Abstract
A fundamental question in microbial oceanography concerns the relationship between prokaryote diversity and biogeochemical function in an ecosystem context. We combined bromodeoxyuridine (BrdU) magnetic bead immunocapture and PCR-denaturing gradient gel electrophoresis (BUMP-DGGE) to examine phylotype-specific growth in natural marine assemblages. We also examined a broad range of marine bacterial isolates to determine their abilities to incorporate BrdU in order to test the validity of the method for application to diverse marine assemblages. We found that 27 of 29 isolates belonging to different taxa could incorporate BrdU. BUMP-DGGE analysis revealed phylogenetic affiliations of DNA-synthesizing, presumably actively growing bacteria across a eutrophic to mesotrophic transect in the Inland Sea of Japan. We found that the BrdU-incorporating (growing) communities were substantially different from the total communities. The majority (34/56) of phylotypes incorporated BrdU and were presumably growing, and these phylotypes comprised 10 alphaproteobacteria, 1 betaproteobacterium, 11 gammaproteobacteria, 11 Cytophaga-Flavobacterium-Bacteroides group bacteria, and 1 unclassified bacterium. All BrdU-responsive alphaproteobacteria were members of the Rhodobacterales, suggesting that such bacteria were dominant in the growing alphaproteobacterial populations in our samples. The BrdU-responsive gammaproteobacteria belonged to the Oceanospirillales, the SAR86 cluster, the Pseudomonadales, the Alteromonadales, and the Vibrionales. Thus, contemporaneous cooccurrence of diverse actively growing bacterial taxa was a consistent pattern in our biogeochemically varied study area.
Bacteria in seawater play important roles in the ocean's food webs. Their activities and responses to organic substrates significantly influence the flux of organic matter and oceanic biogeochemical cycles (3, 4). Because only a small fraction of bacteria in any seawater sample can be isolated and cultivated, culture-independent methods employing molecular approaches have been used to great advantage during the last two decades to study the natural assemblages of marine bacteria (11, 19, 39). These and other studies have revealed unexpected diversity and dynamics of bacterial community structure in seawater (8, 16, 47). However, the great challenge remains to relate diversity to ecological function and biogeochemical activities of bacteria. In recent studies workers have combined fluorescence in situ hybridization (FISH) with microautoradiography and found differences in the uptake of specific organic substrates by different phylogenetic groups of bacteria (7, 8, 40). Stable-isotope probing has enabled PCR-based DNA analysis of bacteria that incorporate specific substrates (45).
Bromodeoxyuridine (BrdU), a halogenated nucleoside that can serve as a thymidine (TdR) analog, has been widely used as an alternative to tritiated TdR ([3H]TdR) incorporation to label proliferating cells in cell biology (2, 59). BrdU incorporated into de novo DNA can be detected using anti-BrdU monoclonal antibody. The applicability of BrdU incorporation as an alternative to [3H]TdR incorporation for bulk measurement of bacterial secondary production in seawater has been demonstrated (22, 38, 50). Employing antibodies conjugated with magnetic beads, BrdU-labeled DNA can be separated from total DNA, and this immunocapture technique and 16S rRNA gene fingerprinting or sequencing have been used to reveal some of the growing bacterial species in both soil and lake water (1, 5, 58, 60). At the single-cell level, actively growing bacteria can be visualized by using anti-BrdU conjugated with a fluorescent label or an enzymatic reporter (23, 42, 58). In one study of coastal seawater, a significant portion of the 4′,6′-diamidino-2-phenylindole (DAPI)-stained bacteria (47%) became BrdU positive in 5 h (23). Pernthaler et al. (42) combined the BrdU immunofluorescence method with 16S rRNA FISH and applied it to North Sea waters. They found that the actively growing community included bacteria belonging to the Roseobacter, Alteromonas, and SAR86 clades. However, detailed community structures and identities of actively growing bacteria in seawater or even in soils and lake water have not been determined yet.
A fingerprinting approach using reverse-transcribed 16S rRNA fragments has been used to infer potentially active bacterial populations in aquatic environments (14, 34, 48, 53, 57). A comparison of DNA- and RNA-based fingerprints provides insight into total bacterial communities and their potentially active fractions. Recent work using DNA- and RNA-derived clone libraries indicated that the RNA-derived library included distinct phylotypes which were not detected in the DNA library, suggesting that RNA-based analysis could be used to characterize actively metabolizing but less abundant bacteria in a community (33). This concept is based on the assumption that the number of ribosomes per cell is a measure of cell activity, including the growth rate. However, the RNA contents of species vary greatly (17, 28), which makes interpretation problematic for natural mixed-species communities. Also, differences between DNA- and RNA-derived fingerprints may be attributed to random priming and cDNA synthesis in reverse transcription-PCR analyses.
In this study, we used combined BrdU magnetic bead immunocapture and PCR-denaturing gradient gel electrophoresis (BUMP-DGGE) to investigate the community structures and phylogenetic identities of actively growing bacteria across a eutrophic to mesotrophic transect. This approach is similar to the method developed by Borneman (5) for soil samples. As one of the essential assumptions for the validity of the method is that all taxa in the microbial communities in a sample are able to incorporate BrdU into DNA, we also examined a broad range of marine bacterial isolates to determine whether they could incorporate BrdU.
MATERIALS AND METHODS
Isolation and phylogenetic identification of marine bacteria.
To determine whether bacteria could incorporate BrdU, marine isolates were obtained from various environments, such as coastal seawater, sea ice, marine macroalgae, and dinoflagellate cultures (Table 1). Cultures were grown and bacteria were isolated using 0.5× ZoBell 2216E medium. Isolates were analyzed by performing PCR with partial 16S rRNA gene sequences, followed by DGGE for phylogenetic typing (37). Isolated strains that yielded PCR amplicons which migrated at the same position on a gel were grouped into one phylotype. The phylogenetic affiliation of each representative isolate was examined by partially sequencing the 16S rRNA gene. Bacterial 16S rRNA genes were amplified by colony PCR using universal forward primer 27F and bacterium-specific reverse primer 1492R. After PCR products were treated with a QIAquick PCR purification kit (QUIAGEN Inc., Valencia, CA), bidirectional sequencing was performed with a Dual CyDye terminator sequencing kit and a Long-Read Tower automated DNA sequencer (Amersham Biosciences Corp., Piscataway, NJ), using primers complementary to the bases at positions 341 to 358 and 517 to 534. Sequences were aligned with known sequences in the DDBJ (DNA Data Bank of Japan) database using BLAST (Basic Local Alignment Search Tool). Since we found no betaproteobacteria, one representative strain of this taxon (Chromobacterium violaceum IAM12470 [= ATCC 12472]) was obtained from the IAM culture collection at the Institute of Molecular and Cellular Biosciences, The University of Tokyo.
TABLE 1.
