Abstract
A carbazole-utilizing bacterium was isolated by enrichment from petroleum-contaminated soil. The isolate, designated Sphingomonas sp. strain XLDN2-5, could utilize carbazole (CA) as the sole source of carbon, nitrogen, and energy. Washed cells of strain XLDN2-5 were shown to be capable of degrading dibenzofuran (DBF) and dibenzothiophene (DBT). Examination of metabolites suggested that XLDN2-5 degraded DBF to 2-hydroxy-6-(2-hydroxyphenyl)-6-oxo-2,4-hexadienic acid and subsequently to salicylic acid through the angular dioxygenation pathway. In contrast to DBF, strain XLDN2-5 could transform DBT through the ring cleavage and sulfoxidation pathways. Sphingomonas sp. strain XLDN2-5 could cometabolically degrade DBF and DBT in the growing system using CA as a substrate. After 40 h of incubation, 90% of DBT was transformed, and CA and DBF were completely removed. These results suggested that strain XLDN2-5 might be useful in the bioremediation of environments contaminated by these compounds.
Heterocyclic compounds (nitrogen-, oxygen-, or sulfur-containing aromatic compounds) such as carbazole (CA), dibenzofuran (DBF), and dibenzothiophene (DBT) together with their degradation products have been detected in groundwater, seawater, sediments, and soil sites contaminated with spills of petroleum and wood-preserving wastes (5, 35). These heterocyclic compound mixtures with other polycyclic aromatic hydrocarbons are widespread because of petroleum application in industrial processes, coal gasification, and wood treatment with creosote. Heterocyclic compounds, DBT and its derivatives in particular, can persist for up to 3 years after an oil spill, while more susceptible compounds would have been biodegraded (7). Moreover, because some of these heterocyclic compounds are mutagenic and carcinogenic (11, 25), ecosystems contaminated with these compounds may elicit serious health risks (26). Fundamental research examining the biodegradation of these recalcitrant compounds and the application of bacteria in bioremediation for reducing their concentrations in the environment has been conducted (22).
CA, an N-heterocyclic compound derived from creosote, crude oil, and shale oil, is known to possess toxic and mutagenic activities (25). Various bacteria have been reported to degrade this recalcitrant compound (5, 11, 16, 17, 24, 25, 30). Most of the isolates degrade CA by following a similar pathway in which CA is initially attacked at the angular position by dioxygenation, followed by the spontaneous conversion of the dihydroxylated intermediate to 2′-aminobiphenyl-2,3-diol, and an extradiol dioxygenase then attacks the hydroxylated ring at the meta position to give 2-hydroxy-6-(2-aminophenyl)-6-oxo-2,4-hexadienoic acid. This meta-cleavage product is hydrolyzed to produce anthranilic acid (AN), which is then completely mineralized. DBF, one of the O-heterocyclic compounds, has been used as an insecticide (35). Several studies of the aerobic biodegradation of DBF have considered DBF as a model for chlorinated DBFs and dibenzo-p-dioxin, which are of greater environmental concern (2, 8, 21, 31). The bacterial degradation pathway of DBF is similar to that of CA, which is initiated by the angular dioxygenase to yield a chemically unstable intermediate that is spontaneously rearomatized to 2,2′,3-trihydroxybiphenyl. The subsequent ring cleavage of 2,2′,3-trihydroxybiphenyl is catalyzed by an extradiol dioxygenase yielding 2-hydroxy-6-(2-hydroxyphenyl)-6-oxo-2,4-hexadienoic acid (2′-OH-HOPDA), which is then hydrolyzed to salicylic acid and 2,4-hexadienoic acid. DBT is widely accepted as a model S-heterocyclic compound that is used for biodegradation and petroleum biodesulfurization, and several reports on biodesulfurization have been published (4, 14, 15, 27, 33, 38, 39). The selective removal of sulfur from DBT through the 4S pathway leaves the carbon skeleton intact. But the end product 2-hydroxybiphenyl is also an environmental contaminant (10), so this pathway cannot be used for bioremediation without some modifications (37). In addition, one limit must be mentioned: sulfate ions and other inorganic sulfurs inhibit the expression of the DBT-degrading enzymes (18, 19). This characteristic suggests that this pathway is not suitable for bioremediation in oil-contaminated environments where the presence of sulfur is not expected to be a growth-limiting factor (19). One of the previously described microbial DBT oxidative pathways, the Kodama pathway (12), transformed the molecule to the final product 3-hydroxy-2-formyl-benzothiophene. The intermediates of the Kodama pathway are colored compounds whose occurrence in the culture is an indication of the microbial activation of this specific pathway (12, 13). Bacterial oxidation pathways of these heterocyclic compounds are different, with the angular dioxygenation of CA and DBF, the lateral dioxygenation of DBT in the Kodama pathway, and the S-oxidation of DBT in the 4S pathway. In the natural environment, it is more likely that bacteria would be exposed to more than one organic compound (20). From a bioremediation point of view, it is imperative to obtain pollutant-degrading bacteria that have a wide substrate range and various types of oxidation abilities. But there are few previously reported bacteria which have versatile oxidation activities. Moreover, few data concerning the simultaneous degradation of CA, DBF, and DBT by growing cells have been described.
