Abstract
The goals of this study were to (i) identify issues that affect the ability of discriminant function analysis (DA) of antimicrobial resistance profiles to differentiate sources of fecal contamination, (ii) test the accuracy of DA from a known-source library of fecal Escherichia coli isolates with isolates from environmental samples, and (iii) apply this DA to classify E. coli from surface water. A repeated cross-sectional study was used to collect fecal and environmental samples from Michigan livestock, wild geese, and surface water for bacterial isolation, identification, and antimicrobial susceptibility testing using disk diffusion for 12 agents chosen for their importance in treating E. coli infections or for their use as animal feed additives. Nonparametric DA was used to classify E. coli by source species individually and by groups according to antimicrobial exposure. A modified backwards model-building approach was applied to create the best decision rules for isolate differentiation with the smallest number of antimicrobial agents. Decision rules were generated from fecal isolates and applied to environmental isolates to determine the effectiveness of DA for identifying sources of contamination. Principal component analysis was applied to describe differences in resistance patterns between species groups. The average rate of correct classification by DA was improved by reducing the numbers of species classifications and antimicrobial agents. DA was able to correctly classify environmental isolates when fewer than four classifications were used. Water sample isolates were classified by livestock type. An evaluation of the performance of DA must take into consideration relative contributions of random chance and the true discriminatory power of the decision rules.
Knowing the source of fecal contamination of surface water is necessary to determine the degree of risk associated with human health and to develop effective control and resource management strategies. One technique that has been reported to be a useful, low-cost screening method is discriminant function analysis (DA) of antimicrobial resistance profiles. DA is a multivariate statistical method designed to separate sets of observations and allocate new observations to previously defined groups (12, 15, 16). DA transforms observations obtained from different populations with overlapping distributions into nonoverlapping distributions. This transformation can then be applied to a set of observations from an unknown source population to determine the most probable population that served as the source for the unknown source observation. DA can be used to determine which variables discriminate between two or more naturally occurring groups and then classify cases into the values of categorical dependent groups (12, 15, 16).
DA has been used successfully to classify the source species for fecal streptococcus, fecal coliforms, and Escherichia coli isolates obtained from surface water samples. When used as a tool for microbial source identification, DA can be applied to antimicrobial resistance profiles from a database of fecal bacterial isolates obtained from various species. This known-source library is used to generate a classification scheme (decision rule). The accuracy of the decision rule is assessed by evaluating the percentage of isolates from the known-source library that are correctly classified by the rule. Once a decision rule with an acceptable correct classification rate is obtained, this model can be applied to bacterial isolates from surface water to identify the most probable source species for the fecal contamination of that surface water.
The use of DA on antimicrobial resistance patterns in fecal streptococci to differentiate between human and animal sources was first described by Wiggins (27), with more than 90 and 84% correct classifications, respectively, when six-species populations were being classified. Several other studies have reported the successful use of this approach to differentiate human versus animal sources of fecal contamination in water using antimicrobial resistance profiles (7, 8, 9, 11, 13, 20, 28, 29), genetic data (3, 5, 21), and carbon source utilization profiles (10) of fecal bacteria. Rates of correct classification using antimicrobial resistance patterns varied from 33 (8) to 90% (27), depending on the classification groups used in the studies.
Using DA to classify bacteria by antimicrobial resistance patterns is an emerging discipline. The wide range of rates of correct classification by DA of antimicrobial resistance patterns of fecal bacteria reported in the literature indicates that there can be great variation in the success of the technique. These studies were conducted in different regions and using different bacterial species, antimicrobial agents, and source species for the definition of the decision rules through DA. Given the various methods and differing results, this method should be used with attention to maximizing the ability of the DA to distinguish between different classification groups for each study's sample population and classification levels.
This study is part of a larger body of research seeking to use DA to classify fecal E. coli isolates from domestic livestock, companion animals, humans, and wildlife by creating a decision rule based upon antimicrobial resistance profiles and then to identify the most probable species source of E. coli isolates obtained from surface water samples from a Michigan watershed. In an earlier phase of this study, we described patterns of antimicrobial susceptibility of E. coli strains from different animal species, human-derived septage, and surface water samples (23). The underlying hypothesis of this portion of the study is that the DA of antimicrobial susceptibility profiles from fecal E. coli isolates obtained from a local database of domestic animals and wildlife can be used to develop a microbial source identification model. The objectives were to (i) identify specific issues that affect the efficiency of the discriminant functions and develop methods to address these issues, (ii) using these methods, develop decision rules from DA of antimicrobial susceptibility profiles from a known-source library of local fecal E. coli isolates from domestic animals and wildlife, (iii) test the accuracy of DA of antimicrobial susceptibility profiles from a known-source library of local fecal E. coli isolates from domestic animals and wildlife with environmental E. coli isolates, and (iv) use this method to identify the most probable source of fecal E. coli contamination of surface water in the Red Cedar Watershed in Michigan.
MATERIALS AND METHODS
Study design. (i) Study area.
The Red Cedar Watershed was chosen as the study area. It encompasses an area of approximately 118,600 ha in Ingham and Livingston counties in Michigan. The watershed area provides residents with numerous recreational activities, including angling, canoeing, kayaking, photography, and bird watching. The river also serves as a source of water for the irrigation of crops throughout the watershed. Swine and dairy cattle are the predominant forms of livestock present in the watershed.
(ii) Enrollment of participating farmers.
Farms were located within the Red Cedar Watershed, and county drain commissioners identified specific farms whose premises drained into the watershed. Through county extension agents, these farmers were sent a letter inviting them to participate in the study. To indicate their willingness to participate, respondents returned a prestamped postcard to the Center for Comparative Epidemiology at Michigan State University. A total of 60 farmers were contacted and asked to participate in the study. Thirty-one farmers agreed to participate, and farm visits were arranged quarterly from winter 2002 to winter 2003.
(iii) Data collection.
Data relating to antimicrobial use and numbers of animals on each farm were collected (during the time of collection of fecal samples) with a questionnaire administered by in-person interviews. Participants were asked about the use of antimicrobial agents for therapy, prophylaxis, and growth promotion during the previous 60 days.
(iv) Sample collection.