BrdU incorporation by marine bacterial isolates
| Most closely allied taxon | Isolate | Source | Accession no. | BrdU antibody detection
|
|
|---|---|---|---|---|---|
| Whole cell | DNA dot blot | ||||
| Alphaproteobacteria | |||||
| Erythrobacter | KUR27 | Seawater | AB266542 | + | + |
| Erythrobacter | OB10 | Seawater | AB266553 | − | + |
| Erythrobacter | ATKR 01 | Dinoflagellate | AB266563 | + | + |
| Sulfitobacter | OB4 | Seawater | AB266552 | + | + |
| Sulfitobacter | OB14 | Seawater | AB266554 | + | + |
| Thalassospira | TAMA2 | Dinoflagellate | AB266548 | + | + |
| Gammaproteobacteria | |||||
| Alteromonas | KUR4 | Seawater | AB266538 | + | + |
| Cobetia | KUR58 | Seawater | AB266544 | − | + |
| Colwellia | SL5 | Sea ice | AB266558 | + | − |
| Colwellia | SL15 | Sea ice | AB266559 | + | + |
| Glaciecola | KUR9 | Seawater | AB266539 | + | + |
| Marinobacter | KUR64 | Seawater | AB266546 | − | + |
| Pseudoalteromonas | MIDOR1 | Green algae | AB266549 | + | + |
| Pseudoalteromonas | CHAKO4 | Brown algae | AB266547 | + | NAa |
| Pseudoalteromonas | OB0 | Seawater | AB266551 | + | + |
| Pseudoalteromonas | KUR2 | Seawater | AB266537 | + | + |
| Pseudomonas | KUR18 | Seawater | AB266541 | − | + |
| Psychrobacter | KUR35 | Seawater | AB266543 | − | + |
| Vibrio | RYO1 | Green algae | AB266555 | + | + |
| Betaproteobacteria | |||||
| Chromobacterium violaceum | IAM12470 (= ATCC 12472) | Freshwater | AE016825 | + | NA |
| CFB group | |||||
| Algibacter | OB15 | Seawater | AB266550 | + | + |
| Algoriphagus | SL1 | Sea ice | AB266556 | + | − |
| Flavobacterium | SL8 | Sea ice | AB266561 | − | − |
| Maribacter | TAMA1 | Dinoflagellate | AB266562 | + | − |
| Maribacter | SL4 | Sea ice | AB266557 | + | + |
| Pibocella | SL14 | Sea ice | AB266560 | − | − |
| Low-G+C-content gram-positive bacteria | |||||
| Bacillus | KUR61 | Seawater | AB266545 | − | + |
| Actinobacteria | |||||
| Microbacterium | KUR17 | Seawater | AB266540 | − | + |
| Microbacterium | ATKR 03-1 | Dinoflagellate | AB266564 | + | + |
NA, not analyzed.
Incubation of isolates with BrdU.
Isolates were grown in SWM medium at room temperature for 1 to 2 days. The SWM medium used for BrdU labeling of cultured isolates was prepared by adding 0.0058% (wt/vol) Casamino Acids, 0.69 mM glucose, 0.30 mM NH4Cl, 0.25 mM NaH2PO4, 0.2 μM ferric citrate, and 0.2 μM EDTA to seawater filtered with Whatman GF/F filters (G. Steward and F. Azam, unpublished data) and was modified by adding 0.001% (wt/vol) Bacto peptone (Difco). The seawater was replaced with distilled water for culturing and labeling of C. violaceum. An aliquot of each culture was inoculated into fresh SWM medium to obtain an optical density at 600 nm of 0.01 to 0.02. After 30 min of incubation at 23°C, 200 nM (final concentration) BrdU was added to the tubes (9 ml). Control tubes received 200 nM TdR. The cultures were incubated for an additional 12 h, and then the contents were pelleted by centrifugation at 20,000 × g for 5 min. The supernatants were discarded, and the pelleted cells were fixed with 4% paraformaldehyde, washed with phosphate-buffered saline (PBS), and resuspended in PBS. Each cell suspension was mixed with an equal volume of 100% ethanol and stored at −15°C.
Detection of incorporated BrdU.
BrdU was detected by two immunochemical methods, one employing immunocytochemistry with whole-cell preparations and the other employing a chemiluminescence immunoassay with genomic DNA dot blots. Immunocytochemical fluorescence staining and assessment by fluorescence microscopy were performed by using the procedure described previously (23). Aliquots (5 μl) of bacterial cell suspensions were spotted onto 6-mm-diameter wells of Teflon printed glass slides (Electron Microscopy Science, Hatfield, PA) and air dried for 1 h. After the cells were permeabilized and the double-stranded DNA was denatured, anti-BrdU antibody was applied to detect incorporated BrdU. The antibody signal was amplified by catalyzed reporter deposition and was observed by using an Olympus BX51 epifluorescence microscope (total magnification, ×1,000; Olympus Corp., Tokyo, Japan). Also, a chemiluminescence immunoassay with genomic DNA dot blots was performed by using the procedure described previously (22). Aliquots (100 μl) of bacterial cell suspensions were filtered through a nylon membrane (pore size, 0.22 μm; MagnaGraph; GE Water & Process Technologies, Trevose, PA) with a 96-well-format dot blotter (Minifold I; Schleicher & Schuell, Dassel, Germany). After the cells were lysed, cross-linked DNA on the membrane was detected with anti-BrdU antibody. The chemiluminescence signal was developed with an enzyme-conjugated antibody and then detected by using a cooled charge-coupled device imaging system (VersaDoc5000; Bio-Rad Laboratories, Hercules, CA).
Field sampling and BrdU labeling.
Coastal seawater was collected at nine stations in the western part of the Inland Sea of Japan from the R/V Toyoshio-Maru in July 2002 and 2003 (ST1 at 34°19′N, 132°22′E; ST2 at 34°09′N, 132°18′E; ST3 at 34°01′N, 132°30′E; ST4 at 33°40′N, 132°23′E; ST5 at 33°34′N, 132°14′E; ST7 at 33°24′N, 132°14′E; ST8 at 33°06′N, 132°14′E; ST9 at 33°17′N, 132°27′E; and ST10 at 32°15′N, 132°00′E). Depth profiles for salinity and temperature at each site were obtained with a Sea-Bird SBE9plus conductivity-temperature-depth profiler (Sea-Bird Electronics, Inc., Bellevue, WA). Seawater was collected from a depth of 5 m with a Van Dorn bottle and was filtered with 100-μm nylon mesh to remove large zooplankton. Samples were diluted (1:3) with 0.22-μm filtered seawater collected from the same site to reduce micro- and nanozooplankton grazing on bacteria during incubation for BrdU labeling. BrdU was added to the seawater at a final concentration of 1 μM, and samples were incubated in a 10-liter dark plastic bottle for 1 or 5 h at the in situ temperature ± 2°C. Bacteria were collected on 0.22-μm cartridge filters (Sterivex GS; Millipore Corp., Billerica, MA).