In this paper, the isolation and characterization of a gram-negative bacterium, Sphingomonas sp. strain XLDN2-5, which could grow on CA as the sole source of carbon and nitrogen, are described. Some products from CA, DBF, and DBT degradation by strain XLDN2-5 were also identified by the investigation of several chemical properties.
MATERIALS AND METHODS
Growth media and cultivation conditions.
Experiments examining the biodegradation of CA were carried out in 500-ml flasks containing 50 ml of mineral salt medium (MSM). This medium had the following composition (per liter of distilled water): 12.0 g K2HPO4, 11.0 g KH2PO4, 0.2 g MgSO4·7H2O, 2.0 g Na2SO4, 2.0 g KCl, and 1.0 ml of trace metals solution, at a pH of 7.0 (17). CA, DBF, and DBT were dissolved in dimethyl sulfoxide (DMSO) (100 mM) and added to the medium at a suitable concentration. Agar plates were prepared by adding 16.0 g of agar per liter of MSM and CA as the sole source of carbon and nitrogen at 0.5 g liter−1. All cultures or cell suspensions were incubated at 30°C on a reciprocal shaker at 180 rpm.
Chemicals.
CA, DBF, DBT, AN, and N,O-bis(trimethylsilyl)trifluoroacetamide were purchased from Sigma-Aldrich. DBT-5,5-dioxide (DBTO2), 2′-hydroxyacetophenone, and salicylic acid were purchased from ACROS Organics. All other commercially available chemicals were of analytical grade.
Isolation and identification of a CA-degrading bacterium.
Soil samples were added to MSM supplemented with CA and incubated aerobically. Every week for 4 weeks, aliquots were transferred to the fresh medium. Pure cultures were obtained by plating the enrichment culture on MSM agar to select colonies that were capable of using CA as the sole growth substrate. One isolate, designated XLDN2-5, was found to use CA as the sole source of carbon, nitrogen, and energy and was subsequently characterized by various taxonomic tests.
Preparation of cells and activity analysis.
For washed-cell experiments, CA-grown cells were harvested by centrifugation (8,000 rpm for 10 min) and washed twice with MSM. The pellet was resuspended in MSM at an optical density at 620 nm (OD620) of 5.0. Each of the test flasks was inoculated with bacterial biomass washed to give an OD620 of 0.1 U. Degradation studies were carried out using 500-ml flasks. Twenty-five milliliters of medium and heterocyclic compounds dissolved in DMSO were added to each flask. In experiments dealing with the degradation of only one of the heterocyclic compounds, the concentrations of CA, DBF, and DBT were 5.0 mM, 1.0 mM, and 1.0 mM, respectively. In experiments dealing with mixtures of CA, DBF, and DBT, the concentrations were 3.0 mM, 0.2 mM, and 0.2 mM, respectively. In addition to the test flasks, two types of controls were prepared. The inactive controls were inoculated with heat-killed cells (autoclaved at 115°C for 20 min). DMSO controls were spiked with 250 μl DMSO and inoculated in the same way as the test flasks. Both controls and test flasks were made in triplicate. For every test, triplicate samples were analyzed, and controls without inocula were monitored.
Metabolite isolation, identification, and analysis.
UV/visible spectrophotometric scans were performed from 220 nm to 600 nm on a 3100 spectrophotometer (Shimadzu, Japan). Supernatants from cultures exposed to CA, DBF, and DBT were obtained by centrifugation at 12,000 rpm for 10 min and then compared spectrophotometrically to identify new peaks formed due to the accumulation of specific metabolites of the different substrates.
The supernatant of each incubation was extracted with a double volume of ethyl acetate (neutral fraction); subsequently, it was adjusted to pH 2 and extracted one more time (acid fraction). The extraction was dried with anhydrous Na2SO4, concentrated, and analyzed by gas chromatography (GC)-mass spectrometry (MS) (Hewlett-Packard GCD 1800C) equipped with a 50-m J&W DB-5MS column (Folsom, CA). In some case, extracts were incubated with N,O-bis(trimethylsilyl)trifluoroacetamide at 80°C for 30 min in order to form trimethylsilyl derivatives.
High-performance liquid chromatography (HPLC) was performed to analyze the aqueous samples using an Agilent 1100 series (Hewlett-Packard) instrument equipped with a variable-wavelength detector and fitted with a reversed-phase C18 column (4.6 mm by 150 mm; Hewlett-Packard). A double volume of ethanol was added to each test flask, followed by centrifugation (12,000 rpm for 20 min) and filtration. Residual concentrations of heterocyclic compounds were determined using a mobile phase of an 80:20 mixture of methanol and deionized water at a flow rate of 0.5 ml min−1.
