Abstract
Gene expression profiles of a wine strain of Saccharomyces cerevisiae PYCC4072 were monitored during alcoholic fermentations with three different nitrogen supplies: (i) control fermentation (with enough nitrogen to complete sugar fermentation), (ii) nitrogen-limiting fermentation, and (iii) the addition of nitrogen to the nitrogen-limiting fermentation (refed fermentation). Approximately 70% of the yeast transcriptome was altered in at least one of the fermentation stages studied, revealing the continuous adjustment of yeast cells to stressful conditions. Nitrogen concentration had a decisive effect on gene expression during fermentation. The largest changes in transcription profiles were observed when the early time points of the N-limiting and control fermentations were compared. Despite the high levels of glucose present in the media, the early responses of yeast cells to low nitrogen were characterized by the induction of genes involved in oxidative glucose metabolism, including a significant number of mitochondrial associated genes resembling the yeast cell response to glucose starvation. As the N-limiting fermentation progressed, a general downregulation of genes associated with catabolism was observed. Surprisingly, genes encoding ribosomal proteins and involved in ribosome biogenesis showed a slight increase during N starvation; besides, genes that comprise the RiBi regulon behaved distinctively under the different experimental conditions. Here, for the first time, the global response of nitrogen-depleted cells to nitrogen addition under enological conditions is described. An important gene expression reprogramming occurred after nitrogen addition; this reprogramming affected genes involved in glycolysis, thiamine metabolism, and energy pathways, which enabled the yeast strain to overcome the previous nitrogen starvation stress and restart alcoholic fermentation.
Alcoholic fermentation is a dynamic process in which Saccharomyces cerevisiae cells are subjected to innumerable environmental stresses, such as progressive nutrient depletion, high osmolarity, increasing alcohol concentration, and temperature variation, which adversely affect cell growth, fermentative activity, and cell viability. In winemaking, nutrient limitation—specifically, nitrogen deficiency—is one of the main reasons for stuck or sluggish fermentation. These major problems have not been clarified in terms of their associated molecular and cellular signaling events (2, 6, 23, 32). One of the ways in which yeast cells adjust to modifications in their environment is by changing gene expression patterns. cDNA microarray technology has emerged as a powerful tool for functional genomics, providing valuable insight into the yeast cellular responses to diverse growth conditions. Studies under laboratory conditions have shown that yeast cells respond to environmental changes with modifications in the expression of hundreds of genes, demonstrating the plasticity of yeast genome (12, 15). This generalized stress response, known as environmental stress response (ESR), includes the repression of most of the genes involved in protein synthesis, including genes required for ribosome synthesis and processing, and others that are functionally related (15). One of the environmental transitions that lead to ESR is starvation, in particular, nitrogen starvation (15). Transcriptomic responses, under nutrient-limiting conditions, have also been studied with cultures growing in chemostats, under highly controlled conditions with a single factor of variation (7, 31, 33, 35, 40). These cultures do not show many of the dramatic changes in gene expression due to starvation because, under these conditions, the cells seem to be “poor” but not “starving” (8). Indeed, the nutrient starvation response is deemed to be different from the nutrient limitation response (26). In the past few years, several transcriptomic (3, 24, 28, 37, 42) and/or proteomic (36, 42) studies have been undertaken under conditions resembling the winemaking process. Their findings indicated that entrance to stationary phase, due to nitrogen depletion, is the landmark event during wine fermentation which results in major changes characterized by a general stress response. In addition, genes involved in carbohydrate metabolism, protein folding and degradation, the oxidative stress response, DNA damage repair, and other cellular processes are induced (12, 15). However, most of these studies were performed using conditions where nitrogen was not a limiting nutrient to ensure complete sugar degradation.
