Abstract
The two peptides (Lcn-α and Lcn-β) of the two-peptide bacteriocin lactococcin G (Lcn) were changed by stepwise site-directed mutagenesis into the corresponding peptides (Ent-α and Ent-β) of the two-peptide bacteriocin enterocin 1071 (Ent), and the potencies and specificities of the various hybrid constructs were determined. Both Lcn and, to a lesser extent, Ent were active against all the tested lactococcal strains, but only Ent was active against the tested enterococcal strains. The two bacteriocins thus differed in their relative potencies to various target cells, despite their sequence similarities. The hybrid combination Lcn-α+Ent-β had low potency against all strains tested, indicating that these two peptides do not interact optimally. The reciprocal hybrid combination (i.e., Ent-α+Lcn-β), in contrast, was highly potent, indicating that these two peptides may form a functional antimicrobial unit. In fact, this hybrid combination (Ent-α+Lcn-β) was more potent against lactococcal strains than wild-type Ent was (i.e., Ent-α+Ent-β), but it was inactive against enterococcal strains (in contrast to Ent but similar to Lcn). The observation that Ent-α is more active against lactococci in combination with Lcn-β and more active against enterococci in combination with Ent-β suggests that the β peptide is an important determinant of target cell specificity. Especially the N-terminal residues of the β peptide seem to be important for specificity, since Ent-α combined with an Ent-β variant with Ent-to-Lcn mutations at positions 1 to 4, 7, 9, and 10 was >150-fold less active against enterococcal strains but one to four times more active against lactococcal strains than Ent-α+Ent-β. Moreover, Ent-to-Lcn single-residue mutations in the region spanning residues 1 to 7 in Ent-β had a more detrimental effect on the activity against enterococci than on that against lactococcal strains. Of the single-residue mutations made in the N-terminal region of the α peptide, the Ent-to-Lcn mutations N8Q and P12R in Ent-α influenced specificity, as follows: the N8Q mutation had no effect on activity against tested enterococcal strains but increased the activity 2- to 4-fold against the tested lactococcal strains, and the P12R mutation reduced the activity >150-fold and only ∼2-fold against enterococcal and lactococcal strains, respectively. Changing residues in the C-terminal half/part of the Lcn peptides (residues 20 to 39 and 25 to 35 in Lcn-α and Lcn-β, respectively) to those found in the corresponding Ent peptides did not have a marked effect on the activity, but there was an ∼10-fold or greater reduction in the activity upon also introducing Lcn-to-Ent mutations in the mid-region (residues 8 to 19 and 9 to 24 in Lcn-α and Lcn-β, respectively). Interestingly, the Lcn-to-Ent F19L+G20A mutation in an Lcn-Ent-β hybrid peptide was more detrimental when the altered peptide was combined with Lcn-α (>10-fold reduction) than when it was combined with Ent-α (∼2-fold reduction), suggesting that residues 19 and 20 (which are part of a GXXXG motif) in the β peptide may be involved in a specific interaction with the cognate α peptide. It is also noteworthy that the K2P and A7P mutations in Lcn-β reduced the activity only ∼2-fold, suggesting that the first seven residues in the β peptides do not form an α-helix.
Lactic acid bacteria (LAB) produce ribosomally synthesized antimicrobial peptides, often referred to as (peptide) bacteriocins (13, 15, 16, 20, 32, 35). There is great interest in these bacteriocins because of the “food-grade quality” and industrial importance of LAB. The LAB peptide bacteriocins are grouped into two main classes. Class I consists of the posttranslationally modified peptide bacteriocins, often referred to as lantibiotics, whereas class II consists of the peptide bacteriocins without modified residues and has been divided into three subclasses, namely, classes IIa, IIb, and IIc. Class IIa consists of the antilisterial one-peptide pediocin-like bacteriocins that have very similar amino acid sequences, whereas non-pediocin-like one-peptide bacteriocins that show no sequence similarity to the pediocin-like bacteriocins are placed in class IIc. Class IIb contains the two-peptide bacteriocins. These bacteriocins are unique in that they consist of two very different peptides and that their optimal antimicrobial activity requires the presence of both peptides in about equal amounts.
Since the isolation and characterization of the first (class IIb) two-peptide bacteriocin, lactococcin G (Lcn), in 1992 (34), more than 10 two-peptide bacteriocins have been identified and characterized (2, 3, 7, 8, 10, 12, 17-20, 25-28, 34, 36-38, 40, 41). All are synthesized with a 15- to 30-residue N-terminal leader sequence of the so-called double-glycine type that is cleaved off at the C-terminal side of two glycine residues by the dedicated ABC transporter upon export of the bacteriocin from cells. Moreover, the genes encoding the two peptides of all two-peptide bacteriocins are adjacent to each other in the same operon, which also contains the gene encoding the immunity protein that protects bacteriocin-producing bacteria from being killed by their own bacteriocins. Near these genes, in some cases in the operon that contains the bacteriocin genes, is a gene encoding a dedicated ABC transporter that transports bacteriocins out of the bacteriocin-producing bacteria and a gene that encodes an accessory protein whose specific role is not known, but which also appears to be required for secretion of bacteriocins. All two-peptide bacteriocins whose mode of action has been studied, including Lcn (30, 31), plantaricins E/F and J/K (29), lactacin F (1), thermophilin 13 (27), and lactocin 705 (9), render membranes permeable to various small molecules. Moreover, it appears that they show some specificity with respect to which small molecules they conduct across membranes and that specificities vary among the various two-peptide bacteriocins.