HPLC-MS was performed using an API 4000 LC-MS-MS system (Applied Biosystems, Foster City, CA). MS analysis was performed using an API 4000 mass spectrometer. The mass spectrometry system was operated under the negative-ion turbo ion spray ionization mode. The liquid chromatography eluent was analyzed using a turbo ion spray with 4,500 V applied to the spray needle. The turbo potential temperature was set at 400°C, with the declustering potential set at 60 V. The mobile phase and flow rate were the same as those described above.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of Sphingomonas sp. strain XLDN2-5 was deposited in GenBank under accession number EF062503.
RESULTS
Isolation and characterization of Sphingomonas sp. strain XLDN2-5.
Strain XLDN2-5 was isolated from petroleum-polluted soil by the enrichment method. The strain was able to utilize CA as the sole source of carbon and nitrogen and degrade DBF and DBT cometabolically. XLDN2-5 was rod shaped and catalase positive. It accumulated poly-β-hydroxybutyrate and gave positive results for citrate and glucose utilization and growth on MacConkey agar. XLDN2-5 gave negative results for the methyl red test, fluorescence on King medium, urease production, Voges-Proskauer test, and oxidase test and yielded negative results for H2S and indole production. The 16S rRNA gene sequence of strain XLDN2-5 showed 99% homology to that of Sphingomonas yanoikuyae. Therefore, strain XLDN2-5 was assigned to the genus Sphingomonas. This strain was deposited at the China Center for Type Culture Collection (CCTCC M205093).
Degradation of CA by strain XLDN2-5.
Strain XLDN2-5 could utilize CA as the sole source of carbon, nitrogen, and energy. The growth accompanied an increase in turbidity (OD620) and a concomitant decrease in CA. Approximately 98% of 5 mM CA was degraded within 18 h. No growth was observed in the control test (with DMSO only). GC-MS and HPLC-MS analyses identified one intermediate, which gave a base peak ion at m/z 145 (C9H7NO) and a fragment ion at m/z 117 (M+-28, CO loss). This metabolite was identified as (1H)-4-quinolinone according to a previous report (28). No peak corresponding to AN was detected in our analysis. However, AN was degraded more efficiently than CA when it was used as the sole source of carbon and nitrogen (data not shown).
Identification of DBF degradation metabolites.
Incubation with DBF showed the accumulation of a yellow compound that had pH-dependent absorption spectra. HPLC-MS analysis showed this compound had a molecular ion at m/z 233 (M-1). Based on this information and according to a previous report, the yellow metabolite was identified as being 2′-OH-HOPDA (31).
Table 1 presents all compounds that could be identified in the course of our studies by GC-MS and high-resolution MS (HR-MS) analyses. 2′-Hydroxyacetophenone and salicylic acid were identified by comparisons of their mass spectral data with authentic samples. 2-Methyl-chroman-one, chromone, 2-(chroman-4-on-2-yl)-ethanol, and monohydroxydibenzofurans were identified by comparisons of their mass spectra with data reported in the literature (2) and the NIST library. In addition to 2-methyl-chroman-one, another two products, designated compounds III and VI, respectively, which had a molecular weight of 162 and similar mass spectra, were also detected. HR-MS of compounds III and VI gave M+ of 162.0686 and 162.0670 (C10H10O2 requires 162.0681), respectively. GC-MS analysis of compound III gave a molecular ion peak at m/z 162. A fragment ion at m/z 145 of compound III corresponds to a loss of hydroxyl from the molecular ion. A high-abundance fragment ion at m/z 121 corresponds to a loss of propenyl from the molecular ion, and the fragment at m/z 93 corresponds to a loss of carbonyl from the m/z 121 fragment ion. Similar to compound III, GC-MS analysis of compound VI also gave a molecular ion peak at m/z 162. The high-abundance fragment ion at m/z 147 corresponds to a loss of methyl from the molecular ion. The fragment ion at m/z 121 corresponds to a loss of vinyl from the ion at m/z 147, and the fragment at m/z 93, 92, and 91 corresponds to a loss of carbonyl plus 0H, 1H, or 2H, respectively, from the m/z 121 fragment ion. Together with HR-MS analysis, compounds III and VI were tentatively identified as being (2-hydroxyphenyl)but-3-en-1-one and (2-hydroxyphenyl)but-2-en-1-one, respectively.
TABLE 1.