To determine the chlorophyll a concentration, duplicate seawater samples were filtered (pressure differential, <100 mm Hg) through Whatman GF/F filters and extracted in the dark with N,N-dimethyl formamide at 4°C for 24 h (52), and the chlorophyll a concentration was determined fluorometrically (25). Particulate matter collected on precombusted (500°C, 2 h) Whatman GF/F filters was analyzed to determine the particulate organic carbon and particulate organic nitrogen contents using a CHN analyzer (Yanaco Co., Ltd., Tokyo, Japan). Seawater filtrates obtained with Whatman GF/F filters were collected for analysis of dissolved organic carbon in precombusted glass ampoules and frozen after flame sealing until analysis with a Shimadzu TOC5000 (Shimadzu Corp., Kyoto, Japan) (51). Seawater samples fixed with 2% (final concentration) formalin were stained with DAPI, filtered onto 0.2-μm-pore-size black polycarbonate filters (Millipore), and mounted on glass slides (44). Bacterial abundance was determined by counting the bacteria in 10 fields using epifluorescence microscopy (Olympus BX-51).
DNA extraction.
DNA was extracted by using the xanthogenate-sodium dodecyl sulfate protocol described by Tillett and Neilan (54), with volume modification. Each Sterivex GS filter unit was thawed and filled with xanthogenate-sodium dodecyl sulfate buffer (1% potassium ethyl xanthogenate [Wako Pure Chemical, Osaka, Japan], 100 mM Tris-HCl [pH 7.4], 20 mM EDTA [pH 8], 1% sodium dodecyl sulfate, 800 mM ammonium acetate). After the filter housing was sealed, the unit was incubated at 70°C for 120 min. Following incubation, the filter unit was vortexed for 10 s. The lysate was eluted into 2-ml microcentrifuge tubes and then placed on ice for 30 min. Cell debris was pelleted by centrifugation at 22,000 × g for 15 min at 4°C. The supernatant was transferred to a clean 1.5-ml microcentrifuge tube and mixed with an equal volume of 100% isopropanol. After incubation at room temperature for 10 min, the precipitated DNA was pelleted by centrifugation at 22,000 × g for 20 min. The pellet was washed once with 70% ethanol, air dried, and resuspended in 50 μl of water.
BrdU magnetic bead immunocapture.
Immunocapture was performed as described by Urbach et al. (58), with slight modifications. In the procedures described below, all preparations were incubated at room temperature. Herring sperm DNA (1.25 mg ml−1 in PBS; Trevigen Inc., Gaithersburg, MD) was boiled for 1 min, quickly frozen in dry-ice ethanol, and thawed. The solution was mixed (9:1) with anti-BrdU monoclonal antibody (diluted 1:10 in PBS; Sigma-Aldrich, St. Louis, MO) and incubated for 30 min. Extracted DNA samples (0.5 to 1 μg) were boiled for 1 min, quickly frozen in dry-ice ethanol, and thawed. Each denatured sample solution was mixed with 10 μl of the herring sperm DNA-antibody mixture and incubated for 30 min. Paramagnetic beads coated with goat anti-mouse immunoglobulin G (Dynabeads; Dynal Biotech, Oslo, Norway) were washed with PBS containing acetylated bovine serum albumin (BSA) (1 mg ml−1; Nacalai Tesque, Kyoto, Japan), using a magnetic concentrator. The beads were resuspended in PBS-BSA at their initial concentration. The bead suspension (25 μl) was mixed with each sample (20 μl). After incubation for 30 min with constant agitation, the beads were washed seven times with 0.5 ml PBS-BSA. BrdU-labeled DNA captured by the beads was eluted by adding 100 μl of 1.7 mM BrdU (in PBS-BSA) and incubating the preparation for 30 min with constant agitation. Glycogen (2 μl; 20 mg ml−1; Roche Diagnostics, Mannheim, Germany) was added to the bead supernatant, and the DNA was collected by two rounds of ethanol precipitation. The precipitate was resuspended in 10 to 20 μl of water and subjected to PCR amplification and DGGE. To evaluate the specificity of immunocapture, a parallel DNA sample without BrdU labeling was processed in the same manner and subjected to PCR amplification as a negative control.
DGGE.
Bacterial 16S rRNA genes were amplified from either immunocaptured or nonimmunocaptured DNA samples by a touchdown PCR protocol (12) and then subjected to bacterial community analysis by DGGE. Nonimmunocaptured samples were analyzed before and after incubation. A bacterial primer complementary to bases at positions 341 to 358 containing a 40-bp GC clamp was used as the forward primer. A universal 17-bp primer complementary to bases at positions 517 to 534 was used as the reverse primer for the samples collected in 2002 (37). A different universal primer complementary to bases at positions 907 to 926 was used for the samples collected in 2003 (6).
The PCR products (200 to 500 ng) were loaded onto 8% polyacrylamide gels in 0.5× TAE (20 mM Tris, 10 mM acetate, 0.5 mM Na2EDTA; pH 8.2) with a denaturing gradient from 20 to 70% from the top to the bottom. Electrophoresis was performed at 85 V for 16 h at 60°C in a hot-bath DGGE unit (CBS Scientific, San Diego, CA) with a running buffer consisting of 0.5× TAE. Gels were stained for 30 min with 1× SYBR gold nucleic acid stain in 0.5× TAE (Molecular Probes, Eugene, OR) and were destained for 10 min with 0.5× TAE. The gels were then visualized and documented using a charge-coupled device camera imaging system (Printgraph AE6911CX; ATTO Corp., Tokyo, Japan). DGGE bands were excised, and the DNA was eluted overnight. The eluants were used for PCR reamplification, followed by cloning with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The presence of inserts was verified by PCR amplification using primers M13F and M13R. Clones with insertions that were the correct size were analyzed by DGGE to confirm the position of the cloned bands relative to the position of the original sample.
Cluster analysis of DGGE banding patterns.
To assess the similarity of community structures (DGGE banding patterns), the Jaccard coefficient was calculated (18), and the distance matrix was analyzed by using the between-group average linkage method for clustering with the software SPSS Base 10.0J (SPSS Inc., Chicago, IL). The presence of DGGE bands in a sample was determined by eye, and the results were scored 1 or 0 to generate a binary matrix. This matrix was used to generate a distance matrix and a dendrogram with SPSS.
Sequencing and phylogenetic analysis.
Bidirectional sequencing was performed with a Dual CyDye terminator sequencing kit using T7 primers and a Long-Read Tower automated DNA sequencer (Amersham Biosciences Corp.). Sequences were aligned with known sequences in the DDBJ database using BLAST. Phylogenetic relationships were inferred by pairwise comparison and the neighbor-joining method using CLUSTAL W (55). Phylogenetic trees were edited using Treeview (41).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA gene determined in this study have been deposited in the DDBJ nucleotide sequence database under accession numbers AB266537 to AB266564. The nucleotide sequences of DGGE bands have been deposited in the DDBJ nucleotide sequence database under accession numbers AB265970 to AB266026.
RESULTS
BrdU incorporation by marine bacterial isolates.
We obtained 98 isolates, which grouped into 28 phylotypes based on DGGE migration of 16S rRNA gene partial fragments. The abilities of 28 marine isolates plus one betaproteobacterium from a culture collection to incorporate BrdU were examined. The phylogenetic groups and affinities with known genera of our isolates were determined by sequencing 16S rRNA genes (Table 1). These isolates included 6 alphaproteobacteria, 13 gammaproteobacteria, 6 Cytophaga-Flavobacterium-Bacteroides (CFB) group bacteria, 2 low-G+C-content gram-positive bacteria, and 1 actinobacterium. Twenty-seven of the 29 isolates incorporated BrdU under the culture conditions that we employed (Table 1).