In the present study, genome-wide expression profiling was used to compare and characterize changes in the wine yeast strain S. cerevisiae PYCC4072 in response to different nitrogen supply regimens. The experiments were carried out by using batch cultures, resembling the stressful conditions that yeast cells have to face in the enological environment. It was found that even under high glucose concentrations yeast cells responded to the challenge of low nitrogen by inducing a great number of genes of respiratory metabolism, those of the tricarboxylic acid cycle and the oxidative phosphorylation pathway. Conversely, yeast cells under low-nitrogen conditions adjusted the expression of genes encoding proteins with functions in ribosome structure or biogenesis and rRNA metabolism by lowering their mRNA levels; it is interesting, however, that all these genes increased in expression during the N-limiting fermentation, a result suggesting an important role for them in cell survival under nitrogen starvation conditions. Globally, the results provide a broad and integrated view of the gene expression changes that may occur under conditions mimicking the enological environment and indicate that the nitrogen availability is an important factor in determining the gene expression profile during fermentation.
MATERIALS AND METHODS
Strain and maintenance conditions.
The wine yeast strain S. cerevisiae PYCC4072 was supplied by the Portuguese Yeast Culture Collection and maintained at 4°C on slants of yeast-peptone-dextrose agar containing glucose (20 g·liter−1), peptone (10 g·liter−1), yeast extract (5 g ·liter−1), and agar (20 g·liter−1). Before use, it was transferred to a new slant of yeast-peptone-dextrose agar for 24 h at 25°C.
Culture medium.
A chemically defined grape juice medium (GJM), similar in composition to typical grape juice and previously described by Henschke and Jiranek (16), was used with minor modifications. This medium contained (per liter): glucose, 200 g; potassium tartrate, 5 g; l-malic acid, 3 g; citric acid, 0.2 g; K2HPO4, 1.14 g; MgSO4·7H2O, 1.23 g; CaCl2·2H2O, 0.44 g; MnCl2·4H2O, 198.2 μg; ZnCl2, 135.5 μg; FeCl2, 32.0 μg; CuCl2, 13.6 μg; H3BO3, 5.7 μg; CO(NO3)2·6H2O, 29.1 μg; NaMoO4·2H2O, 24.2 μg; and KIO3, 10.8 μg); vitamins (myo-inositol, 100 mg; pyridoxine HCl, 2 mg; nicotinic acid, 2 mg; calcium pantothenate, 1 mg; thiamine HCl, 0.5 mg; p-amino benzoic acid, 0.2 mg; riboflavin, 0.2 mg; biotin, 0.125 mg; and folic acid, 0.2 mg); diammonium phosphate was added as the only nitrogen source. The pH was adjusted to 3.7 with NaOH prior to sterile filtration of the medium.
Inoculum and fermentation conditions.
The inoculum was prepared by pregrowing the yeast overnight in 100-ml shake flasks containing 70 ml of medium with the same composition as that used in all assays. The flasks were then incubated overnight at 25°C in an orbital shaker at 150 rpm. This preculture was used to inoculate experimental cultures with an initial viable population of 5 × 105 CFU·ml−1.
Fermentations were carried out in 1,000-ml flasks equipped with cotton stoppers, filled to 2/3 of their volume and maintained at 20°C under permanent but moderate shaking (120 rpm), mimicking real industrial conditions. The effect of nitrogen on yeast performance was studied with GJM containing an initial nitrogen concentration of 267 or 66 mg·liter−1, supplied as diammonium phosphate. In the medium with a lower nitrogen content (66 mg·liter−1), cells were grown until stationary phase (72 h). At that time, the culture was split into two smaller 500-ml flasks, and 200 mg·liter−1 of nitrogen was added to one of the flasks while the other remained as a control. At the indicated time (Fig. 1), culture aliquots were taken for metabolite analysis, cell counting, and RNA preparation. Samples for RNA preparation were taken from the fermentation flasks by rapidly transferring them into centrifuge tubes. The yeast cells were pelleted by centrifugation at 2,205 × g at 4°C for 5 min (Sigma 3K18; rotor 11133), immediately frozen in liquid nitrogen, and stored at −80°C for later RNA isolation. The progress of fermentation was evaluated by determining the amount of glucose consumption and ethanol production as well as ammonium disappearance during the process, as previously described (25).