HR-MS data for some intermediates from cultures of DBF exposed to Sphingomonas sp. strain XLDN2-5
| Metabolite | Molecular formula | Mol wt
|
||
|---|---|---|---|---|
| Calculated | Found | Difference (ppm) | ||
| 2′-Hydroxyacetophenone | C8H8O2 | 136.0524 | 136.0521 | −2.4 |
| Salicylic acid | C7H6O3 | 138.0317 | 138.0303 | −10.1 |
| (2-Hydroxyphenyl)but-3-en-1-one | C10H10O2 | 162.0681 | 162.0686 | 3.2 |
| Chromone | C9H6O2 | 146.0004 | 145.9984 | −13.7 |
| (2-Hydroxyphenyl)but-2-en-1-one | C10H10O2 | 162.0681 | 162.0670 | −6.7 |
| Monohydroxydibenzofuran | C12H8O2 | 184.0524 | 184.0523 | −0.7 |
| Monohydroxydibenzofuran | C12H8O2 | 184.0524 | 184.0529 | 2.6 |
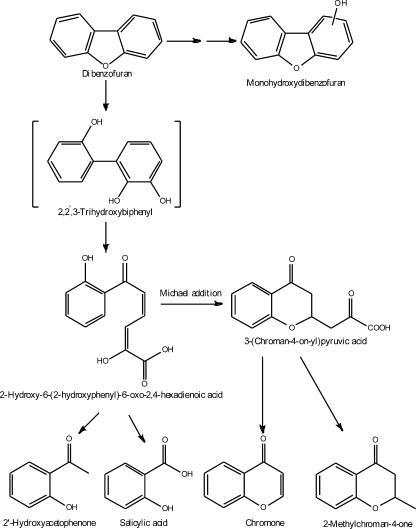
The formation of monohydroxydibenzofurans could be explained by the spontaneous conversion of the corresponding unstable dihydrodiols (23, 26). Our result suggested that XLDN2-5 could catalyze the lateral dioxygenation of DBF. On the basis of the identified metabolites, the metabolic pathway of DBF by strain XLDN2-5 was proposed as shown in Fig. 1.
FIG. 1.
Pathway proposed for the metabolism of DBF by Sphingomonas sp. strain XLDN2-5. The compound in brackets was not detected.
Identification of DBT degradation metabolites.
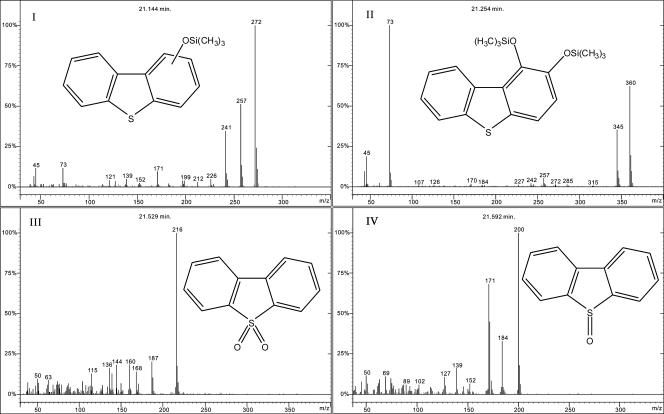
When CA-grown washed cells (OD620 of 5) were incubated with 1.0 mM DBT, the DBT concentration in the culture was decreased by 93% after 24 h of incubation, and the culture turned to an orange color that had a maximal absorbance at 472 nm (A472). The A472 was attributed to trans-4-[2-(3-hydroxy)-thianaphthenyl]-2-oxo-3-butenoic acid, analogous to that reported previously by Kodama et al. (13). The A472 decreased when incubation was allowed for a long time. Two peaks were identified as being monohydroxydibenzothiophene (compound I) (Fig. 2) and dihydroxydibenzothiophene (compound II) (Fig. 2) according to data described in a previous report (23).
FIG. 2.
GC-MS analysis of the metabolites of DBT transformed by Sphingomonas sp. strain XLDN2-5. Compound I, monohydroxydibenzothiophene; compound II, dihydroxydibenzothiophene; compound III, DBTO2; compound IV, DBTO.
A GC-MS chromatogram of the sample from the experiments with DBT showed two major compounds in addition to the two above-described hydroxylated DBTs (Fig. 2). One compound (compound III) (Fig. 2) had an M+ of 216.0246 (C12H8O2S requires 216.0245) and was identified as being DBTO2 by comparison with the GC retention time and mass spectrum of authentic sample. The other compound (compound IV) (Fig. 2) had a mass spectrum comparable to the mass spectrum of DBT-5-oxide (DBTO) (3). HR-MS of compound IV gave an M+ of 200.0300 (C12H8OS requires 200.0296) and was identified as being DBTO. When cells of XLDN2-5 were incubated with DBTO2, no product was detected (data not shown). Therefore, it was concluded that XLDN2-5 could not further oxidize DBTO2.
Cometabolic degradation of DBF and DBT using CA as a carbon source.