Environmental factors.
The near-surface salinity increased along our north-south transect, with lower salinity observed near the Ohta River runoff in Hiroshima Bay in the northern part of the sampling area, while the temperature did not exhibit such a pattern (see Table S1 in the supplemental material). The concentration of chlorophyll a varied from 0.35 to 14 mg m−3, and the concentrations at ST1 were high (11 and 14 mg m−3). The particulate organic carbon and particulate organic nitrogen concentrations were also highest at ST1 and lowest at the southern end of transect. The same pattern was observed for the dissolved organic carbon concentration. The total bacterial counts ranged from 3.3 × 105 to 4.6 × 106 cells ml−1, and a regular pattern was not observed.
BUMP-DGGE analysis.
To confirm the specificity of immunocapture, control samples were processed like the BrdU-treated samples and then subjected to PCR. The control samples were obtained from parallel incubation mixtures to which BrdU was not added, as well as mixtures amended with BrdU. No PCR products were obtained with the control unlabeled samples, while PCR products that were the appropriate size (ca. 200 or 500 bp) were obtained with the BrdU-treated samples.
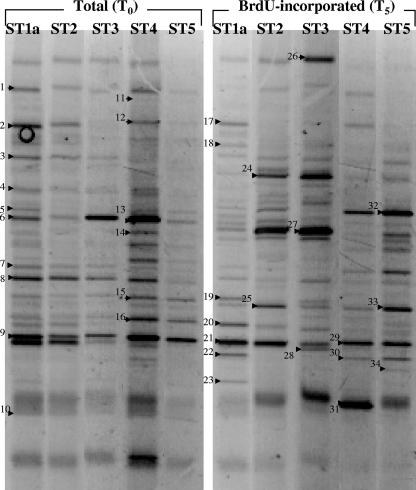
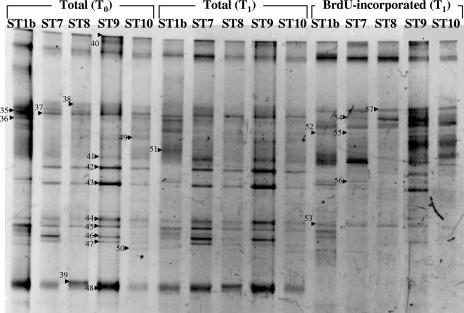
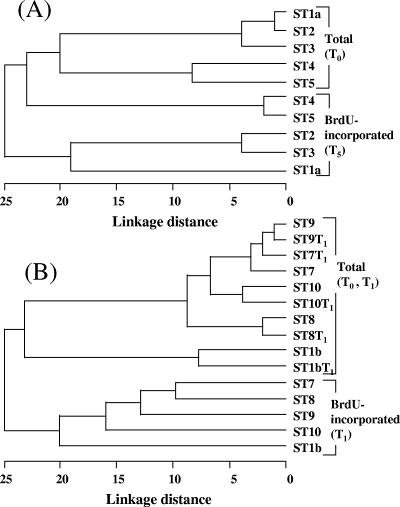
To rapidly compare the growing fraction of the bacterial community with the total bacterial community, we performed a DGGE analysis of the 16S rRNA genes. BrdU labeling and DGGE analysis of 16S rRNA gene fragments produced banding patterns for the BrdU-incorporating bacterial community that were used for comparisons with the total bacterial community for both 2002 and 2003 samples (Fig. 1 and 2). In both transects the bacterial community structure varied, and the variation was greater in 2003 (Fig. 1 and 2). Two distinct communities were observed for the 2002 transect; the communities in Hiroshima Bay (ST1a, ST2, and ST3) and the outer samples (ST4 and ST5) exhibited about 80% similarity, as determined by using Jaccard's index. When the total bacterial community was compared to the BrdU-incorporating phylotypes, the levels of dissimilarity were greater, and the differences between the community at ST1a, ST2, and ST3 and the community at ST4 and ST5 were reinforced. In 2003, a similar pattern was observed; the community at the Hiroshima Bay station (ST1b) was different from the community at the remaining stations. In 2003, the total communities at the beginning and at the end of the incubations were similar (<10% difference) as determined by a pairwise analysis of samples from the same site; however, these communities differed by 25% or more from the BrdU-incorporating communities at the same site (Fig. 2 and 3B). Comparisons of DGGE bands revealed that 15 to 30 phylotypes were present at each location and that 47 to 76% of these phylotypes were present in BrdU-incorporating fractions (Table 2).
FIG. 1.
Structures of total and active bacterial communities in the Inland Sea of Japan in July 2002. Seawater was incubated with 1 μM BrdU for 5 h to label actively growing bacteria. The PCR products amplified from BrdU-containing and immunocaptured DNA [BrdU-incorporated (T5)] were compared with the products amplified from total community DNA [Total (T0)] by DGGE analysis. Each excised, cloned, and sequenced band is numbered. The relationships of excised band sequences to other sequences in the DDBJ database are shown in Table 3.
FIG. 2.
Structures of total and active bacterial communities in the Inland Sea of Japan in July 2003. Seawater was incubated with 1 μM BrdU for 1 h to label actively growing bacteria. The PCR products amplified from BrdU-containing and immunocaptured DNA [BrdU-incorporated (T1)] were compared with the products amplified from total community DNA before [Total (T0)] and after [Total (T1)] incubation by DGGE analysis. Each excised, cloned, and sequenced band is numbered. The relationships of excised band sequences to other sequences in the DDBJ database are shown in Table 3.
FIG. 3.
Relationship between total and active bacterial community structures in the Inland Sea of Japan in 2002 (A) and 2003 (B). The structure of the total community (Total) was compared with the structure of the BrdU-incorporating community (BrdU-incorporated) before (T0) and after (T1 or T5) incubation by DGGE analysis. The tree was constructed by using the between-group average linkage method with the Jaccard coefficient distance matrix for clustering with the software SPSS Base 10.0J (SPSS Inc.).
TABLE 2.
Comparisons of total and BrdU-incorporating operational taxonomic units in the Inland Sea of Japana
| Sampling location | No. of DGGE bands
|
% of BrdU-positive bands
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (T0) | Total (T1) | BrdU | In commonb
|
Uniquec
|
|||||
| Total (T0) and BrdU | Total (T1) and BrdU | Total (T0) and BrdU | Total (T1) and BrdU | Total (T0) and BrdU | Total (T1) and BrdU | ||||
| ST1a | 16 | 17 | 4 | 29 | 59 | ||||
| ST2 | 16 | 16 | 4 | 28 | 57 | ||||
| ST3 | 13 | 17 | 4 | 26 | 65 | ||||
| ST4 | 22 | 15 | 7 | 30 | 50 | ||||
| ST5 | 15 | 16 | 6 | 25 | 64 | ||||
| ST1b | 13 | 11 | 13 | 6 | 7 | 20 | 17 | 65 | 76 |
| ST7 | 10 | 12 | 10 | 4 | 5 | 16 | 17 | 63 | 59 |
| ST8 | 11 | 10 | 7 | 3 | 4 | 15 | 13 | 47 | 54 |
| ST9 | 11 | 11 | 13 | 7 | 7 | 17 | 17 | 76 | 76 |
| ST10 | 12 | 12 | 10 | 3 | 3 | 19 | 19 | 53 | 53 |
Total (T0), total community before incubation; Total (T1), total community after incubation; BrdU, BrdU-incorporating community.