Lcn, produced by a few strains of Lactococcus lactis, is perhaps the best-characterized (class IIb) two-peptide bacteriocin (23, 30, 31, 33, 34). One of the peptides of Lcn contains 39 residues and is termed α (or Lcn-α), and the other peptide contains 35 residues and is termed β (or Lcn-β) (Fig. 1). Both peptides have putative amphiphilic α-helices in their N-terminal and mid-regions (residues 3 to 27 in α and 8 to 25 in β), as judged by displaying their sequences on an Edmundson α-helical wheel (34). The nonpolar amino acids are found almost exclusively on one side of the helix, while the polar residues are found on the opposite side. Circular dichroism (CD) structural studies of these peptides and fragments thereof showed that the peptides are unstructured in water and that they become structured upon exposure to membrane-like entities (23). Amphiphilic α-helices in the N-terminal and mid-regions of both peptides are then formed (23). A recent nuclear magnetic resonance (NMR) analysis of the α peptide in micelles indicated that the α-helical region stretches from about residues 3 to 22, with a possible break in the glycine-rich region between residues 7 and 12 (P. Rogne et al., unpublished data). For the β peptide, the NMR data suggest that the α-helical region includes the proline residue at position 11 and stretches about three to five residues to each side (Rogne et al., unpublished data). Interestingly, CD studies indicate that in the presence of liposomes, the α and β peptides induce additional α-helical structuring in each other (23), indicating that the two complementary peptides interact in a structure-inducing manner upon contact with target membranes, resulting in the formation of an antimicrobial peptide complex with amphiphilic α-helical regions. Similar CD structural studies have also been carried out with two other two-peptide bacteriocins, plantaricin E/F and plantaricin J/K, and basically similar results were obtained (22). The synergistic antimicrobial effect obtained with the two complementary peptides of two-peptide bacteriocins thus appears to be due to interpeptide interactions rather than to the two complementary peptides interacting separately at different sites on the target cell. The interpeptide interaction seems to be specific, since peptides from different two-peptide bacteriocins do not interact to form a functional unit. For instance, the two complementary peptides that constitute Lcn are active at pico- to nanomolar concentrations when combined but show no activity when tested individually at concentrations as high as 50 μM (31) or when combined with either the E or F peptide of plantaricin E/F or the J or K peptide of plantaricin J/K (3). The requirement of both complementary peptides for a potent antimicrobial effect clearly indicates that the two peptides of two-peptide bacteriocins function together as one antimicrobial entity. Also, the facts that (i) the genes encoding the two peptides of two-peptide bacteriocins are found next to each other on the same operon, (ii) there is only one immunity gene for each two-peptide bacteriocin, and (iii) there is a direct physical interaction between complementary peptides when they exert their bactericidal effect indicate that the peptides of two-peptide bacteriocins function together as one unit.
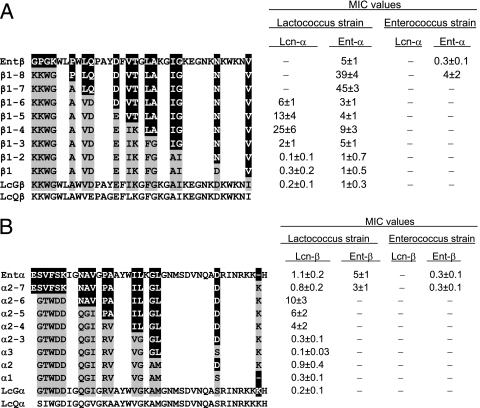
FIG. 1.
Sequences and activities of (A) β peptides of Ent, Lcn (LcGβ), and lactococcin Q (LcQβ; only the sequence is shown for LcQβ) and of variants of these peptides and (B) α peptides of Ent, Lcn (LcGα), and lactococcin Q (LcQα; only the sequence is shown for LcQα) and of variants of these peptides. As indicated in the figures, these peptides were combined with Lcn-α and Ent-α (A) and with Lcn-β and Ent-β (B) for activity measurements. Residues with a gray background are those that are present in Lcn but not in Ent, and residues with a black background are those that are present in Ent but not in Lcn. Only residues that differ in Ent and Lcn are shown for the altered peptides, whose sequences are otherwise the same as those for Ent and Lcn. Activity was quantitated in terms of MICs of the two peptides in nM (the sum of the concentrations of the two complementary peptides at a 1:1 ratio), measured against Lactococcus LMGT-2077 and E. faecalis LMGT-2333. −, the MIC is >50 nM, i.e., no activity was detected. Somewhat similar results to those obtained with Lactococcus LMGT-2077 were also obtained with L. lactis LMGT-2130, LMGT-2063, and IL-1403, and similar results to those obtained with E. faecalis LMGT-2333 were also obtained with E. faecalis NCDO 581.
The two-peptide bacteriocins lactococcin Q, isolated from a Lactococcus lactis strain (41), and enterocin 1071 (Ent), isolated from strains of Enterococcus faecalis (7, 8, 18), show marked sequence similarity to Lcn (about 57% sequence identity between Lcn and Ent, 88% identity between Lcn and lactococcin Q, and 59% identity between lactococcin Q and Ent) (Fig. 1). Despite this sequence similarity, they appear to differ somewhat in their relative potencies to different target cells (36). These three bacteriocins consequently represent an excellent system for correlating the (primary) structure of two-peptide bacteriocins with target cell specificity and potency. In this study, we have changed the α and β peptides (i.e., Lcn-α and Lcn-β) of Lcn into the corresponding Ent peptides (termed Ent-α and Ent-β) by stepwise in vitro site-directed mutagenesis. The potencies and specificities of different combinations of the various peptide constructs were then determined in order to gain insight into which peptide and/or which part of the peptide(s) is of importance in determining target cell specificity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lactobacillus sake Lb790, containing the two-plasmid expression system for production of Lcn, Ent, and mutated variants of these, was grown without agitation at 30°C in MRS (Oxoid) medium. The pSAK20 and pLT100 plasmids in the bacteriocin-producing cells contain markers for chloramphenicol and erythromycin resistance, respectively, and the cells were consequently grown in the presence of both antibiotics, with each at a concentration of 10 μg/ml, when cells were cultured under normal conditions in liquid medium.
The indicator organisms used in the bacteriocin assay were Enterococcus faecalis LMGT-2333, E. faecalis NCDO 581, Lactococcus strain LMGT-2077, Lactococcus lactis LMGT-2130, L. lactis LMGT-2063, L. lactis IL-1403, and L. lactis MG1363. The last five strains were grown without agitation at 30°C in M17 medium supplemented with 0.4% (wt/vol) glucose and 0.1% (vol/vol) Tween 80, whereas the first two strains were grown without agitation at 30°C in MRS medium.
Escherichia coli DH5α (21) was used for production and isolation of the plasmids before transfer of the plasmids to L. sake Lb790. The E. coli strain was grown with vigorous agitation at 37°C in LB medium supplemented with erythromycin to a final concentration of 150 μg/ml.
For growth of cells on agar plates, the medium was solidified by adding 1.5% (wt/vol) agar. LB agar plates containing erythromycin (150 μg/ml) were used for E. coli DH5α, and MRS agar plates containing erythromycin (2 μg/ml) and chloramphenicol (5 μg/ml) were used for L. sake Lb790.
Two-plasmid expression system for production of bacteriocins.