Animal fecal and farm environment samples and human septage samples were taken using culturette swabs, and 100-ml water samples were collected from specific locations in the watershed. Water sampling sites were determined with the help of the Ingham County drain commissioner, based on the direction of the rain flow from every farm enrolled in the study. Bottles for water sampling contained 10 mg sodium thiosulfate to neutralize any residual chlorine in the water. All samples were shipped to the University of Maryland for bacterial isolation, identification, and antimicrobial susceptibility testing.
Fecal samples were obtained from dairy and beef cattle, swine, horses, sheep, goats, poultry, deer, ducks, and wild geese. Fecal samples from livestock (dairy cattle, beef cattle, swine, sheep, goats, and horses) were collected rectally from individual animals using culturette swabs. Samples were collected from fresh manure using culturette swabs on feedlots where individual animal sampling was not feasible. Poultry samples were collected by cloacal swab. Deer samples were collected from freshly voided droppings. Wild goose samples were collected from freshly voided droppings and cloacal swabs by the Michigan Department of Natural Resources. Environmental samples from manure storage facilities (i.e., lagoons, slurry pits, and manure piles) on the farms were collected using culturette swabs. Septage samples from humans were collected from septic tanks (prior to chemical treatment) in the study area with the help of local septic-pumping companies.
(v) Isolation of E. coli from water samples.
The membrane filtration method used by the United States Environmental Protection Agency (6) was used to isolate E. coli from water samples. In this procedure, water samples were filtered through a sterile, white, grid-marked, 47-mm-diameter membrane (pore size, 0.45 ± 0.02 μm) that retained bacteria. After filtration, the membrane containing the bacteria was placed on a selective and differential medium (mTEC) (10) and incubated at 35°C for 2 h to resuscitate the injured or stressed bacteria and then incubated at 44°C for 22 h. The filter was transferred from mTEC agar to a filter pad saturated with urea substrate medium. After 15 to 20 min, yellow, yellow-green, or yellow-brown colonies on mTEC were transferred to urea substrate medium; any non-E. coli colonies turned pink or purple on the medium.
(vi) Identification of E. coli from surface water, fecal, and environmental samples.
Standard methods were used for the enrichment, isolation, identification, and biochemical confirmation of E. coli isolates (1).
Upon arrival at the laboratory, culturette swabs (fecal and farm environment samples and human-derived septage samples) or colonies picked from urea substrate medium (surface water samples) were placed in tubes with tryptic soy broth (TSB) and incubated at 35°C for 24 h. Approximately 10 μl of the turbid broth was streaked onto violet red bile agar and incubated for 18 to 20 h at 35°C. The plates containing violet red bile agar were examined for reddish purple colonies that fluoresced under a black light. Selected colonies were streaked onto MacConkey's agar and incubated at 35°C for 18 to 20 h. The MacConkey plate was examined for red colonies that precipitated bile and had dark red centers. One or two colonies were selected, streaked onto tryptic soy agar (TSA), and incubated for 18 h. The TSA plate was then examined for single colonies that were round, milk-colored, and slightly convex. One single colony was selected and placed in a tube containing TSB and incubated for approximately 3 to 4 h until turbid.
One or two presumptive E. coli colonies were obtained from each fecal, environmental, and water sample. To ensure that antimicrobial resistance profiles were obtained for confirmed E. coli isolates only, each presumptive E. coli isolate was biochemically confirmed using the indole-methyl red-Voges-Proskauer-citrate and triple-sugar iron tests. Isolates that failed to demonstrate biochemical test results consistent with those of typical E. coli strains for any single test were excluded from further analysis. Confirmed isolates were inoculated into a new TSB tube and incubated to the turbidity of a 0.5 McFarland standard (approximately about 2 to 3 h).
(vii) Antimicrobial susceptibility testing.
The standard Kirby-Bauer disk diffusion method was used to develop the antimicrobial susceptibility profile of E. coli isolates (18, 19) for 12 antimicrobial agents (neomycin, gentamicin, streptomycin, chloramphenicol, ofloxacin, nalidixic acid, sulfisoxazole, trimethoprim-sulfamethoxazole, tetracycline, ampicillin, nitrofurantoin, and cephalothin). These antimicrobial agents were chosen on the basis of their importance in treating human or animal E. coli infections or their use as feed additives to promote growth in animals and to provide diversity in representation of different antimicrobial classes (14).
A TSB tube was inoculated with E. coli and incubated to the turbidity of a 0.5 McFarland standard and then swabbed onto a 150-mm Mueller-Hinton plate. Twelve commercially prepared antimicrobial disks were dispensed onto the inoculated plates. The plates were incubated at 35°C for 18 to 20 h. The diameter of the clear zones of growth inhibition around the antimicrobial disks, including the 6-mm disk diameter, was measured in millimeters using precision calipers (18, 19). E. coli isolates from American Type Culture Collection strain 25922 were used for quality control.
(viii) Statistical analysis.
Separate known-species source libraries were developed for fecal and environmental samples. Initially, classification rules were developed for eight species groups with at least 50 isolates (beef cattle, dairy cattle, sheep, swine, poultry, equids, wildlife [wild geese and white-tailed deer], and humans). Species groups were then combined by exposure to antimicrobial use to reduce the number of categories entering the DA. These groups included, in order of increasing scope, all ruminants (cattle and sheep), livestock (all ruminants and swine), and food animals (livestock and poultry).
Exploratory analyses were conducted with the diffusion zone data to determine whether the use of DA was warranted. Descriptive statistics were generated to assess the distributions of the diffusion zones, and simple nonparametric tests (Kruskal-Wallis χ2 tests) were carried out to test for differences in diffusion zones between different species groups. Principal component analysis (PCA) was conducted on the diffusion zones to describe any grouping of populations of isolates, and multivariate analysis of variance was used to determine whether significant differences were found in disk diffusion zone distributions for all species classification groups at a P value of ≤0.05.
We utilized Mahalanobis distances to generate discriminant function models for the different species classification groups (PROC DISCRIM, SAS 9.1.3; SAS Institute, Cary, NC). Since diffusion zone measures were not normally distributed, three different nonparametric DA models were utilized: linear, quadratic, and Epanechnikov density kernel models. The cross-validation method was used for DA development, in which individual isolates were removed from the data set one at a time, the decision rule was developed from the remainder of the data set, and then the removed isolate was classified based on the rule created by those remaining observations. The cross-validation classification table was used to calculate the percentage of misclassified isolates and determine the average rate of correct classification (ARCC) (11, 27).