Number of bands appearing at the same position for the total community and the BrdU-incorporating community samples.
Number of unique bands found at each sampling location.
Sequencing and phylogenetic analysis of 16S rRNA gene fragments.
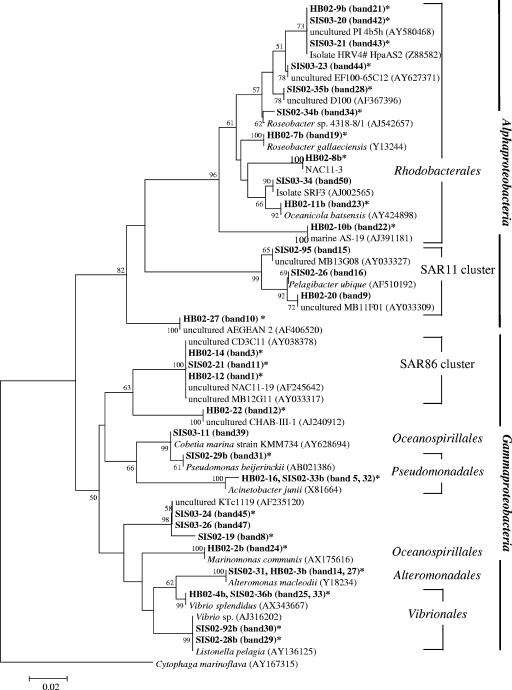
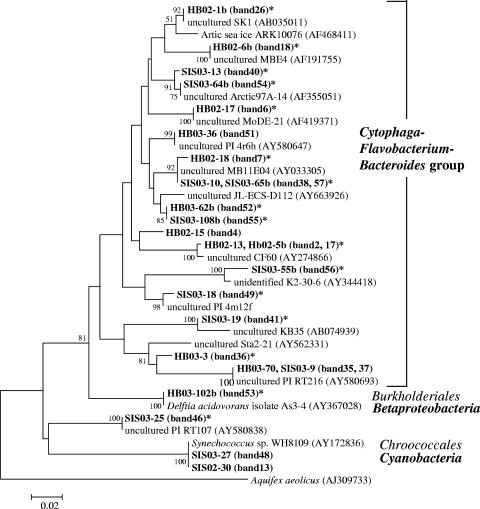
To investigate which groups were represented in the total community fractions and the BrdU-incorporating fractions, we excised 57 DGGE bands or phylotypes (34 bands in 2002 and 23 bands in 2003) (Fig. 1 and 2) from the gels and sequenced them; these phylotypes included 24 phylotypes from the BrdU-incorporating community (Table 3). We found 51 phylotypes related to 15 alphaproteobacteria, 1 betaproteobacterium, 15 gammaproteobacteria, 17 CFB group bacteria, 2 cyanobacteria, and 1 unclassified bacterium (Fig. 4 and 5). Forty of these phylotypes were found in BrdU-incorporating fractions. These phylotypes were related to 11 alphaproteobacteria, 1 betaproteobacterium, 13 gammaproteobacteria, 14 CFB group bacteria, and 1 unclassified bacterium. All of the phylotypes that were related to the Alphaproteobacteria except one unclassified phylotype were clustered in two major subgroups, the Rhodobacterales and SAR11 clusters, and all of the BrdU-incorporating phylotypes were clustered in the Rhodobacterales. The phylotypes that were related to the Gammaproteobacteria were clustered in five major subgroups, the Oceanospirillales, the SAR86 cluster, the Pseudomonadales, the Alteromonadales, and the Vibrionales, and BrdU-positive phylotypes were present in every cluster. In particular, the phylotype corresponding to Vibrio splendidus (Fig. 1, bands 25 and 33) was present at all locations from ST1a to ST5, and the signals of this phylotype at ST2 and ST5 were intense. Also, the two phylotypes corresponding to uncultured CFB group bacteria (Fig. 2, bands 40, 38, and 57) were present at all locations from ST1b to ST10, as determined by BrdU-positive bands. Almost the same phylotype (band 7) as the band 38 and 57 phylotype was found to be present at ST1, ST4, and ST5, as determined by BrdU-positive bands (Fig. 1). Eleven phylotypes did not appear in BrdU-incorporating fractions. These phylotypes included three SAR11 group bacteria, one bacterium belonging to the Rhodobacterales, one bacterium belonging to the Oceanospirillales, one unclassified gammaproteobacterium, three CFB group bacteria, and two cyanobacteria (Table 3).
TABLE 3.