The Lcn-α and -β peptides and mutated variants of these peptides, including Ent-α and Ent-β, were produced by a two-plasmid bacteriocin expression system consisting of pSAK20 and pLT100 introduced into the bacteriocin-deficient strain L. sake Lb790 (termed L. sake Lb790/pSAK20 when it contains pSAK20). pSAK20 is a pVS2-derived plasmid that contains the orf4-sapKRTE operon from L. sake Lb706 (5, 6, 14). The orf4-sapKRTE operon contains genes encoding proteins necessary for activation of the bacteriocin-specific promoters and for processing and secretion of prebacteriocins that have an N-terminal leader derived from sakacin A, a class IIa bacteriocin (5, 6, 14). pLT100 is a pLPV111-derived E. coli-Lactobacillus shuttle vector in which the Lcn immunity gene and the gene encoding either Lcn-α (this plasmid is termed pLT100α) or -β (this plasmid is termed pLT100β) have been placed under the control of a bacteriocin-specific promoter derived from the sakacin A producer L. sake Lb706. The genes for Lcn-α and -β in pLT100 encode preforms of the peptides that have an N-terminal leader sequence derived from sakacin A. The preforms of Lcn-α and -β are consequently recognized and secreted by the sakacin A export machinery encoded by the genes on pSAK20. The genes introduced into pLT100 were constructed by use of PCR with DNA isolated from L. lactis LMGT-2081, a natural Lcn-producing strain (34). One megaprimer for the PCR contained a sequence recognized by the restriction enzyme MluI followed by the sequence that encodes the N-terminal leader of sakacin A, which in turn was followed by the sequence that encodes the N-terminal region of either Lcn-α or -β (sppA-α or sppA-β) (Table 1; also see the footnote in Table 1 describing how the megaprimers were constructed by use of PCR). The other primer contained the sequence that encodes the C-terminal region of either Lcn-α or -β followed by the sequence that encodes the N-terminal region of the Lcn immunity gene (lcnGαC or lcnGβC) (Table 1). PCR with these primers yielded a new megaprimer that encoded either Lcn-α or -β with an N-terminal leader sequence derived from sakacin A, followed by the N-terminal region of the Lcn immunity protein. Using this megaprimer in PCR (with DNA isolated from L. lactis LMGT-2081), together with a primer that contained the sequence encoding the C-terminal part of the Lcn immunity protein followed by a sequence recognized by the restriction enzyme ClaI (lcnGimmC) (Table 1), yielded a DNA segment that contained both the Lcn immunity gene and the gene for either Lcn-α or -β with a sakacin A N-terminal leader. After digestion with the proper restriction enzymes (ClaI and MluI), the DNA segment was ligated into pLPV111, forming pLT100.
TABLE 1.
End primers used in PCR SOEing and primers used to construct the plasmids pLT100α and pLT100β, containing the Lcn-α and -β genes, respectively
| Primer | Sequence (5′-3′) |
|---|---|
| sppA-αa | ACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTATTTAGGTGACACTATAGAATACTCAAGCTATGCATCCAACGCGTGAATTCCCCTGTTTAGGAATGATTTCTGTAGGCTTCAAGAAGTTATGCCACGTGATCAAAGAAATCTTGTTATACTTCACTCGTACAAAAAATAATAACAGAGGAGATTCTTAGTTATGGAAAAGTTTATTGAATTATCTTTAAAAGAAGTAACAGCAATTACAGGTGGAGGAACTTGGGATGATATTG |
| sppA-βa | ACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTATTTAGGTGACACTATAGAATACTCAAGCTATGCATCCAACGCGTGAATTCCCCTGTTTAGGAATGATTTCTGTAGGCTTCAAGAAGTTATGCCACGTGATCAAAGAAATCTTGTTATACTTCACTCGTACAAAAAATAATAACAGAGGAGATTCTTAGTTATGGAAAAGTTTATTGAATTATCTTTAAAAGAAGTAACAGCAATTACAGGTGGAAAAAAATGGGGCTGGCTAGCTTG |
| sakPB | ACACTTTATGCTTCCGGCTCGTATGTTGTGT |
| lcnGαA | CAATATCATCCCAAGTTCCTCCACCTGTAATTGCTGTTACTTC |
| lcnGβA | CAAGCTAGCCAGCCCCATTTTTTTCCACCTGTAATTGCTGTTACTTC |
| lcnGαC | CTACTTGCCTTATTTAATCTAACTTAGTGTTTCTTTTTTCTATTA |
| lcnGβC | CTACTTGCCTTATTTAATCTAACTCAGATATTTTTCCACTTATC |
| lcnGimmCb | CCATCGATGGCTATACAAGCTACTCCACAATC |
| sakP2 | AGCTATGACCATGATTACGCCAAG |
The megaprimers were constructed by use of PCR and the plasmid pSPP2 (6) as the template. The primers used for this PCR were sakPB and a primer that contained the C-terminal end of the sakacin A leader sequence and the sequence encoding the N-terminal end of either Lcn-α (primer lcnGαA; resulted in sppA-α) or -β (primer lcnGβA; resulted in sppA-β). The recognition site of MluI is underlined.
The recognition site of ClaI is underlined.
Plasmid isolation, preparation of competent cells, and cell transformation.
Plasmids were isolated from E. coli DH5α and L. sake Lb790 by using a QIAprep Spin Minipreps DNA purification system (QIAGEN). To ensure lysis of L. sake, lysozyme was added to the cell resuspension to a final concentration of 5 mg/ml.
For preparation of competent E. coli DH5α cells, the cells were cultured in LB medium to an optical density at 600 nm of about 0.3. The culture was cooled on ice for 10 min, and the cells were then washed with ice-cold 0.1 M CaCl2 and suspended in ice-cold 0.1 M CaCl2 containing 15% (vol/vol) glycerol. The cells were ready for transformation after incubation on ice for 45 min and were transformed by use of heat shock (42°C, 90 s).
L. sake Lb790/pSAK20 cells were made competent as described previously (4). Briefly, the cells were cultured to an optical density at 600 nm of between 0.5 and 0.6 in MRS broth containing 10 μg/ml chloramphenicol and 2% (wt/vol) glycine and, thereafter, washed with 1 mM MgCl2 and then with 30% (wt/vol) polyethylene glycol 1500 (molecular weight range, 1,300 to 1,600) prior to electroporation. L. sake Lb790/pSAK20 cells were transformed by electroporation by using a Gene Pulser and Pulse Controller unit (Bio-Rad Laboratories) as described previously (4).
Site-directed mutagenesis and DNA sequencing.