To develop the most efficient decision rules for each species classification group, an approach similar to backward model building for regression models was undertaken. First, a “full” discriminant function model using all 12 antimicrobial agents was generated. Next, agents were removed from the full model one at a time, based on the results of PCA and univariable analyses, and the resulting 12 models were compared. The model with the best performance (highest rate of correct classification) was selected as the base model for the next level of model building, and the process of elimination/selection was repeated. These steps were repeated until the removal of additional agents did not improve the performance of the DA or until only one agent remained in the discriminant function model.
Once developed from known-source fecal isolates, the decision rules were applied to a set of isolates from environmental samples where the species of animals housed in the environment were known. This served as a test of the reliability of the decision rules for use in classifying environmental samples where the fecal sources were known. Finally, DA rules derived from known-source fecal isolates were applied to a set of isolates from water samples to classify each water isolate into the most probable source species population.
RESULTS
A total of 1,247 fecal, environmental, and septage samples were collected, and data from 954 fecal- and septage-origin E. coli isolates were used to develop the discriminant function models (Table 1). The disk diffusion zones were not normally distributed. Bimodal distributions were seen for neomycin, gentamicin, trimethoprim-sulfamethoxazole, tetracycline, ampicillin, nalidixic acid, nitrofurantoin, cephalothin, and sulfisoxazole. Given the bimodal nature of these distributions, isolates were classified as belonging to either the lower or higher bimodal distribution for descriptive purposes (Table 2). The breakpoints suggested by the bimodal distribution of the majority of antimicrobial agents in this study did not coincide with currently established resistance breakpoints (19) (Table 2). The only agents for which the CLSI (formerly NCCLS)-defined breakpoints were comparable to the natural breakpoints seen in this study were streptomycin, tetracycline, and sulfisoxazole.
TABLE 1.
Numbers of different samples, by species and species groups
| Species group | No. of samples |
|---|---|
| Species group | |
| Beef cattle | 184 |
| Dairy cattle | 228 |
| Sheep | 155 |
| Swine | 175 |
| Poultry | 85 |
| Equids | 60 |
| Wildlife | 64 |
| Humans | 3 |
| Total | 954 |
| Combined groups | |
| Ruminantsa | 567 |
| Livestockb | 742 |
| Food animalsc | 827 |
The ruminant species group includes beef and dairy cattle and sheep.
The livestock species group includes beef and dairy cattle, sheep, and swine.
The food animal species group includes beef and dairy cattle, sheep, swine, and poultry.
TABLE 2.
Distributions of disk diffusion zones of 951 fecal isolates to 12 antimicrobial agents
| Agent | Overall
|
Lower peak
|
Upper peak
|
CLSI breakpointa | |||
|---|---|---|---|---|---|---|---|
| Mean | Median | Range | Median | Range | Median | ||
| Neomycin | 17.7 | 16.3 | 6.0-18.7 | 15.4 | 19.0-26.2 | 22.0 | 12 |
| Streptomycin | 15.2 | 14.5 | 3.2-16.7 | 13.5 | 17.0-26.0 | 20.0 | 17 |
| Tetracycline | 18.6 | 21.7 | 6.0-13.9 | 6.0 | 15.0-35.0 | 23.7 | 14 |
| Ampicillinb | 19.3 | 19.0 | 13 | ||||
| TMP/SMZc | 26.7 | 26.1 | 6.0-29.9 | 24.5 | 30.0-38.0 | 33.0 | 10 |
| Cephalothin | 17.6 | 16.1 | 6.0-17.9 | 14.4 | 18.0-32.0 | 22.0 | 14 |
| Sulfisoxazole | 20.6 | 22.1 | 6.0-7.8 | 6.0 | 12.0-35.0 | 22.8 | 12 |
| Gentamicin | 21.0 | 19.6 | 6.0-20.9 | 18.3 | 21.0-30.0 | 25.0 | 12 |
| Chloramphenicol | 25.6 | 25.4 | 6.0-26.9 | 24.3 | 27.0-35.0 | 28.0 | 12 |
| Ofloxacinb | 29.3 | 29.4 | 12 | ||||
| Nalidixic acid | 24.2 | 22.9 | 6.0-25.9 | 21.0 | 26.0-36.0 | 29.0 | 13 |
| Nitrofurantoin | 21.0 | 19.5 | 6.0-20.8 | 18.1 | 21.0-30.0 | 25.0 | 14 |
Maximum diffusion zone (mm) breakpoint for the determination of antimicrobial resistance (17).
Not bimodally distributed; lower and upper peak ranges and medians not reported.
TMP/SMZ, trimethoprim-sulfamethoxazole.
Tests were conducted to determine whether the data warranted the use of DA, and results of the Kruskall-Wallis χ2 analysis and multivariate analysis of variance found significant differences in all disk diffusion zone distributions for all species classification groups (with the Kruskal-Wallis χ2 test, P was <0.0001; with the Wilks lambda test, P was <0.001). PCA was also conducted on the 951 fecal isolates by using all 12 antimicrobial agents. The first, second, and third principal components accounted for 55.2, 19.4, and 10.2% of total variance, respectively. When loadings for the components were examined, the first principal component was representative of generally susceptible isolates (all positive loadings for diffusion zone diameters), the second component was representative of isolates with low susceptibility to tetracycline (−0.67 loading for tetracycline), and the third component was representative of isolates with low susceptibility to tetracycline and high susceptibility to sulfisoxazole (loadings of −0.53 for tetracycline and 0.73 for sulfisoxazole).