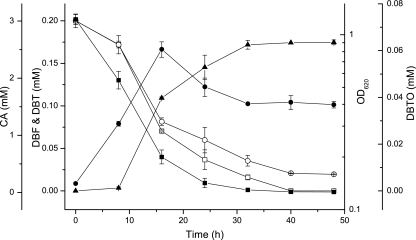
XLDN2-5 could cometabolically degrade DBF and DBT simultaneously using CA as a substrate (Fig. 3). After 48 h of incubation, the concentration of CA and DBF decreased from 3.0 mM and 0.2 mM to below the detection limit. Likewise, the concentration of DBT decreased from 0.2 mM to 0.02 mM. No significant decrease of CA, DBF, and DBT was seen in the controls. Over 90% of the DBT was cometabolized by strain XLDN2-5, but only 30 to 35% was transformed into DBTO; the other 55 to 60% degradation did not result in any detectable transformation product.
FIG. 3.
Cometabolic degradation of CA, DBF, and DBT and accumulation of DBTO by growing cells of Sphingomonas sp. strain XLDN2-5. The initial concentrations of CA, DBF, and DBT were 3.0 mM, 0.2 mM, and 0.2 mM, respectively. •, OD620 (presents on log scale in a semilog plot); ▪, CA; □, DBF; ○, DBT; ▴, DBTO. Values are means of three replicates ± standard deviations.
DISCUSSION
In the present study, the isolation and characterization of Sphingomonas sp. strain XLDN2-5, which could utilize CA as the sole source of carbon and energy, were investigated. The chemical structures of the degradation products of CA, DBF, and DBT were determined, and the degradation pathways of these compounds were proposed. The (1H)-4-quinolinone detected in the degradation of CA had a structure that was similar to that of the chromone identified in DBF degradation by XLDN2-5. This result suggested that angular dioxygenation might have occurred when XLDN2-5 degraded CA and DBF. Although AN was not detected as a metabolite in the degradation of CA by this bacterium, isolate XLDN2-5 did use AN as a growth substrate, as previously reported for other CA-degrading bacteria (5, 24). Thus, it is likely that AN was also an intermediate compound in the CA degradation pathway for strain XLDN2-5. From these results, we proposed that XLDN2-5 might degrade CA through a similar angular dioxygenation pathway reported previously by other researchers (16, 24, 25).
Metabolites detected from DBF degradation suggested that XLDN2-5 degrades DBF via a pathway similar to that of Sphingomonas sp. strain HH69 (previously identified as Pseudomonas sp.) (2). The presence of 2′-OH-HOPDA could be assumed from the bright yellow color of the culture medium due to the production of a metabolite at A446 (2). 2′-OH-HOPDA was further metabolized to give salicylic acid and 2-hydroxypenta-2,4-dienoic acid. 3-(Chroman-4-on-2-yl)pyruvic acid formed by a straightforward intramolecular Michael addition of the phenolic OH group to the α,β-unsaturated ketone side chain of 2′-OH-HOPDA (31), together with its derivatives 2-methyl-chroman-one, chromone, and 2-(chroman-4-on-2-yl)-ethanol, had been observed in the culture supernatant of several DBF-degrading bacteria (2, 31, 36). Salicylic acid is a key metabolite in the bacterial degradation of DBF. In the DBF degradation pathway reported for Sphingomonas sp. strain HH69 (2), Sphingomonas sp. strain RW1 (34), and Staphylococcus auriculans DBF63 (21), the metabolite salicylic acid was branched into catechol and gentisic acid degradation pathways. 2′-Hydroxyacetophenone was also found in the culture medium of DBF-grown Sphingomonas sp. strain HH69. Moreover, 2′-hydroxyacetophenone was found to be formed by the decarboxylation of 3-(2-hydroxyphenyl)-3-oxopropionic acid by Sphingomonas sp. strain HH69 when 2-hydroxydibenzofuran served as a substrate (9). A satisfactory mechanism of acetophenone nonenzymatic formation is from the meta-cleavage compound of 2,3-dihydroxybiphenyl. It is reasonable to suggest that the formation of 2′-hydroxyacetophenone is in accordance with the mechanism proposed previously by Seah et al. (29). The detection of (2-hydroxyphenyl)but-3-en-1-one and (2-hydroxyphenyl)but-2-en-1-one in our study suggested that further transformation like decarboxylation or β-oxidation of 2′-OH-HOPDA would occur. Furthermore, the cooccurrence of monohydroxydibenzofurans and 2-methyl-chroman-4-one strongly suggested that strain XLDN2-5 degraded DBF through different DBF degradation pathways.