Affiliations of excised DGGE band sequences with other sequences in the DDBJ database
| DGGE banda | Clone | Accession no.b | DNA fractionc | BrdUd | Phylogenetic group |
|---|---|---|---|---|---|
| From ST1a to ST5 in 2002 | |||||
| 1 | HB02-12 | AB265970 | Total | + | Gammaproteobacteria, SAR86 |
| 2 (17) | HB02-13 | AB265971 | Total | + | CFB |
| 3 | HB02-14 | AB265972 | Total | + | Gammaproteobacteria, SAR86 |
| 4 | HB02-15 | AB265973 | Total | − | CFB |
| 5 (32) | HB02-16 | AB265974 | Total | + | Gammaproteobacteria, Pseudomonadales |
| 6 | HB02-17 | AB265975 | Total | + | CFB |
| 7 | HB02-18 | AB265976 | Total | + | CFB |
| 8 | HB02-19 | AB265977 | Total | + | Gammaproteobacteria |
| 9 | HB02-20 | AB265978 | Total | − | Alphaproteobacteria, SAR11 |
| 10 | HB02-27 | AB265979 | Total | + | Alphaproteobacteria |
| 11 | SIS02-21 | AB265980 | Total | + | Gammaproteobacteria, SAR86 |
| 12 | HB02-22 | AB265981 | Total | + | Gammaproteobacteria |
| 13 | SIS02-30 | AB265982 | Total | − | Cynnobacteria |
| 14 (27) | SIS02-31 | AB265983 | Total | + | Gammaproteobacteria, Alteromonadales |
| 15 | SIS02-95 | AB265984 | Total | − | Alphaproteobacteria, SAR11 |
| 16 | SIS02-26 | AB265985 | Total | − | Alphaproteobacteria, SAR11 |
| 17 (2) | HB02-5b | AB265986 | BrdU | + | CFB |
| 18 | HB02-6b | AB265987 | BrdU | + | CFB |
| 19 | HB02-7b | AB265988 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| 20 | HB02-8b | AB265989 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| 21 | HB02-9b | AB265990 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| 22 | HB02-10b | AB265991 | BrdU | + | Alphaproteobacteria |
| 23 | HB02-11b | AB265992 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| 24 | HB02-2b | AB265993 | BrdU | + | Gammaproteobacteria, Oceanospirillales |
| 25 (33) | HB02-4b | AB265994 | BrdU | + | Gammaproteobacteria, Vibrionales |
| 26 | HB02-1b | AB265995 | BrdU | + | CFB |
| 27 (14) | HB02-3b | AB265996 | BrdU | + | Gammaproteobacteria, Alteromonadales |
| 28 | SIS02-35b | AB265997 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| 29 | SIS02-28b | AB265998 | BrdU | + | Gammaproteobacteria, Vibrionales |
| 30 | SIS02-92b | AB265999 | BrdU | + | Gammaproteobacteria, Vibrionales |
| 31 | SIS02-29b | AB266000 | BrdU | + | Gammaproteobacteria, Pseudomonadales |
| 32 | SIS02-33b | AB266001 | BrdU | + | Gammaproteobacteria, Pseudomonadales |
| 33 (25) | SIS02-36b | AB266002 | BrdU | + | Gammaproteobacteria, Vibrionales |
| 34 | SIS02-34b | AB266003 | BrdU | + | Alphaproteobacteria, Rhodobacterales |
| From ST1b to ST10 in 2003 | |||||
| 35 (37) | HB03-70 | AB266004 | Total | − | CFB |
| 36 | HB03-3 | AB266005 | Total | + | CFB |
| 37 (35) | SIS03-9 | AB266006 | Total | − | CFB |
| 38 (57) | SIS03-10 | AB266007 | Total | + | CFB |
| 39 | SIS03-11 | AB266008 | Total | − | Gammaproteobacteria, Oceanospirillales |
| 40 | SIS03-13 | AB266009 | Total | + | CFB |
| 41 | SIS03-19 | AB266010 | Total | + | CFB |
| 42 | SIS03-20 | AB266011 | Total | + | Alphaproteobacteria |
| 43 | SIS03-21 | AB266012 | Total | + | Alphaproteobacteria |
| 44 | SIS03-23 | AB266013 | Total | + | Alphaproteobacteria, Rhodobacterales |
| 45 | SIS03-24 | AB266014 | Total | + | Gammaproteobacteria |
| 46 | SIS03-25 | AB266015 | Total | + | Unclassified |
| 47 | SIS03-26 | AB266016 | Total | − | Gammaproteobacteria |
| 48 | SIS03-27 | AB266017 | Total | − | Cyanobacteria |
| 49 | SIS03-18 | AB266018 | Total | + | CFB |
| 50 | SIS03-34 | AB266019 | Total | − | Alphaproteobacteria, Rhodobacterales |
| 51 | HB03-36 | AB266020 | Total | − | CFB |
| 52 | HB03-62b | AB266021 | BrdU | + | CFB |
| 53 | HB03-102b | AB266022 | BrdU | + | Betaproteobacteria |
| 54 | SIS03-64b | AB266023 | BrdU | + | CFB |
| 55 | SIS03-108b | AB266024 | BrdU | + | CFB |
| 56 | SIS03-55b | AB266025 | BrdU | + | CFB |
| 57 | SIS03-65b | AB266026 | BrdU | + | CFB |
The number in parentheses indicates a band that had the same sequence.
Accession numbers for sequences obtained in this study.
Origin of the excised and sequenced bands. Total, total community DNA fraction; BrdU, BrdU-incorporating community DNA fraction.
Presence (+) or absence (−) of the band in samples representing BrdU-incorporating fractions.
FIG. 4.
Distribution of phylotypes found in the Inland Sea of Japan: phylogenetic tree showing the similarities of sequences from bands (bold type) to sequences from members of the Alpha- and Gammaproteobacteria. The tree was inferred by the neighbor-joining method using CLUSTAL W. The bands found in lanes representing the BrdU-incorporating community are indicated by asterisks. The scale bar indicates the number of base pair substitutions per nucleotide position.
FIG. 5.
Distribution of phylotypes found in the Inland Sea of Japan: phylogenetic tree showing the similarities of sequences from bands (bold type) to sequences from members of the CFB group bacteria, cyanobacteria, and Betaproteobacteria. The tree was inferred by the neighbor-joining method using CLUSTAL W. The bands found in lanes representing the BrdU-incorporating community are indicated by asterisks. The scale bar indicates the number of base pair substitutions per nucleotide position.
DISCUSSION
A fundamental question in microbial oceanography concerns the relationship between prokaryote diversity and biogeochemical function in an ecosystem context. While culture-independent methods for determining diversity are well established, it is still methodologically challenging to determine phylotype-specific in situ activities of bacteria that have not been cultured yet. Phylotype-specific growth performance and phenome expression in natural marine assemblages are influenced by physicochemical and nutritional conditions, as well as inter- and intraspecies interactions. Insight at the broad phylogenetic group level has been obtained by combining [3H]TdR incorporation and FISH (8, 9, 40). RNA-based DGGE fingerprints (reverse transcription-PCR-DGGE) have revealed phylotypes with active gene expression and whether a gene of interest was expressed (13, 57).
BUMP-DGGE complements these methods by addressing phylotype-specific growth (growing organisms versus nongrowing organisms). It allows workers to determine phylogenetic affiliations of DNA-synthesizing, presumably actively growing bacteria in seawater, as in this study. Thus, it is possible to determine the phylotypes contributing to bacterial production and biogeochemical cycles at a given time and locale. As suggested by Borneman (5), this approach could also help determine phylotype-specific functional gene expression (e.g., genes important in biogeochemical cycles) in a given ecological scenario, although this was not tested here. Furthermore, if pelagic archaeon-specific primers are used, the protocol should reveal archaeal growth and help address currently cutting-edge questions (e.g., whether some or all shallow-water or deep-sea Archaea are chemoautotrophic) (24, 26). This approach should also guide the development of enrichment or pure cultures of bacteria and archaea that have not been cultured yet.
In laboratory-based studies workers have found that not all bacteria incorporate TdR (27), so the bacteria that are not responsive to TdR may also not incorporate BrdU because of the lack of a transporter or thymidine kinase (43). In previous studies researchers found that 34/36 isolates were responsive to BrdU (23, 42, 58). We tested 29 additional isolates, and only 2 were not responsive to BrdU (Table 1). Cumulatively then, 61/65 isolates belonging to diverse phylogenetic groups (i.e., Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria, CFB group, low-G+C-content gram-positive bacteria, and Actinobacteria) incorporated BrdU. Thus, the BUMP-DGGE approach is potentially broadly applicable to phylogenetically diverse assemblages in pelagic marine environments; however, three of the four negative bacterial isolates were affiliated with the CFB group, which reportedly can dominate bacterial assemblages in seawater environments and especially associate with organic particles. The possible significance of bacteria that are not responsive to BrdU must not be dismissed, and this phenomenon needs to be examined further. Also, there is a lower limit for retrieving and detecting BrdU-labeled DNA, mainly because of a loss of labeled DNA during immunocapture. Hence, some bacteria may be overlooked due to their very low densities in a community even if they are actively growing and incorporating BrdU.