Mutations in the Lcn-α and -β genes, cloned into pLT100α and pLT100β, respectively, were made by using a QuikChange site-directed mutagenesis kit (Stratagene). PCR was performed with a Gene-Amp 2400 PCR system (Perkin-Elmer) by using Pfu Turbo DNA polymerase (Stratagene). The 50-μl reaction mixtures each contained about 40 ng plasmid template, 125 ng of each oligonucleotide primer (Eurogentec), each deoxynucleoside triphosphate (Stratagene) at a final concentration of 0.05 mM, and 2.5 U Pfu Turbo DNA polymerase. After a 2-min hot start at 95°C, 16 cycles of the following program were run: denaturation for 45 seconds at 95°C, primer annealing for 1 min at 48°C, and polymerization for 7 min at 68°C. The PCR products were digested for 1 h at 37°C with the restriction enzyme DpnI (Stratagene) to eliminate the original template and thereby increase the mutation efficiency. The resulting mutated plasmids contained a nick which was sealed by DNA repair systems in E. coli DH5α after transformation.
The QuikChange site-directed mutagenesis method was used to introduce all of the mutations except for the multiple amino acid changes in the N-terminal ends of Lcn-α and -β (mutations introduced into α2-6 and β1-8) (Fig. 1). These N-terminal mutations were introduced using the PCR splicing by overhang elongation method as described previously (24). The method entails the use of three PCR steps. The first and third steps used 25 amplification cycles (denaturation for 45 seconds at 95°C, primer annealing for 1 min at 48°C, and 1 min of polymerization at 68°C), while the second step used 10 amplification cycles (with the same conditions as in steps 1 and 3). LcnGimmC and sakP2 (Table 1) were used as end primers for both α and β mutations. The fragments containing the desired mutations and the vector (pLT100) were cut with the restriction enzymes ClaI and MluI, and the fragments were ligated into the vector by using T4 DNA ligase. The DNA sequences of all mutated plasmids were verified by automated DNA sequence determination by using an ABI Prism 377 DNA sequencer and an ABI Prism Ready Reaction Dye Terminator cycle sequencing kit (Perkin-Elmer).
Purification of Lcn, Ent, and mutated variants.
Lcn-α and -β and mutated variants of these peptides, including the Ent peptides, were purified to >80% purity basically according to a previously described method (39). Briefly, the cells in 1-liter overnight cultures were pelleted by centrifugation, and the supernatant was applied to a 5- to 6-ml SP Sepharose Fast Flow (GE Healthcare) cation-exchange column that had been equilibrated with 50 ml 20 mM sodium phosphate buffer, pH 6. The column was then washed with 100 ml of the sodium phosphate buffer, and the peptides were subsequently eluted with 40 ml of the sodium phosphate buffer supplemented with NaCl to a final concentration of 1 M. Trifluoroacetic acid and 2-propanol were added to the eluent to final concentrations of 0.1 and 5% (vol/vol), respectively, and the eluent was then applied to a reverse-phase column (Resource RPC; GE Healthcare). The peptides were eluted from the reverse-phase column with a linear propanol gradient in 0.1% trifluoroacetic acid.
The primary structures of the peptides were confirmed by determining the molecular mass with a Voyager-DE RP matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (Perseptive Biosystems), using α-cyano-4-hydroxycinnamic acid as the matrix. Typically, the errors in the masses that were determined were 1 Da or less. The purities of the bacteriocins were verified to be >80% by analytical reverse-phase chromatography by using a μRPC SC 2.1/10 C2/C18 column (GE Healthcare) in a SMART chromatography system (GE Healthcare).
The concentrations of purified peptides were determined by measuring the UV absorption at 280 nm, and the values were converted to protein concentrations by using molar extinction coefficients calculated from the contributions of individual amino acid residues.
Bacteriocin assay.
The antimicrobial activities of different combinations of the α and β peptides of Lcn and Ent as well as variants of these peptides were quantified in a microtiter plate assay system essentially as described previously (31, 34). Each well of a microtiter plate contained 200 μl M17 broth supplemented with 0.4% (wt/vol) glucose and 0.1% (vol/vol) Tween 80 (when lactococcal strains were used as indicator organisms) or MRS medium (when enterococcal strains were used as indicator organisms), a combination of the α and β peptides and variants of these peptides (in fractions at twofold dilutions), and the indicator organism. Stationary-phase cultures of the lactococcal strains were diluted 1:50 before being added to the microtiter plates, whereas stationary-phase cultures of the enterococcal strains were diluted 1:200. The microtiter plates with lactococcal strains were incubated for 4 h at 30°C, and the plates with enterococcal strains were incubated overnight at 30°C. Growth inhibition was measured spectrophotometrically at 600 nm (Tecan microplate reader). The MIC was defined as the total amount of peptides (the sum of both peptide concentrations [at a 1:1 ratio]) that inhibited growth by 50%.
RESULTS AND DISCUSSION
Ent-α is more active against lactococci in combination with Lcn-β and more active against enterococci in combination with Ent-β.
Presumably due to high sequence similarities, Lcn and Ent represent an exception to the generalization that only combinations of complementary peptides from one and the same two-peptide bacteriocin interact and exert antimicrobial activity, since activity was obtained when individual Lcn and Ent peptides were combined and tested against an L. lactis strain that was highly—and about equally—sensitive to Lcn and Ent (strain LMGT-2130) (Table 2). In fact, the hybrid combination Ent-α+Lcn-β had a potency that was equal to or greater than those of the wild-type combinations (i.e., Lcn-α+Lcn-β and Ent-α+Ent-β), indicating that Ent-α and Lcn-β interact readily to form a functional antimicrobial unit. The reciprocal hybrid combination (Lcn-α+Ent-β), however, was about 100-fold less potent, indicating that Lcn-α and Ent-β do not interact optimally. The other lactococcal strains that were tested were also sensitive to both Lcn and, to a somewhat lesser extent, Ent (Table 2). They were also sensitive to the Ent-α+Lcn-β hybrid combination but not to the Lcn-α+Ent-β combination (Table 2), consistent with the conclusion that Lcn-α and Ent-β do not interact optimally. Interestingly, when tested against a lactococcal strain (LMGT-2077) that was nearly 30 times more sensitive to Lcn than to Ent, the Ent-α+Lcn-β hybrid combination was about fourfold more potent than the wild-type Ent-α+Ent-β combination. Similarly, when tested against lactococcal strains (LMGT-2063 and MG1363) that were about three to seven times more sensitive to Lcn than Ent, the hybrid Ent-α+Lcn-β combination had a potency that was the same as or greater than that of the wild-type Ent-α+Ent-β combination (Table 2). It thus seems that Lcn-β is especially important for rendering Lcn-α and Ent-α highly potent against lactococcal strains, suggesting that the β peptide may be particularly important in determining the target cell specificities of these two two-peptide bacteriocins.