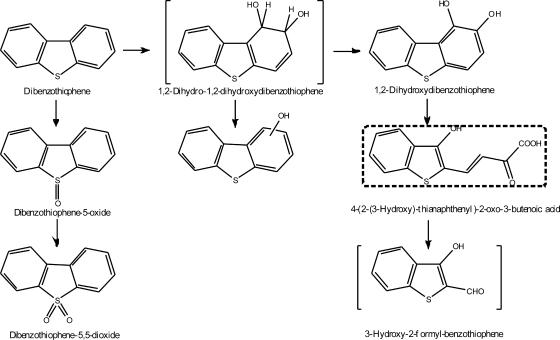
The detection of hydroxydibenzothiophenes indicated that lateral dioxygenation of DBT occurred. Moreover, an orange compound with a maximal A472 was produced in the medium. The appearance of colored metabolites during bacterial degradation of DBT has been described previously (3, 12, 13). These results suggested that the ring cleavage pathway occurred. To the best of our knowledge, this is the first reported CA-utilizing strain capable of degrading DBT via the ring cleavage pathway. In addition to catalyzing angular dioxygenation of DBF and lateral dioxygenation of DBF and DBT, XLDN2-5 also catalyzed the sulfoxidation of DBT to DBTO2. Over 90% of the DBT was cometabolized by strain XLDN2-5, but only 30 to 35% was transformed into DBTO and DBTO2; the other 55 to 60% degradation did not result in detectable transformation products. Therefore, it is assumed that there are two pathways at work that result in DBT degradation. One pathway leads to the formation of DBTO and DBTO2, which cannot be further metabolized, while the other is more prevalent because a greater percentage of DBT degradation (55 to 60%) resulted. The formation of DBTO agrees with previous findings (23). Carbazole 1,9a-dioxygenase of Pseudomonas sp. strain CA10 catalyzed the sulfoxidation of DBT to DBTO as a dominant reaction (35 to 55% of DBT was transformed, resulting in the formation of 40 to 60% of DBTO), while no more than 3% of lateral dioxygenation products were detected. Moreover, no DBTO2 was detected when DBT was exposed to strain CA10. The DBT metabolites DBTO and DBTO2 produced by strain XLDN2-5 were probably formed by consecutive oxygenations of the sulfur atom of DBT and accumulated in a relative concentration ratio of 50:1. Most of the isolated strains utilizing DBT as the sole sulfur source degrade DBT to 2-hydroxybiphenyl via DBTO and DBTO2 (4, 14, 38). On the other hand, DBT is presumed to be converted to DBTO and DBTO2 by dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1 (1) and Pseudomonas sp. strain F247 (6). However, these strains also seem to lack the ability to cleave the sulfur-containing ring of DBTO2, resulting in the accumulation of DBTO and DBTO2. Although strain XLDN2-5 could not further degrade DBTO2, it also had the ability of lateral dioxygenation of DBT, which led to the ring cleavage compounds. The metabolic pathways of DBT by Sphingomonas sp. strain XLDN2-5 are summarized in Fig. 4.
FIG. 4.
Pathway proposed for the metabolism of DBT by Sphingomonas sp. strain XLDN2-5. The compound in brackets was not detected, and the compound in the square dotted box was tentatively identified.
In the experiment where CA, DBF, and DBT were added as a mixture, the degradation of CA, DBF, and DBT began simultaneously. As no salicylic acid was accumulated, the results indicated that the CA-grown cells could also degrade salicylic acid. However, DBT, DBF, and salicylic acid could not support the growth of strain XLDN2-5. If strain XLDN2-5 was not pregrown with CA, the cells had no ability to degrade DBT or DBF, and subsequently, no salicylic acid was produced. This may be due to fact that CA is a real inducer of the expression of degradation enzymes, while DBT, DBF, or salicylic acid is not. Therefore, it is concluded that the metabolic pathways were controlled by CA-degrading enzymes and that enzyme expression was induced by CA only.
In conclusion, our identification of the degradation metabolites of CA, DBF, and DBT suggested that strain XLDN2-5 catalyzed angular dioxygenation, lateral dioxygenation, and monooxygenation, as described above. These results indicated that XLDN2-5 has a broad substrate range and catalyzes diverse oxygenation. Nojiri et al. (23) and Takagi et al. (32) previously reported that carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10 and dibenzofuran 4,4a-dioxygenase from Staphylococcus auriculans DBF63 catalyzed these three modes of oxygenation. The substrate specificity of strain XLDN2-5 seems to be similar to that of strain CA10, but strain CA10 was not reported to catalyze the sulfoxidation of DBT to DBTO2 and the fission of DBT via the ring cleavage pathway. This study has demonstrated the versatility of Sphingomonas sp. strain XLDN2-5 in the transformation of the planar-structure compounds of CA, DBF, and DBT. From a bioremediation point of view, it is imperative to obtain pollutant-degrading bacteria that have a wide substrate range and various types of oxidation abilities. The versatile oxidation activities of strain XLDN2-5 should be a great advantage for bioremediation in the natural environment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 20590368, 20377026, and 20577031).
We gratefully acknowledge the partially financial support and GC-MS analysis by Shanghai Apple Flavor & Fragrance Co., Ltd., and Shanghai Jiao Tong University (People's Republic of China).