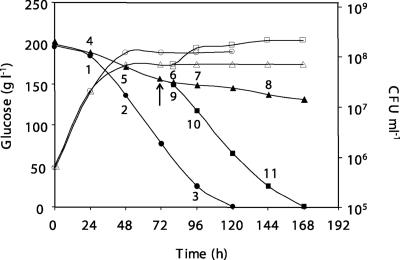
FIG. 1.
Fermentation (filled symbols) and growth patterns (open symbols) of S. cerevisiae PYCC4072 in synthetic GJM, under different nitrogen concentrations (•, control fermentation [267 mg of N ·liter−1]; ▴, N-limiting fermentation [66 mg of N·liter−1]; and ▪, refed fermentation [66 + 200 mg of N ·liter−1]), supplied as ammonium phosphate, at 20°C and pH 3.7. The arrow indicates the time of nitrogen addition. In the control fermentation, cells were collected at mid-exponential phase (24 h), at entry into stationary phase (48 h), and at the final stage of alcoholic fermentation (96 h), corresponding to time points 1, 2, and 3, respectively. In the N-limiting fermentation, samples were collected at 24 h (time point 4), when cells were still actively growing despite the fact that nitrogen was already exhausted from the medium; at 48 h (time point 5), when cells entered into stationary phase; and at 80, 96, and 144 h during stationary phase, corresponding to time points 6, 7, and 8, respectively. Samples were also taken 8, 24, and 72 h after nitrogen addition, corresponding to time points 9, 10, and 11, respectively.
Nucleic acid extraction and labeling.
Total RNA extraction and labeling by random priming using [α-33P]dCTP (3,000 Ci/mmol; 10 μCi/μl) was performed as described by Alberola et al. (1). The labeled cDNAs were purified by using a G50 MicroSpin column (Amersham Biosciences). Between 3 × 106 and 5 × 106 dpm/ml of labeled cDNA was used for filter hybridization. Prehybridization, hybridization, and washing were carried out according to published protocols (1).
Estimation of total RNA and mRNA.
For further normalizations, the RNA amount obtained from a fixed amount of cells during the experiment was estimated, using five different cell aliquots taken at each of the sampling times from a mock experiment (standard deviation, <2%). Based on the known RNA amount in a fixed number of cells, the RNA amount obtained in the real experiment was expressed as number of cells, as determined by using a particle count and size analyzer (Z2; Coulter, Inc.). In addition, a dot blot procedure was used to estimate the proportion of poly(A) mRNA in the total RNA (14). Using these data, the proportion of poly(A) mRNA per microgram of total RNA per cell, at each of the time points, was determined.
Data generation, correction, and normalization.
Global gene expression analysis was performed by hybridization of nylon filters (fabricated by the DNA chips section of the Servicio Central de Soporte a la Investigación Experimental of the Universitat de València, València, Spain; http://scsie.uv.es/chipsdna) containing PCR-amplified whole open reading frame sequences as probes (1), and signal was measured in a phosphorimager scanner (FLA-3000; FujiFilm). Membranes were hybridized with total yeast genomic DNA labeled by random priming before the set of cDNA hybridizations. In addition, for a better homogenization of the quantified signals and to minimize differences between filters, a swap of the membranes was done among the different sampling points. Each replicate was, accordingly, represented by hybridizations done with three independent membranes. Hybridization signals were quantified using ArrayVision 7.0 software (Imaging Research, Inc.), taking the artifact-removed median density (with the corresponding subtracted background) as signal. Poor or inconsistent signals were not considered for further analysis. Genomic hybridization signals were used to normalize cDNA signals in each respective filter in order to eliminate differences in membrane fabrication. Normalization between different hybridizations is usually done by assuming that the overall amount of mRNA per cell in each sample is constant. However, it is not possible to assume that condition when cells are subjected to external perturbations. Thus, the total amount of poly(A) mRNA per cell was evaluated as described in the previous section. This datum was used to normalize the different hybridizations of cDNA.