TABLE 2.
Activities of Lcn (i.e., Lcn-α+Lcn-β), Ent (i.e., Ent-a+Ent-β), and combinations of complementary peptides from Lcn and Ent (i.e., Lcn-α+Ent-β and Lcn-β+Ent-α) against various indicator organisms
| Indicator organism | MIC (nM)a
|
|||
|---|---|---|---|---|
| Lcn-α+Lcn-β | Ent-α+Ent-β | Lcn-α+Ent-β | Ent-α+Lcn-β | |
| L. lactis LMGT-2130 | 0.2 ± 0.1 | 0.3 ± 0.2 | 6 ± 3 | 0.07 ± 0.03 |
| Lactococcus LMGT-2077 | 0.2 ± 0.1 | 5 ± 1 | − | 1.2 ± 0.2 |
| L. lactis LMGT-2063 | 0.06 ± 0.02 | 0.4 ± 0.3 | − | 0.2 ± 0.1 |
| L. lactis MG1363 | 0.10 ± 0.05 | 0.4 ± 0.1 | − | 0.3 ± 0.1 |
| E. faecalis NCDO 581 | − | 0.3 ± 0.1 | − | − |
| E. faecalis LMGT-2333 | − | 0.3 ± 0.1 | − | − |
Activity was quantitated in terms of the MICs of the two peptides in nM (the sum of the concentrations of the two complementary peptides at a 1:1 ratio), measured against the indicated strains. −, no activity was detected; the MIC must be >50 nM. Data are means ± standard errors of the means.
The enterococcal strains that were tested were sensitive to Ent (i.e., Ent-α+Ent-β) but not to Lcn (i.e., Lcn-α+Lcn-β) or the Ent-α+Lcn-β hybrid combination (Table 2). Thus, Ent-α could not sensitize enterococcal strains to Lcn-β. The strains were also insensitive to the reciprocal hybrid combination (Lcn-α+Ent-β), consistent with the apparent low ability of these two peptides to function optimally together.
Stepwise conversion of Lcn-β to Ent-β.
To investigate which parts of the β peptides are of special importance for shaping the preferences that Lcn and Ent have for lactococci and enterococci, respectively, Lcn-β was transformed stepwise by site-directed mutagenesis into Ent-β, and the potencies of the peptide constructs against various target strains were determined. Eight of the altered Lcn-β peptides (in addition to Lcn-β and Ent-β) are shown in Fig. 1A.
Changing the residues near the C-terminal end of Lcn-β (D30N and I35V, i.e., peptides β1 and β1-2 in Fig. 1A) to those found in Ent-β did not have a marked effect on potency when the peptides were combined with Lcn-α or Ent-α (Fig. 1A). There was, however, a marked reduction (5- to 10-fold) in the potency against the lactococcal strains upon also introducing the A23I-plus-I24G mutation (β1-3) (Fig. 1A) and an additional marked reduction upon also introducing the F19L-plus-G20A mutation (β1-4) (Fig. 1A). Interestingly, there was a >10-fold reduction in activity (compared to the activity of β-3) when β1-4 was combined with Lcn-α but only about a twofold reduction in activity when it was combined with Ent-α (Fig. 1). Against lactococcal strains, β1-4, β1-5, β1-6, β1-7, β1-8, and Ent-β were consequently more potent in combination with Ent-α than with Lcn-α, whereas Lcn-β, β1, β1-2, and β1-3 were more potent when combined with Lcn-α (Fig. 1A). It thus appears that residues 19 and 20 in Lcn-β and Ent-β may be involved in specific interactions with Lcn-α and Ent-α, respectively. It is noteworthy that residues 19 and 20 are part of a GXXXG motif (two glycine residues separated by three residues) in both Lcn-β and Ent-β. This motif is often involved in helix-helix interactions (11), suggesting a possible specific helix-helix interaction between complementary α and β peptides in this region.
The mutations (E14D and I16V plus K17T) introduced upon transforming β1-4 into β1-6 partly neutralized the detrimental mutations introduced upon generating β1-4, since both β1-5 and β1-6 were more potent than β1-4 (Fig. 1A). β1-6 is identical to Ent-β except for seven Ent-to-Lcn substitutions in the N-terminal region, and it is thus especially noteworthy that β1-6 in combination with Ent-α was ≥150-fold less active against the tested enterococcal strain but ∼2 times more active against the lactococcal strain than Ent-β (Fig. 1A). Moreover, in combination with Lcn-α, β1-6 was also ≥10 times more active against the lactococcal strain than Ent-β (Fig. 1). The N-terminal residues of the β peptides thus seem to be especially important for the relative potencies of these two-peptide bacteriocins against lactococci and enterococci.
Stepwise conversion of Lcn-α to Ent-α.
Lcn-α was also transformed stepwise to the corresponding Ent peptide (i.e., Ent-α) in order to identify residues or regions in the α peptides that are of importance for the potency of Lcn and Ent. Eight of the altered Lcn-α peptides (in addition to Lcn-α and Ent-α) are shown in Fig. 1B. Like the case with Lcn-β, changing the residues in the C-terminal half of Lcn-α (α1, α2, α3, and α2-3) (Fig. 1B) to those in Ent-α did not have a marked effect on potency (Fig. 1B). The most marked effect was the S31D mutation (α2) (Fig. 1B), which reduced the activity two- to fourfold but which had a less negative effect when combined with the A20G-plus-M21L mutation (α2-3) (Fig. 1B). The latter mutation alone seemed, in fact, to slightly improve the potency (α3) (Fig. 1B). None of the peptides (Lcn-α, α1, α2, α3, or α2-3) was active against lactococcal strains when combined with Ent-β or against the enterococcal strains in combination with either Lcn-β or Ent-β (Fig. 1B). There was a marked reduction (10-fold) in potency against the lactococcal strains upon transforming α2-3 into α2-4 by introducing the V17I-plus-G18L mutation into α2-3 (α2-4) (Fig. 1B). This mutation destroys a GXXXG motif (two glycine residues separated by three residues) (11) that is present in Lcn-α but not in Ent-α and is consequently expected to be very detrimental if it interferes with helix-helix interactions between Lcn-α and another polypeptide (such as Lcn-β).