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Bünz, P. V., and A. M. Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J. Bacteriol. 175:6467-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortnagel, P., H. Harms, R. M. Wittich, S. Krohn, H. Meyer, V. Sinnwell, H. Wilkes, and W. Francke. 1990. Metabolism of dibenzofuran by Pseudomonas sp. strain HH69 and the mixed culture HH27. Appl. Environ. Microbiol. 56:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frassinetti, S., L. Setti, A. Corti, P. Farrinelli, P. Montevecchi, and G. Vallini. 1998. Biodegradation of dibenzothiophene by a nodulating isolate of Rhizobium meliloti. Can. J. Microbiol. 44:289-297. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher, J. R., E. S. Olson, and D. C. Stanley. 1993. Microbial desulfurization of dibenzothiophene: a sulfur-specific pathway. FEMS Microbiol. Lett. 107:31-35. [DOI] [PubMed] [Google Scholar]
- 5.Gieg, L. M., A. Otter, and P. M. Fedorak. 1996. Carbazole degradation by Pseudomonas sp. LD2: metabolic characteristics and the identification of some metabolites. Environ. Sci. Technol. 30:575-585. [Google Scholar]
- 6.Grifoll, M., S. A. Selifonov, and P. J. Chapman. 1995. Transformation of substituted fluorenes and fluorene analogs by Pseudomonas sp. strain F274. Appl. Environ. Microbiol. 61:3490-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gundlach, E. R., P. D. Boehm, M. Marchand, R. M. Atlas, D. M. Ward, and D. A. Wolfe. 1983. The fate of Amoco Cadiz oil. Science 221:122-129. [DOI] [PubMed] [Google Scholar]
- 8.Happe, B., L. D. Eltis, H. Poth, R. Hedderich, and K. N. Timmis. 1993. Characterization of 2,2′,3-trihydroxybiphenyl dioxygenase, an extradiol dioxygenase from the dibenzofuran- and dibenzo-p-dioxin-degrading bacterium Sphingomonas sp. strain RW1. J. Bacteriol. 175:7313-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms, H., H. Wilkes, R. M. Wittich, and P. Fortnagel. 1995. Metabolism of hydroxydibenzofurans, methoxydibenzofurans, acetoxydibenzofurans, and nitrodibenzofurans by Sphingomonas sp. strain HH69. Appl. Environ. Microbiol. 61:2499-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichinose, H., H. Wariishi, and H. Tanaka. 1999. Bioconversion of recalcitrant 4-methyldibenzothiophene to water-extractable products using lignin-degrading basidiomycete Coriolus versicolor. Biotechnol. Prog. 15:706-714. [DOI] [PubMed] [Google Scholar]
- 11.Jensen, A. M., K. W. Finster, and U. Karlson. 2003. Degradation of carbazole, dibenzothiophene, and dibenzofuran at low temperature by Pseudomonas sp. strain C3211. Environ. Toxicol. Chem. 22:730-735. [PubMed] [Google Scholar]
- 12.Kodama, K., S. Nakatani, K. Umehara, K. Shimizu, Y. Minoda, and K. Yamada. 1970. Microbial conversion of petro-sulfur compounds. Part III. Isolation and identification of products from dibenzothiophene. Agric. Biol. Chem. 34:1320-1324. [Google Scholar]
- 13.Kodama, K., K. Umehara, K. Shimizu, S. Nakatani, Y. Minoda, and K. Yamada. 1973. Identification of microbial products from dibenzothiophene and its proposed oxidation pathway. Agric. Biol. Chem. 37:45-50. [Google Scholar]
- 14.Li, F. L., P. Xu, C. Q. Ma, L. L. Luo, and S. S. Wang. 2003. Deep desulfurization of hydrodesulfurization-treated diesel oil by a facultative thermophilic bacterium Mycobacterium sp. X7B. FEMS Microbiol. Lett. 223:301-307. [DOI] [PubMed] [Google Scholar]
- 15.Li, F. L., P. Xu, J. H. Feng, L. Meng, Y. Zheng, L. L. Luo, and C. Q. Ma. 2005. Microbial desulfurization of gasoline in a Mycobacterium goodii X7B immobilized-cell system. Appl. Environ. Microbiol. 71:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., P. Xu, and H. D. Blankerspoor. 2004. Degradation of carbazole in the presence of non-aqueous phase liquids by Pseudomonas sp. Biotechnol. Lett. 26:581-584. [DOI] [PubMed] [Google Scholar]
- 17.Li, L., Q. G. Li, F. L. Li, Q. Shi, B. Yu, F. R. Liu, and P. Xu. 2006. Degradation of carbazole and its derivatives by a Pseudomonas sp. Appl. Microbiol. Biotechnol. 73:941-948. [DOI] [PubMed] [Google Scholar]
- 18.Li, M. Z., C. H. Squires, D. J. Monticello, and J. D. Childs. 1996. Genetic analysis of the dsz promoter and associated regulatory regions of Rhodococcus erythropolis IGTS8. J. Bacteriol. 178:6409-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, J., T. Nakajima-Kambe, T. Shigeno, A. Ohbo, N. Nomura, and T. Nakahara. 1999. Biodegradation of dibenzothiophene and 4,6-dimethyldibenzothiophene by Sphingomonas paucimobilis strain TZS-7. J. Biosci. Bioeng. 88:293-299. [DOI] [PubMed] [Google Scholar]