The use of the same DNA chip for successive cDNA hybridization improved the comparisons between values for each gene. cDNA hybridizations were normalized within each experiment replicate by the global mean procedure. Reproducibility of the replicates was tested by ArrayStat software (Imaging Research, Inc.), considering the data as independent and allowing the program to take a minimum number of two valid replicates in order to calculate the mean and standard deviation values for every gene (only one of the three replicates was allowed to be a removable outlier). Normalization between sampling points was done using the previously calculated amount of mRNA/cell to give values of mRNA copies/cell for each gene at every time point. These values were used for cluster analysis and comparisons. To estimate significantly differentially expressed genes in all possible time courses, an F-test for multiple conditions was applied. For pairwise comparisons, a z-test was used to determine differential gene expression. False discovery rate was the method used for false-positive error correction.
Clustering procedures.
For clustering, the Gene Expression Pattern Analysis Suite (GEPAS) v1.0, included in the website of the CIPF Bioinformatic Unit (http://gepas.org), was used. Log-transformed data were preprocessed (18) to remove genes with missing values of more than 80%. The K nearest-neighbor impute option was used to impute missing values. Flat patterns were filtered according to their standard deviations by using a threshold of 0.5. Preprocessed data was used for cluster analysis by transferring it to the SOTA tree server (17) using the linear correlation coefficient as the distance between genes. The tree was allowed to grow to seven clusters as training conditions.
Functional searches.
Functional analysis of the expression data was done using the FuncAssociate tool (http://llama.med.harvard.edu/cgi/func/funcassociate) for finding statistically significant overrepresented functional classes. The Saccharomyces Genome Database (http://www.yeastgenome.org) and the MIPS Comprehensive Yeast Genome Database (http://mips.gsf.de/genre/proj/yeast/) were used to retrieve information about specific gene function and biological process. Supplemental research data accompanying this article are available through the website http://scsie.uv.es/chipsdna.
Semiquantitative RT-PCR assays.
The expression of some genes was also analyzed by semiquantitative reverse transcription (RT)-PCR according to the protocol described by Zuzuarregui et al. (42). Table 1 includes the sequences of the oligonucleotides used in these amplification reactions, the number of cycles, and the hybridization temperatures. The PDA1 gene was used for normalization of the data. This gene encodes the E1 alpha subunit of the pyruvate dehydrogenase complex, and it shows a constitutive expression in batch and chemostat cultures in the presence of various carbon sources (39).
TABLE 1.
Gene-specific primers for RT-PCR assays
| Gene name | Primer 1 | Primer 2 | Temp (°C) | No. of cycles |
|---|---|---|---|---|
| SPI1 | TTGTCTAACGCTAAGCTCCT | AAGCATCATAACTGCACCAG | 55 | 22 |
| YGP1 | ACTTTGCCGGCCTGGAATG | GTACTCCGGTGTCTTCAC | 45 | 22 |
| CHA1 | TTACGTCAATTCTTCCCCGG | ACCACGACTGTACATGGTAG | 50 | 25 |
| DUT1 | AATTGCGCTCAGCAAG | AGTGCTACCAAAGCCA | 60 | 25 |
| ARG1 | GGGAAAAGTTTGTTTGGCTT | CTTCACCTTTGGTTTTTTTGG | 45 | 22 |
| ALD3 | CCACTCATCTTAAATCCGCC | CTTACAAGATACTATGCGGG | 45 | 22 |
| PDA1 | GCTTCATTCAAACGCCAACC | TCCCTAGAGGCAAAACCTTG | 45 | 22 |
Microarray data accession number.