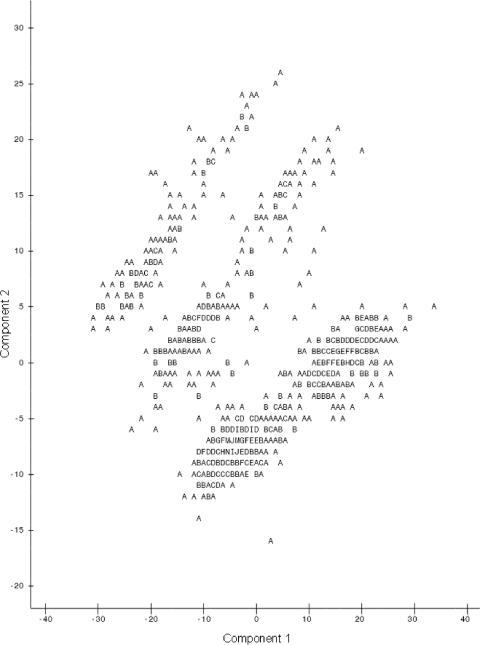
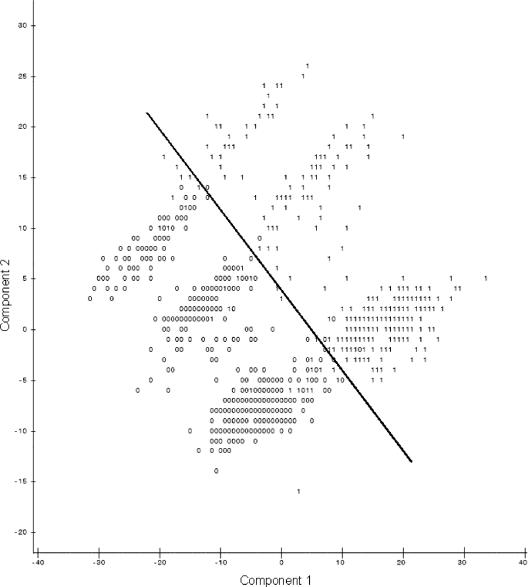
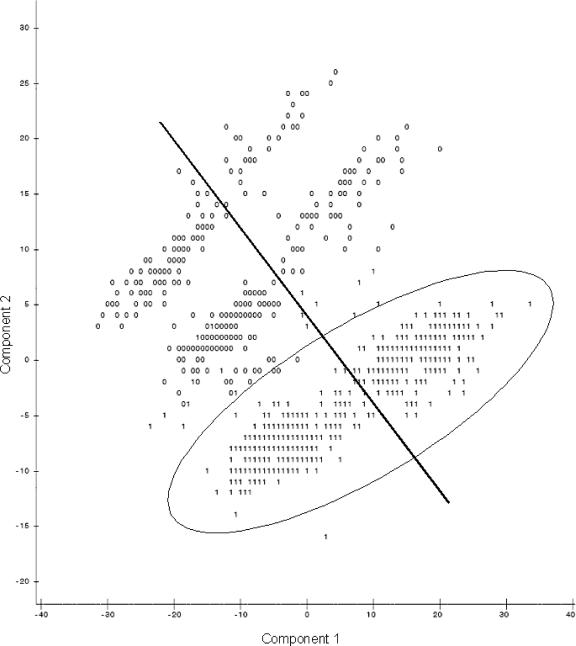
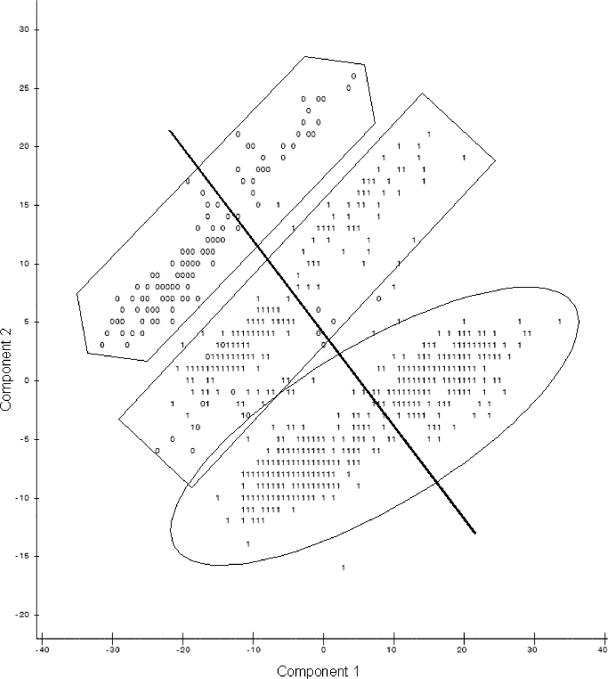
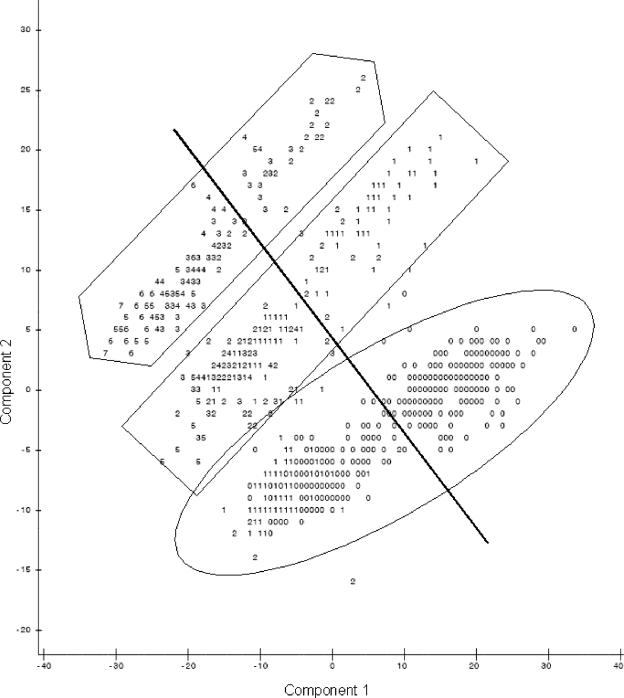
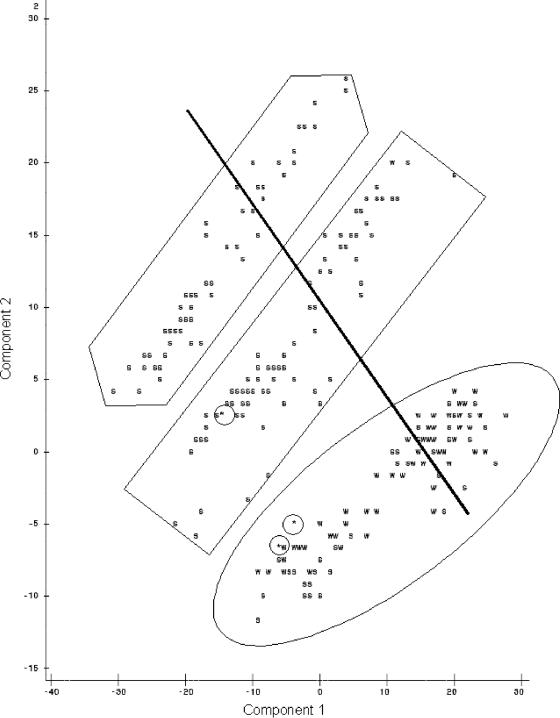
When the first and second principal components were plotted against each other (Fig. 1), observations were present in three groups. Identifying data points as being resistant or susceptible according to CLSI breakpoints (Table 2) showed no pattern of distribution of resistant isolates. However, when the plots identified isolates from the naturally observed high or low bimodal distribution groups, distinctions were seen between high and low susceptibility to the majority of antimicrobial agents (Fig. 2) and isolates with low susceptibilities to tetracycline (Fig. 3) and sulfisoxazole (Fig. 4). It appears that there were three distinct populations: high susceptibility (high-diffusion-zone-diameter group members) to tetracycline and sulfisoxazole, low susceptibility (low-diffusion-zone-diameter group members) to tetracycline and sulfisoxazole, and low susceptibility to sulfisoxazole alone. Each of these three groups could be further divided into two subgroups: high and low susceptibilities to the remaining antimicrobial agents. Grouping was also evident for multidrug resistance when observations were labeled with the numbers of antimicrobial agents to which resistance was present (Fig. 5). When points were labeled by source species, the clearest distinctions in the distributions of points were seen for wildlife, swine, and humans (Fig. 6), with wildlife isolates present in the high-susceptibility groups and human and swine isolates in the low-susceptibility groups.