The bacterial community structure at low-salinity stations was quite different from that at other stations. ST1 was located near an area where there was river runoff, which presumably supported a community with a high concentration of chlorophyll a (see Table S1 in the supplemental material). Changes in physicochemical properties of estuarine waters, including salinity, have been found to be associated with community shifts (7, 10). Micro-FISH previously showed that [3H]TdR assimilation due to major bacterial phylogenetic groups changed along a salinity gradient from 0.1 to 29 practical salinity units (7). In our study, the DGGE patterns at low-salinity stations ST1, ST2, and ST3 were generally different from those at ST4 and ST5. One might expect the presence of freshwater phylotypes in BUMP-DGGE profiles at low-salinity stations. While the salinity was not very low (31.5 practical salinity units), freshwater influence was reflected in communities that incorporated BrdU, and one band obtained in 2003 (ST1b) was closely related to Delftia acidovorans belonging to the Betaproteobacteria, the dominant group in freshwater environments.
Two incubation times (1 and 5 h) were employed for BrdU labeling in this study. Although a shorter incubation time is preferable to avoid bias due to protistan grazing or viral lysis, 5 h was chosen to achieve enough labeling for immunocapture in 2002; however, a shorter incubation (1 h) was found to be enough in 2003. DGGE banding patterns for 1- and 5-h incubations were compared in 2003, and no significant differences were observed (not shown). When the method is used for coastal seawater samples, 1 h of incubation is recommended for BrdU labeling.
Micro-FISH studies have shown that the correlation between abundance and growth (assimilation of TdR) was not significant for alphaproteobacteria, gammaproteobacteria, and CFB group bacteria in the Delaware estuary (9). The results implied that the impact of mortality was greater than the impact of growth in structuring the bacterial community. We found that BrdU-incorporating (growing) communities were largely different than the “total” communities. The percentages of overlapping bands for the total and BrdU-labeled communities were ≤40% (Table 2). Indeed, many bands (86/134) for growing phylotypes were not detectable in the gels for the “total” community. The difference could not be explained by a shift in the total community since the 1-h incubation time was too short to significantly change the “total” community banding (Fig. 3). It is tempting to propose that BUMP-DGGE was able to detect a number of rapidly growing but low-abundance (hence undetected in “total” community gels) phylotypes. However, the minimum BrdU incorporation intensity for positive immunocapture is not known, and further work is needed to test this hypothesis. One motivation for a future test of this hypothesis is that phylotypes that are present at low levels but have high growth rates can make large contributions to community bacterial production and organic matter fluxes. Rapidly growing bacteria might be subject to intense grazing by protists or viral lysis and thus prevented from becoming abundant enough to be detected in PCR-DGGE.
The Roseobacter and SAR11 clusters are two major subgroups of Alphaproteobacteria found in marine environments (19). In our study, these two clusters exhibited contrasting characteristics with respect to BrdU incorporation. We found 11 phylotypes related to Roseobacter but no phylotypes related to the SAR11 cluster in BrdU-incorporating fractions. We thought that since Roseobacter-related phylotypes were reportedly rapid growers, they might dominate actively growing alphaproteobacterial populations in our samples. Gonzalez et al. (20) reported that Roseobacter and SAR11 were numerically important in the heterotrophic bacterial community (average, >20% of the 16S rRNA genes sampled) and important in cycling of organic sulfur compounds during a North Atlantic algal bloom. The importance of these clusters was also shown by bacterial production and dimethylsulfoniopropionate consumption during an algal bloom in the North Sea (61). Members of the Roseobacter cluster are commonly found in association with algal cells and have relatively high growth rates in nutrient-rich conditions (21, 47). As suggested previously (20, 31, 32, 35, 61), Roseobacter-related bacteria are well adapted to nutrient-rich conditions, such as coastal waters, algal blooms, and microscale “hot spots” in oceanic waters.
In contrast, SAR11 bacteria might be relatively slow growers in coastal waters, because they did not respond well to BrdU even though at least three representatives were present. This hypothesis is consistent with the results of culture studies of Pelagibacter ubique, the cultured representative of the SAR11 group, which grows in a pristine seawater medium at very low rates (46). However, it is inconsistent with the conclusion reached by Malmstrom et al. (30). These workers examined in situ assimilation of leucine by SAR11 bacteria in the Northwest Atlantic Ocean and suggested that the specific growth rates of SAR11 bacteria either equaled or exceeded the growth rates of the total prokaryotic community. One possible explanation for these findings is that there are metabolically different subpopulations of SAR11 cluster bacteria, because this cluster is a phylogenetically, and probably metabolically, diverse group of bacteria (19). Alternatively, members of the SAR11 cluster may not be able to incorporate BrdU and thus may have escaped detection as growing bacteria in this study. Methodology needs to be developed to test for the possible presence of uncultured bacteria that are growing but are not responsive to BrdU.
A phylotype similar to V. splendidus was found in BrdU-incorporating fractions at all sampling locations from ST1 to ST5. Quantification of the V. splendidus population by quantitative PCR in a previous study (56) indicated that it was consistently present as a member of the coastal bacterioplankton community. Vibrio species, including many pathogenic strains, are widely distributed in marine environments. This group can comprise ∼10% of the readily culturable bacteria, whereas culture-independent molecular analyses suggested that it accounts for <1% of total bacterial populations (15). However, the molecularly based estimates to date do not account for the difference between active and inactive populations. Our previous work examining BrdU incorporation at the single-cell level in 1-h incubations suggested that ∼20% of the total bacteria in coastal waters are robustly growing (23). Hence, Vibrio spp. that are active growers in coastal waters may contribute disproportionately more to bacterial production than the amount expected from calculations based on their biomass.
In conclusion, this study revealed the consistent cooccurrence of diverse actively growing bacterial taxa in rather small seawater samples in all natural assemblages examined over biogeochemically varied areas in coastal waters. While cooccurrence of diverse taxa is commonly reported, there is only a limited amount of culture-independent data on contemporaneous growth of diverse bacterial taxa. Microscale patchiness of organic matter distribution might be invoked as a mechanism creating microniches in the pelagic realm (29). Experimental perturbation experiments involving BUMP-DGGE might be used to determine the environmental mechanisms for maintenance and regulation of the diversity and the phylogenetic identity of the growing taxa. Further application of the approach to various marine environments can help determine key players in bacterium-mediated biogeochemical processes, as well as bacterial productivity. There may be some universal active growers, considering the wide distribution of particular types of bacteria, such as SAR11, RCA, and Silicibacter-like bacteria (35, 36, 49). Once the key players in a particular environment are determined, specific probes can be designed and used to obtain quantitative estimates of their dynamics by FISH and/or quantitative PCR of immunocaptured DNA. Such analyses should lead to linkages of microbial community dynamics and microbial metabolic activity in ocean biogeochemical modeling.
Supplementary Material
Acknowledgments
We thank the officers and crew of the R/V Toyoshio-maru for their assistance and support during the cruise.
