination of 16S ribosomal sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leon, O., L. Quintana, G. Peruzzo, and J. Carlos Slebe. 1992. Purification and properties of an extracellular agarase from Alteromonas sp. strain C-1. Appl. Environ. Microbiol. 58:4060-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, C., C. Shi, J. Li, Y. Gu, Y. Ma, Y. Chu, F. Han, Q. Gong, and W. Yu. 2007. Molecular cloning and characterization of a novel β-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J. Biol. Chem. 282:3747-3754. [DOI] [PubMed] [Google Scholar]
- 18.Ohta, Y., Y. Hatada, Y. Nogi, Z. Li, S. Ito, and K. Horikoshi. 2004. Cloning, expression, and characterization of a glycoside hydrolase family 86 beta-agarase from a deep-sea Microbulbifer-like isolate. Appl. Microbiol. Biotechnol. 66:266-275. [DOI] [PubMed] [Google Scholar]
- 19.Ohta, Y., Y. Hatada, Y. Nogi, M. Miyazaki, Z. Li, M. Akita, Y. Hidaka, S. Goda, S. Ito, and K. Horikoshi. 2004. Enzymatic properties and nucleotide and amino acid sequences of a thermostable beta-agarase from the novel marine isolate, JAMB-A94. Biosci. Biotechnol. Biochem. 68:1073-1081. [DOI] [PubMed] [Google Scholar]
- 20.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potin, P., C. Richard, C. Rochas, and B. Kloareg. 1993. Purification and characterization of the α-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur. J. Biochem. 214:599-607. [DOI] [PubMed] [Google Scholar]
- 22.Ravn, P., J. Arnau, S. M. Madsen, A. Vrang, and H. Israelsen. 2000. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 242:347-356. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Schroeder, D. C., M. A. Jaffer, and V. E. Coyne. 2003. Investigation of the role of a β (1-4) agarase produced by Pseudoalteromonas gracilis B9 in eliciting disease symptoms in the red alga Gracilaria gracilis. Microbiology 149:2919-2929. [DOI] [PubMed] [Google Scholar]
- 25.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugano, Y., T. Matsumoto, H. Kodama, and M. Noma. 1993. Cloning and sequencing of agaA, a unique agarase 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl. Environ. Microbiol. 59:3750-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugano, Y., T. Matsumoto, and M. Noma. 1994. Sequence analysis of the agaB gene encoding a new β-agarase from Vibrio sp. strain JT0107. Biochim. Biophys. Acta 1218:105-108. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki, H., Y. Sawai, T. Suzuki, and K. Kawai. 2003. Purification and characterization of an extracellular β-agarase from Bacillus sp. MK03. J. Biosci. Bioeng. 95:328-334. [PubMed] [Google Scholar]
- 29.Syn, C. K., and S. Swarup. 2000. A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal. Biochem. 278:86-90. [DOI] [PubMed] [Google Scholar]
- 30.Tashiro, K., H. Tada, R. Heilker, M. Shirozu, T. Nakano, and T. Honjo. 1993. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science 261:600-603. [DOI] [PubMed] [Google Scholar]
- 31.Wang, J., H. Mou, X. Jiang, and H. Guan. 2005. Characterization of a novel beta-agarase from marine Alteromonas sp. SY37-12 and its degrading products. Appl. Microbiol. Biotechnol. 24:1-7. [DOI] [PubMed] [Google Scholar]
- 32.Wang, J. X., X. L. Jiang, H. J. Mou, and H. S. Guan. 2004. Anti-oxidation of agar oligosaccharides produced by agarase from a marine bacterium. J. Appl. Phycol. 16:333-340. [Google Scholar]
- 33.Weinberger, F., C. Richard, B. Kloareg, Y. Kashman, H. Hoppe, and M. Friedlander. 2001. Structure-activity relationships of oligoagar elicitors towards Gracilaria conferta. J. Phycol. 37:418-426. [Google Scholar]
- 34.Yu, C. S., Y. C. Chen, C. H. Liu, and J. K. Hwang. 2006. Predition of protein subcellur localization. Proteins 64:643-651. [DOI] [PubMed] [Google Scholar]