FIG. 1.
Plot of the first two principal components resulting from PCA of all 12 antimicrobial agents (A indicates one observation, B indicates two observations, etc.), demonstrating the grouping of data points into three groups.
FIG. 2.
Plot of the first two principal components resulting from PCA of all 12 antimicrobial agents, with points from the high (1) and low (0) distribution peaks for nalidixic acid (the line indicates the division between high- and low-susceptibility groups). Isolate distribution patterns for neomycin, streptomycin, ampicillin, trimethoprim-sulfamethoxazole, cephalothin, gentamicin, chloramphenicol, ofloxacin, nalidixic acid, and nitrofurantoin were very similar; nalidixic acid was chosen as a representative case for graphical purposes.
FIG. 3.
Plot of the first two principal components resulting from PCA of all 12 antimicrobial agents, with points from the high (1) and low (0) distribution peaks for tetracycline (the line indicates the division between low- and high-susceptibility groups for nalidixic acid, and the oval indicates the high-tetracycline-susceptibility group).
FIG. 4.
Plot of the first two principal components resulting from PCA of all 12 antimicrobial agents, with points from the high (1) and low (0) distribution peaks for sulfisoxazole (the line indicates the division between low and high groups for nalidixic acid, the oval indicates the high-tetracycline susceptibility group, and the rectangle indicates high sulfisoxazole susceptibility alone, and the hexagon indicates the low-susceptibility group).
FIG. 5.
Plot of the first two principal components resulting from PCA on all 12 antimicrobial agents, with points labeled by the number of antimicrobial agents the isolate expressed resistance to (0 to 12) (the line indicates the division between low and high groups for nalidixic acid, the oval indicates the high-tetracycline susceptibility group, the rectangle indicates high sulfisoxazole susceptibility alone, and the hexagon indicates the low-susceptibility group).
FIG. 6.
Plot of the first two principal components resulting from PCA of all 12 antimicrobial agents for species selected to demonstrate differences in PCA, with points labeled as wildlife (w), swine (s), and human (*) (the line indicates the division between low- and high-susceptibility groups for nalidixic acid, the oval indicates the high-tetracycline-susceptibility group, the rectangle indicates high sulfisoxazole susceptibility alone, and the hexagon indicates the low-susceptibility group).
DA was executed for the different species groups (three-, four-, five-, and eight-species groups), using the three DA approaches (Table 3). The ARCC values for each DA model were higher than those based purely on random chance and increased as the number of classification groups decreased. When assessed by overall ARCCs, the linear and Epanechnikov models performed better than did the quadratic models. When the models' performance in identifying specific species groups was assessed, the quadratic method was the most efficient at identifying wildlife sources over all species groups (Table 3). The linear method was able to correctly identify swine and combined species groups containing swine at least 50% of the time, while the Epanechnikov model correctly identified human isolates (Table 3).
TABLE 3.
Overall rates of correct classification (ARCC) for different reductions in numbers of drugs and species classifications for three DA methods: results from cross-validation analysis
| Approach (probability due to random chance) | % of isolates correctly classified by DA method, best group for identificationa
|
||
|---|---|---|---|
| Linear | Quadratic | Epanechnikov | |
| Twelve drugs; eight species (12.5%) | 28.7,* swine (49) | 25.6, wildlife (75) | 27.5, humans (67) |
| Reduced drugs; eight species (12.5%) | 30.8, swine (50) | 27.7, wildlife (75) | 37.3,* wildlife (89) |
| Twelve drugs; six species (16.7%) | 35.0,* swine (59) | 33.1, wildlife (80) | 33.8, humans (67) |
| Reduced drugs; six species (16.7%) | 37.1, swine (57) | 35.0, wildlife (78) | 54.5,* humans (100) |
| Twelve drugs; five species (20%) | 33.6, livestockb (67) | 32.7, wildlife (83) | 42.3,* humans (100) |
| Reduced drugs, five species (20%) | 36.3, livestockb (67) | 35.4, wildlife (80) | 51.5,* wildlife (89) |
| Twelve drugs; four species (25%) | 32.6, food animalsc (82) | 36.8, wildlife (84) | 48.7,* humans (100) |
| Reduced drugs; four species (25%) | 39.0, food animalsc (80) | 40.7, wildlife (81) | 67.9,* humans (100) |
*, best model by approach; the value in parentheses following the best group for identification is the percentage of isolates correctly identified in the group.
The livestock group is a combination of beef and dairy cattle, sheep, and swine.
The food animal group is a combination of beef and dairy cattle, sheep, swine, and poultry.