There appeared to be a slight additional reduction in activity (∼2-fold) upon transforming α2-4 further into α2-5 and α2-6 by introducing the A12P-plus-V13 and Q8N-plus-G9A-plus-I10V mutations, respectively (Fig. 1B). Interestingly, α2-7 in combination with either Lcn-β or Ent-β was much more potent than α2-4, α2-5, and α2-6 (Fig. 1B). The 0E-plus-G1S-plus-T2V-plus-W3F-plus-D4S-plus-D5K mutation thus largely neutralized the negative effects of the mutations introduced upon generating α2-4, α2-5, and α2-6 and also enabled the peptide to function together with Ent-β and thereby be active against enterococcal strains (Fig. 1B). The α2-7+Ent-β combination was as potent as Ent-α+Ent-β (Fig. 1B), indicating that the extra lysine residue (present in Lcn-α but not in Ent-α) near the C terminus of α2-7 did not affect the activity.
Point mutations in the N-terminal region of the β peptide.
The results obtained with the Lcn-Ent hybrid peptides indicated that the N-terminal residues of Lcn-β and Ent-β are important for the target cell specificities of Lcn and Ent. Single-residue Lcn-to-Ent and Ent-to-Lcn substitutions were consequently made in the N-terminal regions (residues 1 to 13) of Lcn-β and Ent-β, respectively, and the potencies of the resulting peptide variants were measured (Table 3). The conservative mutations V9L and L9V in Lcn-β and Ent-β, respectively, were not analyzed, however, nor was the G3W mutation in Ent-β due to difficulties in producing this variant.
TABLE 3.
Activities of variants of the β peptide, in combination with α peptides, toward various indicator strains
| Peptide variant | MIC (nM)a
|
|||
|---|---|---|---|---|
|
Lactococcus LMGT-2077
|
E. faecalis NCDO 581 Ent-α | E. faecalis LMGT-2333 Ent-α | ||
| Lcn-α | Ent-α | |||
| Lcn-β | 0.2 ± 0.1 | 1.2 ± 0.2 | − | − |
| Lcn-β[K1G] | 0.10 ± 0.08 | 1.1 ± 0.4 | − | − |
| Lcn-β[K2P] | 0.5 ± 0.2 | 2.3 ± 0.9 | − | − |
| Lcn-β[W3G] | 0.6 ± 0.2 | 1.3 ± 0.4 | − | − |
| Lcn-β[G4K] | 0.3 ± 0.1 | 0.7 ± 0.3 | − | − |
| Lcn-β[A7P] | 0.3 ± 0.1 | 5 ± 3 | − | − |
| Lcn-β[D10Q] | 5 ± 3 | 16 ± 6 | − | − |
| Ent-β | − | 5 ± 1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| Ent-β[G1K] | − | 2.7 ± 0.8 | 1.7 ± 0.3 | 0.7 ± 0.3 |
| Ent-β[P2K] | 28 ± 11 | 5 ± 1 | 21 ± 17 | 1.2 ± 0.3 |
| Ent-β[K4G] | − | 38 ± 10 | 1.0 ± 0.3 | 1.0 ± 0.3 |
| Ent-β[P7A] | − | 23 ± 6 | 27 ± 10 | 26 ± 15 |
| Ent-β[Q10D] | − | 8 ± 2 | 0.3 ± 0.1 | 0.3 ± 0.1 |
Activity was quantitated in terms of the MICs of the two peptides in nM (the sum of the concentrations of the two complementary peptides at a 1:1 ratio), measured against the indicated strains. All combinations with Lcn-α were inactive (MIC, >50) against the enterococcal strains. −, no activity was detected; the MIC must be >50 nM. Data are means ± standard errors of the means.
Except for the D10Q mutation, single-residue Lcn-to-Ent mutations in the region spanning residues 1 to 13 of Lcn-β did not have a marked effect on the activity (Table 3). Even introducing helix-disrupting proline residues at positions 2 and 7 reduced the activity only ∼2-fold (Table 3), indicating that activity does not depend on α-helical structuring up to residue 7, consistent with recent NMR structural studies indicating that α-helical structuring in Lcn-β starts at about residue 8 (Rogne et al., unpublished data). Replacing Trp at position 3 with Gly had no apparent effect when the altered peptide was assayed with Ent-α but reduced the activity about threefold when assayed with Lcn-α (Lcn-β[W3G] in Table 3), suggesting that this part of the β peptide might interact with its cognate α peptide.
In contrast to the case for Lcn-β, most of the single-residue Ent-to-Lcn mutations made in the N-terminal region spanning residues 1 to 13 of Ent-β had a marked negative effect, especially when tested against enterococcal strains. The Q10D mutation was an exception, since this mutation was well tolerated (Table 3). Interestingly, a negative charge at position 10 is important in Lcn-β but not in Ent-β (D10Q and Q10D mutants in Table 3), apparently because Ent compensates for the lack of a negative charge at position 10 in Ent-β with a negative charge at position 1 in Ent-α. The latter negative charge is absent in Lcn-α (see discussion below in connection with Table 4). The most detrimental mutation in the N-terminal region of Ent-β was the P7A mutation. The mutation reduced the activity of the peptide (in combination with Ent-α) almost 100-fold against the two enterococcal strains but only ∼5-fold against the lactococcus strain (Ent-β[P7A] in Table 3). Also, replacement of Pro in position 2 (P2K) had a detrimental effect on the activities against the two enterococcal strains, with about 70- and 4-fold reductions against strains NCDO 581 and LMGT-2333, respectively, but it did not affect the activity against the lactococcal strain (Ent-β[P2K] in Table 3). The G1K mutation reduced the activity against the two enterococcal strains 2- to 6-fold, whereas it increased the activity against the lactococcal strain ∼2-fold. These results indicate that the residues in positions 1, 2, and 7 in Ent-β are important for the relative potencies of Ent against lactococci and enterococci. The K4G mutation reduced the activities against both the enterococcal and lactococcal strains three- to sevenfold (Table 3).
TABLE 4.