- 20.Meade, J. D., J. Hellou, and T. R. Patel. 2002. Aerobic co-metabolism of sulfur, nitrogen and oxygen heterocycles by three marine bacterial consortia. J. Basic Microbiol. 42:19-36. [DOI] [PubMed] [Google Scholar]
- 21.Monna, L., T. Omori, and T. Kodama. 1993. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl. Environ. Microbiol. 59:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller, J. G., S. E. Lantz, D. Ross, R. J. Colvin, D. P. Middaugh, and P. H. Pritchard. 1993. Strategy using bioreactors and specially selected microorganisms for bioremediation of groundwater contaminated with creosote and pentachlorophenol. Environ. Sci. Technol. 27:691-698. [Google Scholar]
- 23.Nojiri, H., J. W. Nam, M. Kosaka, K. I. Morii, T. Takemura, K. Furihata, H. Yamane, and T. Omori. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouchiyama, N., S. Miyachi, and T. Omori. 1998. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J. Gen. Appl. Microbiol. 44:57-63. [DOI] [PubMed] [Google Scholar]
- 25.Ouchiyama, N., Y. Zhang, T. Omori, and T. Kodama. 1993. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci. Biotechnol. Biochem. 57:455-460. [DOI] [PubMed] [Google Scholar]
- 26.Resnick, S. M., and D. T. Gibson. 1996. Regio- and stereospecific oxidation of fluorene, dibenzofuran, and dibenzothiophene by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62:4073-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saftić, S., P. M. Fedorak, and J. T. Andersson. 1993. Transformations of methyldibenzothiophenes by three Pseudomonas isolates. Environ. Sci. Technol. 27:2577-2584. [Google Scholar]
- 28.Schneider, J., R. J. Grosser, K. Jayasimhulu, W. Xue, B. Kinkle, and D. Warshawsky. 2000. Biodegradation of carbazole by Ralstonia sp. RJGII.123 isolated from a hydrocarbon contaminated soil. Can. J. Microbiol. 46:269-277. [DOI] [PubMed] [Google Scholar]
- 29.Seah, S. Y. K., G. Labbé, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 30.Shepherd, J. M., and G. Lloyd-Jones. 1998. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem. Biophys. Res. Commun. 247:129-135. [DOI] [PubMed] [Google Scholar]
- 31.Strubel, V., K. H. Engesser, P. Fischer, and H. J. Knackmuss. 1991. 3-(2-Hydroxyphenyl)catechol as substrate for proximal meta ring cleavage in dibenzofuran degradation by Brevibacterium sp. strain DPO 1361. J. Bacteriol. 173:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi, T., H. Nojiri, T. Yoshida, H. Habe, and T. Omori. 2002. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol. Lett. 24:2099-2106. [Google Scholar]
- 33.Tao, F., B. Yu, P. Xu, and C. Q. Ma. 2006. Biodesulfurization in biphasic systems containing organic solvents. Appl. Environ. Microbiol. 72:4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittich, R. M., H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, P., B. Yu, F. L. Li, X. F. Cai, and C. Q. Ma. 2006. Microbial degradation of sulfur, nitrogen, and oxygen heterocycles. Trends Microbiol. 14:398-405. [DOI] [PubMed] [Google Scholar]
- 36.Yamazoe, A., O. Yagi, and H. Oyaizu. 2004. Degradation of polycyclic aromatic hydrocarbons by a newly isolated dibenzofuran-utilizing Janibacter sp. strain YY-1. Appl. Microbiol. Biotechnol. 65:211-218. [DOI] [PubMed] [Google Scholar]
- 37.Yu, B., C. Q. Ma, W. J. Zhou, S. S. Zhu, Y. Wang, J. Y. Qu, F. L. Li, and P. Xu. 2006. Simultaneous biodetoxification of S, N, and O pollutants by engineering a carbazole-degrading gene cassette in a recombinant biocatalyst. Appl. Environ. Microbiol. 72:7373-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, B., P. Xu, Q. Shi, and C. Q. Ma. 2006. Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl. Environ. Microbiol. 72:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, B., P. Xu, S. S. Zhu, X. F. Cai, Y. Wang, L. Li, F. L. Li, X. Y. Liu, and C. Q. Ma. 2006. Selective biodegradation of S and N heterocycles by a recombinant Rhodococcus erythropolis strain containing carbazole dioxygenase. Appl. Environ. Microbiol. 72:2235-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]