A stepwise model-building approach was applied to select antimicrobial agents from the three different DA methods (Table 4) by using the ARCC as the criterion for the retention of agents. The efficiency of DA was improved by the removal of agents for models using eight-, five-, four-, and three-species groups (Table 3). The agents present in all reduced models included tetracycline, trimethoprim-sulfamethoxazole, nitrofurantoin, and cephalothin. The Epanechnikov models performed better than the linear and quadratic models did. The performance of these models in identifying specific species sources was similar to that of the full models using 12 agents, with the exception of the Epanechnikov models, which were able to correctly identify wildlife samples at rates higher than those for human samples for the eight- and five-species group models.
TABLE 4.
Antimicrobial agents present after drug reduction process for three DA models
| Antimicrobial agent | Status of agent in reduced modela
|
||
|---|---|---|---|
| Linear | Quadratic | Epanechnikov | |
| Tetracycline | X | X | X |
| Trimethoprim-sulfamethoxazole | X | X | X |
| Nitrofurantoin | X | X | X |
| Cephalothin | X | X | X |
| Neomycin | X | X | |
| Sulfisoxazole | X | X | |
| Gentamicin | X | X | |
| Chloramphenicol | X | X | |
| Ofloxacin | X | X | |
| Nalidixic acid | X | X | |
| Ampicillin | X | ||
| Streptomycin | X | ||
An “X” indicates the presence of the agent in the model.
Decision rules generated from fecal isolates were applied to E. coli isolates from 230 environmental samples with known sources (Table 5). The linear models performed better than the quadratic and Epanechnikov models. In general, when they were used for environmental samples, there was little difference between the performance of the 12-agent DA models and that of the reduced agent models.
TABLE 5.
Rates of correct classification (ARCC) of environmental samples, based on known source locations, for different reductions in numbers of species classifications and drugs for three DA methods: results from cross-validation analysis using DA training rules from fecal samples
| Approach | % of isolates classified in agreement, best group for identificationa
|
||
|---|---|---|---|
| Linear | Quadratic | Epanechnikov | |
| Twelve drugs; eight species | 26.0, swine (39) | 18.7, swine (42) | 22.5, dairy cattle (39) |
| Reduced drugs; eight species | 25.4, swine (34) | 19.8, swine (37) | 17.1, dairy cattle (39) |
| Twelve drugs; six species | 37.9, ruminantsb (56) | 28.9, swine (50) | 31.0, ruminantsb (66) |
| Reduced drugs; six species | 36.9, ruminantsb (58) | 30.2, swine (50) | 30.0, swine (50) |
| Twelve drugs; five species | 40.2, livestockc (70) | 26.3, equine (29) | 33.6, livestockc (56) |
| Reduced drugs; five species | 39.8, livestockc (69) | 27.2, livestockc (31) | 30.4, equine (41) |
| Twelve drugs; four species | 57.2, food animalsd (85) | 32.7, food animalsd (36) | 43.1, food animalsd (57) |
| Reduced drugs; four species | 57.4, food animalsd (85) | 35.4, food animalsd (35) | 38.1, equine (41) |
The value in parentheses following the best group for identification is the percentage of isolates correctly identified in the group.
The ruminant group is a combination of beef and dairy cattle and sheep.
The livestock group is a combination of beef and dairy cattle, sheep, and swine.
The food animal group is a combination of beef and dairy cattle, sheep, swine, and poultry.
Finally, reduced model decision rules for the different species groups were applied to 26 surface water samples (Table 6). The majority of isolates were classified as being from food-producing animals when linear DA was used, while the majority of isolates were classified as wildlife isolates by quadratic and Epanechnikov DA. There were three (11.5%) water samples that were consistently identified as wildlife in origin, regardless of the DA method or number of species groups, and one (3.9%) sample was identified as equine by the linear and quadratic methods. When eight-species groups were used, dairy cattle were the most commonly identified food-producing animals regardless of DA method, with one individual isolate classified as being from dairy cattle by all three methods. No samples were classified as human in origin.
TABLE 6.
Rates of classification of 26 water samples, for eight species classifications and reduced drugs for three DA methods: results from cross-validation analysis
| Species group | Species | Percent classified by method
|
||
|---|---|---|---|---|
| Linear | Quadratic | Epanechnikov | ||
| Eight | Beef | 19.2 | 0 | 0 |
| Dairy | 26.9 | 7.7 | 15.4 | |
| Sheep | 19.2 | 3.9 | 0 | |
| Swine | 3.9 | 0 | 0 | |
| Poultry | 7.7 | 0 | 3.9 | |
| Equids | 11.5 | 11.5 | 11.5 | |
| Wildlife | 11.5 | 76.9 | 69.2 | |
| Humans | 0 | 0 | 0 | |
| Six | Ruminantsa | 65.4 | 3.9 | 3.9 |
| Swine | 3.9 | 0 | 0 | |
| Poultry | 7.7 | 0 | 3.9 | |
| Equids | 11.5 | 15.4 | 11.5 | |
| Wildlife | 11.5 | 80.8 | 80.8 | |
| Humans | 0 | 0 | 0 | |
| Five | Livestockb | 73.1 | 3.9 | 0 |
| Poultry | 3.9 | 0 | 3.9 | |
| Equids | 11.5 | 15.4 | 11.5 | |
| Wildlife | 11.5 | 80.8 | 84.6 | |
| Humans | 0 | 0 | 0 | |
| Four | Food animalsc | 76.9 | 3.9 | 3.9 |
| Equids | 11.5 | 15.4 | 11.5 | |
| Wildlife | 11.5 | 80.8 | 84.6 | |
| Humans | 0 | 0 | 0 | |
The ruminant group is a combination of beef and dairy cattle and sheep.
The livestock group is a combination of beef and dairy cattle, sheep, and swine.
The food animal group is a combination of beef and dairy cattle, sheep, swine, and poultry.
DISCUSSION
In this study, DA was performed with the goal of optimizing decision rules for use in classifying fecal E. coli isolates by species group, using data on antimicrobial susceptibility. As the results of this study indicate, this technique can be used for this purpose, but the methods used to develop the DA result in decision rules that differ significantly in their performance.