Activities of variants of the α peptide, in combination with β peptides, toward various indicator strains
| Peptide variant | MIC (nM)a
|
||||||
|---|---|---|---|---|---|---|---|
|
Lactococcus LMGT-2077
|
E. faecalis NCDO 581
|
E. faecalis LMGT-2333
|
|||||
| Lcn-β | Ent-β | Ent-β[Q10D] | Ent-β | Ent-β[Q10D] | Ent-β | Ent-β[Q10D] | |
| Lcn-α | 0.2 ± 0.1 | − | − | − | − | − | − |
| Lcn-α[+E0] | 0.2 ± 0.1 | − | − | − | − | − | − |
| Lcn-α[Q8N] | 0.2 ± 0.1 | − | − | − | − | − | − |
| Lcn-α[R12P] | 1.0 ± 0.2 | − | − | − | − | − | − |
| Ent-α | 1.2 ± 0.2 | 5 ± 1 | 8 ± 2 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| Ent-α[ΔE0] | 1.3 ± 0.3 | 21 ± 3 | 7 ± 5 | 2.8 ± 0.3 | 0.2 ± 0.1 | 0.5 ± 0.2 | 0.1 ± 0.1 |
| Ent-α[N8Q] | 0.7 ± 0.3 | 2.4 ± 0.5 | 2.2 ± 1.3 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.3 ± 0.1 |
| Ent-α[P12R] | 2.6 ± 1.1 | 13 ± 3 | 14 ± 8 | − | − | − | − |
Activity was quantitated in terms of the MICs of the two peptides in nM (the sum of the concentrations of the two complementary peptides at a 1:1 ratio), measured against the indicated strains. All combinations with Lcn-β were inactive (MIC, >50) against the enterococcal strains. −, no activity was detected; the MIC must be >50 nM. Data are means ± standard errors of the means.
Point mutations in the N-terminal region of the α peptide.
Some N-terminal mutations were also introduced into Lcn-α and Ent-α. More specifically, the importance of the N-terminal Glu residue in Ent-α (which is not present in Lcn-α) and the Q8N and R12P mutations in Lcn-α and reciprocal mutations in Ent-α were analyzed. Neither the addition of an N-terminal Glu residue to Lcn-α nor the reciprocal mutation in Ent-α (i.e., the removal of the N-terminal Glu in Ent-α) had an effect on the activity when the altered peptides, Lcn-α[+E0] and Ent-α[ΔE0], were combined with Lcn-β, but the Ent-α[ΔE0] peptide had a two- to ninefold reduced activity when combined with Ent-β (Table 4). The N-terminal Glu residue in Ent-α is thus required for the peptide to function optimally together with Ent-β but is not required for it to function optimally with Lcn-β. This difference in interactions with Ent-β and Lcn-β seems (at least partly) to be due to a negative charge at position 10 in Lcn-β which is absent in Ent-β (Fig. 1A), since introducing a negative charge at position 10 in Ent-β neutralized the negative effect of removing the N-terminal negative charge in Ent-α; wild-type activity was obtained with the Ent-α[ΔE0]+Ent-β[Q10D] combination but not with the Ent-α[ΔE0]+Ent-β combination (Table 4). It thus seems that Ent compensates for the absence of a negatively charged residue at position 10 in its β peptide by instead having a negatively charged residue at position 1 in its α peptide. The N-terminal negative charge of the α peptide of Ent is consequently functionally equivalent to the negative charge at position 10 of the β peptide of Lcn. This result also underscores the point that the α and β peptides interact and act as one functional unit.
Also, in the α peptide, mutations (at positions 8 and 12) in the N-terminal part influenced the relative potencies toward lactococci and enterococci. The N8Q Ent-to-Lcn mutation in Ent-α increased the activity two- to fourfold against the lactococcal strain but had no effect on the activity against the enterococcal strains, whereas the P12R Ent-to-Lcn mutation inactivated the peptide activity against the two enterococcal strains but resulted in only a twofold reduction in activity against the lactococcal strain (Table 4). The effects of the reciprocal Q8N and R12P Lcn-to-Ent mutations in Lcn-α were not as striking; the former mutation had no apparent effect, while the latter reduced the activity about fivefold. The detrimental effect of the latter mutation was not unexpected, since introducing a Pro residue at position 12 interferes with the putative helical structuring in this region of Lcn-α (23).
Concluding remarks.
Earlier studies with Lcn revealed that activity is detected when target cells are first pretreated with one peptide, followed by extensive washing, and then treated with the complementary peptide (30). In contrast, no activity is detected when cells treated with one peptide are mixed with cells treated with the complementary peptide. Thus, Lcn-α alone and Lcn-β alone are both—independent of the other peptide—able to bind stably to a docking site on the cell surface and remain in an idle state until the complementary peptide is added, whereupon the peptides become active. It is consequently not surprising that residues in both the α and β peptides influenced the target cell specificity, although the results with wild-type peptides (Table 2) suggested that residues in β were especially important. Although the results do not exclude the possibility that residues in regions other than the N-terminal part may also be important for target cell specificity, residues in the C-terminal parts of α and β (residues 20 to 39 and 25 to 35, respectively) do not seem to influence specificity, since mutations in this region did not have a marked effect on activity. The C-terminal region is the region which is most similar in Ent, Lcn, and lactococcin Q (Fig. 1), and it is consequently not likely to be responsible for the differences in specificity between these bacteriocins. We are presently analyzing the NMR structure of Lcn, and the structural information obtained, combined with data on genetically modified Lcn and Ent, should give additional insight into structural features that are important for the modes of action and potencies of these two-peptide bacteriocins, which will be invaluable for future rational design of more optimal bacteriocin variants and for optimizing their use in biotechnological applications.