This research was supported by grants-in-aid 15688006 and 16651007 from JSPS and by an Environmental Research Grant from the Showa Shell Sekiyu Foundation to K.H. F.A. was supported by NSF grants ANT04-44134 and OCE04-28900. R.A.L. was supported by NSF grant MCB 0453877.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Artursson, V., and J. K. Jansson. 2003. Use of bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl. Environ. Microbiol. 69:6208-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asquith, B., C. Debecq, D. Macallan, L. Willems, and C. Bangham. 2002. Lymphocyte kinetics: the interpretation of labeling data. Trends Immunol. 23:596-601. [DOI] [PubMed] [Google Scholar]
- 3.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 4.Azam, F., and R. Long. 2001. Sea snow microcosms. Nature 414:495-498. [DOI] [PubMed] [Google Scholar]
- 5.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specific stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casamayor, E., H. Schafer, L. Baneras, C. Pedros-Alio, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrell, M., and D. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 8.Cottrell, M., and D. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell, M., and D. Kirchman. 2004. Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware estuary. Aquat. Microb. Ecol. 34:139-149. [Google Scholar]
- 10.Crump, B., C. Hopkinson, M. Sogin, and J. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influence of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. 2002. Microbial population genomics and ecology. Curr. Opin. Microbiol. 5:520-524. [DOI] [PubMed] [Google Scholar]
- 12.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebie, Y., N. Noda, H. Miura, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Comparative analysis of genetic diversity and expression of amoA in wastewater treatment processes. Appl. Microbiol. Biotechnol. 64:740-744. [DOI] [PubMed] [Google Scholar]
- 14.Eichler, S., R. Ghristen, C. Holtje, P. Westphal, J. Botel, I. Brettar, A. Mehling, and M. Hofle. 2006. Comparison and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl. Environ. Microbiol. 72:1858-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilers, H., J. Pernthaler, F. Glockner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fandino, L., L. Riemann, G. Steward, R. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23:119-130. [Google Scholar]
- 17.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari, V., and J. Hollibaugh. 1999. Distribution of microbial assemblages in the central Arctic ocean basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia 401:55-68. [Google Scholar]
- 19.Giovannoni, S., and M. Rappe. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 20.Gonzalez, J., R. Simo, R. Massana, J. Covert, E. Casmayor, C. Pedros-Alio, and M. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossart, H., F. Levold, M. Allgaier, M. Simon, and T. Binkhoff. 2005. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7:860-873. [DOI] [PubMed] [Google Scholar]
- 22.Hamasaki, K. 2006. Comparison of bromodeoxyuridine immunoassay with tritiated thymidine radioassay for measuring bacterial productivity in oceanic waters. J. Oceanogr. 62:793-799. [Google Scholar]
- 23.Hamasaki, K., R. Long, and F. Azam. 2004. Individual cell growth rates of marine bacteria, measured by bromodeoxyuridine incorporation. Aquat. Microb. Ecol. 35:217-227. [Google Scholar]
- 24.Herndl, G. J., T. Reinthaler, E. Teira, H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm-Hansen, O., C. J. Lorenzen, R. W. Holems, and J. D. H. Strickland. 1965. Fluorometric determination of chlorophyll. J. Cons. Perm. Int. Explor. Mer 30:3-15. [Google Scholar]
- 26.Ingalls, A. E., S. R. Shah, R. L. Hansman, L. I. Aluwihare, G. M. Santos, E. R. M. Druffel, and A. Pearson. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. USA 103:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffrey, W., and J. Paul. 1990. Thymidine uptake, thymidine incorporation, and thymidine kinase activity in marine bacterium isolates. Appl. Environ. Microbiol. 56:1367-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S., and P. Kemp. 1994. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limmnol. Oceanogr. 39:869-879. [Google Scholar]
- 29.Long, R., and F. Azam. 2001. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat. Microb. Ecol. 26:103-113. [Google Scholar]
- 30.Malmstrom, R., M. Cottrell, H. Elifantz, and D. Kirchman. 2005. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71:2979-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malmstrom, R., R. Kiene, M. Cottrell, and D. Kirchman. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malmstrom, R., R. Kiene, and D. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 33.Moeseneder, M., J. Arrieta, and G. Herndl. 2005. A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol. Ecol. 51:341-352. [DOI] [PubMed] [Google Scholar]
- 34.Moeseneder, M., C. Winter, and G. Herndl. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 46:95-107. [Google Scholar]
- 35.Moran, M. A., A. Buchan, J. M. Gonzalez, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 36.Morris, R., M. Rappe, S. Connon, K. Vergin, W. Siebold, C. Carlson, and S. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., E. Del Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, C. E., and C. Carlson. 2005. A nonradioactive assay of bacterial productivity optimized for oligotrophic pelagic environments. Limnol. Oceanogr. Methods 3:211-220. [Google Scholar]
- 39.Olsen, G. j., D. J. Lane, S. J. Giovanonni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 40.Ouverney, C., and J. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page, R. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Pernthaler, A., J. Pernthaler, M. Schattenhofer, and R. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard, P., and D. Moriarty. 1984. Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Appl. Environ. Microbiol. 48:1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 45.Radajewski, S., P. Ineson, N. Parekh, and J. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 46.Rappe, M., S. Connon, K. Vergin, and S. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 47.Riemann, L., G. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 49.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 50.Steward, G., and F. Azam. 1999. Bromodeoxyuridine as an alternative to 3H-thymidine for measuring bacterial productivity in aquatic samples. Aquat. Microb. Ecol. 19:57-66. [Google Scholar]
- 51.Sugimura, Y., and Y. Suzuki. 1988. A high-temperature catalytic oxidation method for the determination of non-volatile dissolved organic carbon in seawater by direct injection of a liquid sample. Mar. Chem. 24:105-131. [Google Scholar]
- 52.Suzuki, R., and T. Ishimaru. 1990. An improved method for the determination of phytoplankton chlorophyll using N,N-dimethylformamide. J. Oceanogr. 46:190-194. [Google Scholar]
- 53.Teske, A., C. Wawer, G. Muyzer, and N. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tillett, D., and B. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251-258. [Google Scholar]
- 55.Thompson, J., D. Higgins, and T. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple-sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 57.Troussellier, M., H. Schafer, N. Batailler, L. Bernard, C. Courties, P. Lebaron, G. Muyzer, P. Servais, and J. Vivas-Rego. 2002. Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat. Microb. Ecol. 28:13-24. [Google Scholar]
- 58.Urbach, E., K. Vergin, and S. Giovanoni. 1999. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl. Environ. Microbiol. 65:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Furth, R., and T. Van Zwet. 1988. Immunocytochemical detection of 5-bromo-2-deoxyuridine incorporation in individual cells. J. Immunol. Methods 108:45-51. [DOI] [PubMed] [Google Scholar]
- 60.Yin, B., D. Crowley, G. Sparovek, W. De Melo, and J. Borneman. 2000. Bacterial functional redundancy along a soil reclamation gradient. Appl. Environ. Microbiol. 66:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zubkov, M., B. Fuchs, S. Archer, R. Kiene, R. Amann, and P. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.