Cost is an important consideration in developing a library of known-source bacterial isolates from which to derive decision rules based on patterns of antimicrobial resistance. It was cost prohibitive to obtain the recommended six to seven isolates per sample for determining the representative E. coli population within a sample. The purpose of using the known-source samples was not to determine the representative E. coli population within each fecal or environmental sample but to determine the representative E. coli population within the species group. Taking a single confirmed E. coli isolate from a very large number of individuals in the population will achieve this objective better than will obtaining multiple isolates from a very limited number of members of the population and thereby enhance the validity of our findings. If the goal of using DA with antimicrobial resistance profiles of enteric bacteria is to identify a source of fecal pollution, it is important to ensure that bacterial isolates are collected from all potential sources of fecal pollution. In this phase of our study, we were unable to collect sufficient numbers of isolates from humans or companion animals (23), so the current version of our DA is limited for use with surface water samples. Despite this limitation, the exercise of refining the DA process for the isolates available in this study yielded insights into the development of optimized decision rules.
Geographic and temporal variations in antimicrobial resistance must be taken into consideration when samples are collected for use with DA. Antimicrobial resistance patterns are known to differ by location and time due to differences in selection pressure (21), and these differences may have contributed to the reduced ARCCs found in this study. It has been demonstrated with samples from multiple regions that there is little variation in patterns of antimicrobial resistance within a 12-month period, so any comparison of resistance patterns within a year are valid (29). In other studies, lower ARCC values were reported when samples were taken from larger numbers of species from more diverse locations, with consequently lower homogeneity levels of study populations (11). Given these results, it is recommended that DA decision rules not be applied outside a study's sampling area and time frame, and comparisons of different decision rules generated from different locations at different times cannot determine whether one analysis is more accurate than another. Applying the methods used in this study to another known-source library would provide additional information on the utility of the development process for this DA model, and results from those libraries could be compared to determine the degree of difference between locations and times. This form of analysis will provide us with the information needed to make assessments of the generalizability of results gained from this approach. The use of molecular techniques, including the ribotyping of E. coli isolates, has been suggested for use in DA for the determination of fecal pollution sources (3, 5, 21), as genetic profiles are less susceptible to localized selection pressures than are antimicrobial resistance patterns (21), which may make decision rules developed with these data more useful on a broader geographic and temporal scale.
The bimodal distribution of diffusion zones has been reported by other investigators (2, 4, 25) and has been attributed to various causes, including a titration effect when dilution techniques are used (2), genetic differences in populations reflecting the presence of new bacterial strains (4), and the acquisition of resistance (17). Since these samples were not collected prospectively from clearly identified sources, it was not possible to determine whether any active shifting of susceptibility is occurring in the regional E. coli population. Breaks in the observed bimodal distribution of the majority of antimicrobial agents in this study did not coincide with currently established resistance breakpoints, with the NCCLS breakpoints (19) falling in the lower peak ranges of these bimodal distributions. While these resistance breakpoints have clinical significance and provide a clear standard with which tests can be interpreted, our results suggest that the simple identification of an isolate as resistant or susceptible may not provide sufficient information for DA or other multivariate tools for the identification of isolate sources.
The PCA of the E. coli isolates from this study demonstrated significant patterns in the antimicrobial disk diffusion zone data, and the groupings of data points labeled by source species (Fig. 6) indicated that DA would be capable of distinguishing sources based on antimicrobial susceptibility. The clear patterns seen for wildlife (in the generally susceptible group), humans (in the generally reduced-susceptibility group), and swine (in both tetracycline and tetracycline/sulfisoxazole reduced-susceptibility groups) were reflected in the power of different DA approaches to distinguish these species (alone or in species groups) from other species. In studies comparing levels of resistance between species, swine have been reported to have higher levels of resistant isolates than other livestock species (30). On the other hand, wildlife should have far lower levels of exposure to antimicrobial agents than intensively managed food-producing livestock and, consequently, should harbor lower levels of resistant bacteria.
Different levels of classification were used for animal species groups in this study, based on potential exposure to antimicrobial agents through common livestock husbandry practices. One of the underlying hypotheses that allows patterns of antimicrobial resistance to be used to identify source species of enteric bacteria is that exposure to antimicrobial agents has been associated with the development of resistance (22, 24) and the different management practices for different species of domestic animals (e.g., dairy cattle, swine, and horses) will influence the types of antimicrobial agents a given species is exposed to. One important area of difference is the type of agents that are legal for use with a given species; for example, the use of nitrofuran is prohibited for food animals, but it can be used for companion animals. How certain antimicrobial agents are used is also important: the use of some agents at subtherapeutic levels (for growth promotion) is believed by some researchers to enhance the selection of resistant bacteria more than the therapeutic use of antimicrobials in response to clinical disease does (26). A reduction in the number of potential classification groups in the discriminant function improved the rates of correct classification, which has also been reported by other investigators (8, 28). However, users of the system need to weigh the benefits of improved model performance (ARCC) versus loss of source specificity, given the low rates of species-specific correct classification.
While DA may not be capable of identifying specific sources of fecal contamination, it can be used to improve current methods of source tracking by focusing more expensive source tracking methods on specific bacterial isolates. One approach would be to use DA in a “serial testing” approach by using the DA models with reduced species groups to include or exclude whole species groups (e.g., by using the five-species-group reduced-agents Epanechnikov model [Table 3] to identify wildlife sources). The results of this testing, coupled with additional information known about the area (e.g., the presence or absence of different kinds of animals in the area where the sample was collected), can be evaluated to determine whether more specific methods should be used. In this way, expensive yet precise tools, such as ribotyping (3, 5, 21), can be applied only to isolates where there is a higher suspicion that they come from a specific animal species. When the method is used in this way, the minimum acceptable correct classification rate for a source would not need to be extremely high, and models with ARCC values of over 50% would still be of use as a screening tool.
The differences in the performance of different types of DA (linear, quadratic, and nonparametric) may be reflective of the nature of the distributions of the diffusion zones in this study. One of the requirements of DA is that the data entering the analysis be normally distributed (15), which is not characteristic of our data. Nonparametric DA was used, and different techniques (linear, quadratic, and Epanechnikov) were utilized to determine how each performed with our bimodal distribution data. In general, the linear models were better at identifying isolates from swine and species groups containing swine (livestock and food animals), while the quadratic models were better at identifying wildlife samples. The Epanechnikov model using all 12 antimicrobial agents was better at identifying human isolates, while the models with reduced antimicrobial agents were better at identifying human and wildlife samples. Given these differences, the choice of a DA model may depend on the ultimate purpose of susceptibility testing: if the purpose of testing unknown samples is to determine whether they are from human sources, the Epanechnikov model would provide the best performance. The quadratic model would be most useful for identifying wildlife as a source of fecal contamination. However, if the purpose of testing is to determine whether the source of fecal contamination comes from a specific domestic animal species, such as swine, the linear models would be preferred. Expanding the source library of isolates to enter into the DA should also improve model performance and may highlight the strengths of different model approaches for the identification of different species groups.