Acknowledgments
This work was supported by the Norwegian Research Council.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Abee, T., T. R. Klaenhammer, and L. Letellier. 1994. Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexes in the cytoplasmic membrane. Appl. Environ. Microbiol. 60:1006-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., C. Fremaux, and T. R. Klaenhammer. 1994. Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J. Bacteriol. 176:2235-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderssen, E. L., D. B. Diep, I. F. Nes, V. G. H. Eijsink, and J. Nissen-Meyer. 1998. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricin EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 64:2269-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aukrust, T. W., M. B. Brurberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. [DOI] [PubMed] [Google Scholar]
- 5.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake LB706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson, L., T. Katla, M. Bjørnslett, V. G. H. Eijsink, and A. Holck. 1998. A system for heterologous expression of bacteriocins in Lactobacillus sake. FEMS Microbiol. Lett. 168:137-143. [DOI] [PubMed] [Google Scholar]
- 7.Balla, E., L. M. T. Dicks, M. Du Toit, M. J. van der Merwe, and W. H. Holzapfel. 2000. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl. Environ. Microbiol. 66:1298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balla, E., and L. M. T. Dicks. 2005. Molecular analysis of the gene cluster involved in the production and secretion of enterocins 1071A and 1071B and of the genes responsible for the replication and transfer of plasmid pEF1071. Int. J. Food Microbiol. 99:33-45. [DOI] [PubMed] [Google Scholar]
- 9.Cuozzo, S. A., P. Castellano, F. J. M. Sesma, G. M. Vignolo, and R. R. Raya. 2003. Differential roles of the two-component peptides of lactocin 705 in antimicrobial activity. Curr. Microbiol. 46:180-183. [DOI] [PubMed] [Google Scholar]
- 10.Cuozzo, S. A., F. Sesma, J. M. Palacios, A. P. de Ruiz Holgado, and R. R. Raya. 2000. Identification and nucleotide sequence of genes involved in the synthesis of lactocin 705, a two-peptide bacteriocin from Lactobacillus casei CRL 705. FEMS Microbiol. Lett. 185:157-161. [DOI] [PubMed] [Google Scholar]
- 11.Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412-417. [DOI] [PubMed] [Google Scholar]
- 12.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drider, D., G. Fimland, Y. Héchard, L. M. McMullen, and H. Prévost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fimland, G., L. Johnsen, L. Axelsson, M. B. Brurberg, I. F. Nes, V. G. H. Eijsink, and J. Nissen-Meyer. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J. Bacteriol. 182:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fimland, G., L. Johnsen, B. Dalhus, and J. Nissen-Meyer. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688-696. [DOI] [PubMed] [Google Scholar]
- 16.Fimland, G., and J. Nissen-Meyer. 2007. Genetically modified bacteriocins, p. 43-66. In M. A. Riley and O. Gillor (ed.), Bacteriocins: current research and applications. Horizon Scientific Press, Norwich, United Kingdom.
- 17.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 18.Franz, C. M. A. P., A. Grube, A. Herrmann, H. Abriouel, J. Stärke, A. Lombardi, B. Tauscher, and W. H. Holzapfel. 2002. Biochemical and genetic characterization of the two-peptide bacteriocin enterocin 1071 produced by Enterococcus faecalis FAIR-E 309. Appl. Environ. Microbiol. 68:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fremaux, C., C. Ahn, and T. R. Klaenhammer. 1993. Molecular analysis of the lactacin F operon. Appl. Environ. Microbiol. 59:3906-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577-592. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hauge, H. H., D. Mantzilas, V. G. H. Eijsink, and J. Nissen-Meyer. 1999. Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J. Bacteriol. 181:740-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauge, H. H., J. Nissen-Meyer, I. F. Nes, and V. G. H. Eijsink. 1998. Amphiphilic α-helices are important structural motifs in the α and β peptides that constitute the bacteriocin lactococcin G: enhancement of helix formation upon α-β interaction. Eur. J. Biochem. 251:565-572. [DOI] [PubMed] [Google Scholar]
- 24.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez-Diaz, R., J. L. Ruiz-Barba, D. P. Cathcart, H. Holo, I. F. Nes, K. H. Sletten, and P. J. Warner. 1995. Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Appl. Environ. Microbiol. 61:4459-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado, A., J. L. Ruiz-Barba, and R. Jiménez-Diaz. 2003. Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69:383-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marciset, O., M. C. Jeronimus-Stratingh, B. Mollet, and B. Poolman. 1997. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272:14277-14284. [DOI] [PubMed] [Google Scholar]
- 28.McCormick, J. K., A. Poon, M. Sailer, Y. Gao, K. L. Roy, L. M. McMullen, J. C. Vederas, M. E. Stiles, and M. J. van Belkum. 1998. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64:4757-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moll, G., E. van den Akker, H. H. Hauge, J. Nissen-Meyer, I. F. Nes, W. N. Konings, and A. J. M. Driessen. 1999. Complementary and overlapping selectivity of the two-peptide bacteriocin EF and JK. J. Bacteriol. 181:4848-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moll, G., H. H. Hauge, J. Nissen-Meyer, I. F. Nes, W. N. Konings, and A. J. M. Driessen. 1998. Mechanistic properties of the two-component bacteriocin lactococcin G. J. Bacteriol. 180:96-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moll, G., T. Ubbink-Kok, H. H. Hauge, J. Nissen-Meyer, I. F. Nes, W. N. Konings, and A. J. M. Driessen. 1996. Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J. Bacteriol. 178:600-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nes, I. F., H. Holo, G. Fimland, H. H. Hauge, and J. Nissen-Meyer. 2002. Unmodified peptide-bacteriocins (class II) produced by lactic acid bacteria, p. 81-115. In C. J. Dutton, M. A. Haxell, H. A. I. McArthur, and R. G. Wax (ed.), Peptide antibiotics: discovery, modes of action and application. Marcel Decker, Inc., New York, NY.
- 33.Nes, I. F., L. S. Håvarstein, and H. Holo. 1995. Genetics of non-lantibiotic bacteriocins, p. 645-651. In J. J. Ferretti, M. S. Gilmore, T. R. Klaenhammer, and F. Brown (ed.), Genetics of streptococci, enterococci and lactococci, developments in biological standards, vol. 85. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 34.Nissen-Meyer, J., H. Holo, L. S. Håvarstein, K. Sletten, and I. F. Nes. 1992. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J. Bacteriol. 174:5686-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nissen-Meyer, J., and I. F. Nes. 1997. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 167:67-77. [PubMed] [Google Scholar]
- 36.Oppegård, C., P. Rogne, L. Emanuelsen, P. E. Kristiansen, G. Fimland, and J. Nissen-Meyer. The two-peptide class II bacteriocins: structure, production, and mode of action. J. Mol. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 37.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephens, S. K., B. Floriano, D. P. Cathcart, S. A. Bayley, V. F. Witt, R. Jiménez-Díaz, P. J. Warner, and J. L. Ruiz-Barba. 1998. Molecular analysis of the locus responsible for production of plantaricin S, a two-peptide bacteriocin produced by Lactobacillus plantarum LPCO10. Appl. Environ. Microbiol. 64:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uteng, M., H. H. Hauge, I. Brondz, J. Nissen-Meyer, and G. Fimland. 2002. Rapid two-step procedure for large-scale purification of pediocin-like bacteriocins and other cationic antimicrobial peptides from complex culture medium. Appl. Environ. Microbiol. 68:952-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Belkum, M. J., B. J. Hayema, R. E. Jeeninga, J. Kok, and G. Venema. 1991. Organization and nucleotide sequence of two lactococcal bacteriocin operons. Appl. Environ. Microbiol. 57:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zendo, T., S. Koga, Y. Shigeri, J. Nakayama, and K. Sonomoto. 2006. Lactococcin Q, a novel two-peptide bacteriocin produced by Lactococcus lactis QU 4. Appl. Environ. Microbiol. 72:3383-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]