The model-building approach was undertaken to attempt to improve the performance of the decision rules by reducing the number of agents entering the analysis (15, 27) and did result in better model performance for the identification of fecal isolates (Table 3). Reducing the numbers of feature variables can be helpful to the DA process, since including too many variables can harm the performance of the DA in situations with smaller sample sizes (3, 15). In a study by Beharav and Nevo (3), a stepwise model-building approach was conducted by using each variable's Wilks lambda statistic at a P value of ≤0.05 as the criterion for factor inclusion and/or retention. This approach was tested with our data, but the resulting models had lower ARCC values than did models generated by using ARCC as the criterion for factor inclusion/retention. As found in this earlier study (3), removing agents from the models improved the ARCC for all species classifications under cross-validation (Table 3). Given the improvement in ARCC found when the number of antimicrobial agents used in the analysis was reduced, agents entered into the DA should be selected for their abilities to distinguish between source species groups. By reducing the number of tests being conducted, limiting the number of agents will also reduce the costs of antimicrobial resistance testing. Further research is needed to select a panel of antimicrobials that best distinguishes between source species.
This study found that linear DA of antimicrobial resistance profiles assigned the majority of surface water samples in this study to dairy cattle and food-producing animals (Table 6), which is supported by other studies that have reported that the majority of surface water isolate sources classified through DA were from cattle (27). The reduced ability of DA to correctly classify environmental samples (Table 5) and the differences in results seen between the linear models and the quadratic and Epanechnikov models suggest caution in the interpretation of these results regarding cattle. However, the consistency in the classification of the one equine and three wildlife samples, regardless of method and number of agents, indicates a higher level of confidence in the classification of these isolates.
Conclusions.
Based upon the findings of this study, DA of antimicrobial resistance profiles can be used as a valid technique for microbial source identification as long as decision rules generated in the process are developed carefully with the goal of improving ARCC for the known-source isolates. The first consideration before performing the DA is to minimize the imbalance of numbers between different classification groups in the known-source library. This can be achieved through the use of targeted sampling to ensure that numbers of bacterial isolates entering the DA will be balanced. Next, the rates of correct classification by the DA should be viewed in terms of the relative contributions of random chance and the true discriminatory power of the DA. Any methods applied to the DA to improve the ARCC should be conducted to specifically increase the true discriminatory power of the DA, rather than simply improving the overall ARCC. Finally, selectively reducing the number of potential species classifications and the number of antimicrobial agents entering the analysis can improve the performance of DA.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.American Public Health Association. 1998. Standard methods for examination of water and waste water, 20th ed. American Public Health Association, Inc., Washington, DC.
- 2.Babini, G. S., M. Yuan, L. M. C. Hall, and D. M. Livermore. 2003. Variable susceptibility to piperacillin/tazobactam amongst Klebsiella spp. with extended-spectrum β-lactamases. J. Antimicrob. Chemother. 51:605-612. [DOI] [PubMed] [Google Scholar]
- 3.Beharav, A., and E. Nevo. 2003. Predictive validity of discriminant analysis for genetic data. Genetica 119:259-267. [DOI] [PubMed] [Google Scholar]
- 4.Blanc, D. S., M. J. Struelens, A. Deplano, R. De Ryck, P. M. Hauser, C. Petignat, and P. Francioli. 2001. Epidemiological validation of pulsed-field gel electrophoresis patterns for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson, A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dufour, A. P., E. R. Strickland, and V. J. Cabelli. 1981. Membrane filter method for enumerating Escherichia coli. Appl. Environ. Microbiol. 41:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graves, A. K., C. Hagedorn, A. Teetor, M. Mahal, A. M. Booth, and R. B. Reneau, Jr. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31:1300-1308. [DOI] [PubMed] [Google Scholar]
- 8.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discrimination among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagedorn, C., S. L. Roberson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagedorn, C., J. B. Crozier, K. A. Mentz, A. M. Booth, A. K. Graves, N. J. Nelson, and R. B. Reneau, Jr. 2003. Carbon source utilization profiles as a method to identify sources of faecal pollution in water. J. Appl. Microbiol. 94:792-799. [DOI] [PubMed] [Google Scholar]
- 11.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, R. A., and D. W. Wichern. 1992. Applied multivariate statistical analysis, 3rd ed. Prentice-Hall, Englewood Cliffs, NJ.
- 13.Kaspar, C. W., and J. L. Burgess. 1990. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 36:891-894. [DOI] [PubMed] [Google Scholar]
- 14.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLachlan, G. J. 1992. Discriminant analysis and statistical pattern recognition. Wiley, New York, NY.
- 16.Morrison, D. F. 1990. Multivariate statistical methods, 3rd ed. McGraw-Hill Publishing Co., New York, NY.
- 17.Moyaert, H., E. M. De Graef, F. Haesebrouk, and A. Decostere. 2006. Acquired antimicrobial resistance in the intestinal microbiota of diverse cat populations. Res. Vet. Sci. 81:1-7. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6. NCCLS, Wayne, PA.
- 19.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing; 9th informational supplement. NCCLS document M100-S9. NCCLS, Wayne, PA.
- 20.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple antibiotic resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescott, J. F., J. D. Baggot, and R. D. Walker. 2000. Antimicrobial therapy in veterinary epidemiology, 3rd ed. Iowa State University Press, Ames, IA.
- 23.Sayah, R., J. B. Kaneene, Y. Johnson, and R. Miller. 2005. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic and wild animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, T. M, J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, P., and M. Hiney. 2005. Towards setting breakpoints for oxolinic acid susceptibility of Aeromonas salmonicida using distribution of data generated by standard test protocols. Aquaculture 250:22-26. [Google Scholar]
- 26.Van den Bogaard, A. E., N. London, C. Driessen, and E. E. Stobberingh. 2001. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47:763-771. [DOI] [PubMed] [Google Scholar]
- 27.Wiggins, B. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburh, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, Z., F. C. Michel, G. Hansen, T. Wittum, and M. Morrison. 2005. Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 71:6926-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]