The array data determined in this study have been submitted to the GEO data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE5842.
RESULTS
Establishment of fermentation and sampling conditions.
The aim of this study was to determine how the wine yeast strain S. cerevisiae PYCC4072 responds to the progressive depletion of nitrogen in batch culture, by using macroarray technology. This was done by monitoring gene expression throughout the alcoholic fermentation in a synthetic grape juice mimicking an enological environment. Three main fermentation conditions with 200 g·liter−1 of glucose were established by manipulating the nitrogen concentration in the culture medium (25) (i) with 267 mg N·liter−1, the nitrogen concentration required by the yeast strain to complete fermentation (control fermentation), (ii) with 66 mg N·liter−1, the nitrogen concentration that leads to sluggish fermentation (N-limiting fermentation), and (iii) by adding 200 mg of N·liter−1 into the N-limiting fermentation after 72 h, coinciding with 48 h of nitrogen depletion (refed fermentation).
A global view of the fermentation and growth patterns associated with each of the three test conditions is presented in Fig. 1. In Table 2, the number of viable cells; glucose, ethanol, and nitrogen levels; and total RNA and poly(A) signal per 108 cells for each sampling point are indicated. The results show that in the control fermentation, ammonium was consumed after 48 h, which coincided with the time of cell growth arrest. In the N-limiting fermentation, all ammonium was exhausted after 24 h. However, according to the CFU determinations, both fermentations reached stationary phase on the second day. Furthermore, the level of total RNA strongly decreased and the amount of poly(A) mRNA also decreased although, in this case, only by approximately half of the initial value. Nitrogen addition, during stationary phase, to the N-limiting fermentation had a significant effect on yeast growth, altering the fermentation profile. However, after only a lag period, yeast cells started to grow and sugar began to be used efficiently. In the 24 h that followed the addition, nitrogen was almost completely exhausted, and the glucose consumption was completed with a 48-h delay, coincident with the time of nitrogen deprivation, in comparison to the control fermentation. From the above results, eleven samples were collected at the indicated times from three biological replicates for transcriptome analysis as indicated in Fig. 1. In the total of 5,890 genes whose expression could be scored at the eleven sampling points, the expression of 4,116 genes, roughly 70% of transcriptome, was significantly changed (P < 0.05) (see Table S1 in the supplemental material).
TABLE 2.
Numbers of viable cells; glucose, ethanol, and nitrogen concentrations; and total RNA and poly(A) signal per 108 cells for each sampling pointa
| Expt | Time (h) | Sampling point | Glucose (g·liter−1) | Nitrogen (mg·liter−1) | Ethanol (%) (vol/vol) | CFU (106)·ml−1 | Total RNA/108 cells (μg) | Poly(A) signal/108 cells |
|---|---|---|---|---|---|---|---|---|
| CF | 24 | 1 | 183.8 ± 0.6 | 178.1 ± 6.4 | 0.6 ± 0.3 | 21.0 ± 9.7 | 48.9 ± 1.7 | 2,530 ± 474 |
| 48 | 2 | 136.1 ± 0.3 | 2.2 ± 2.4 | 3.9 ± 0.7 | 122.5 ± 26.3 | 19.2 ± 1.4 | 1,252 ± 130 | |
| 96 | 3 | 24.7 ± 4.0 | 0.0 ± 0.0 | 9.0 ± 0.7 | 122.5 ± 22.2 | 14.4 ± 3.2 | 1,051 ± 508 | |
| LF | 24 | 4 | 189.1 ± 0.7 | 2.3 ± 0.2 | 0.4 ± 0.1 | 20.3 ± 6.1 | 51.7 ± 2.8 | 2,325 ± 245 |
| 48 | 5 | 171.5 ± 5.7 | 0.0 ± 0.0 | 1.4 ± 0.2 | 69.4 ± 7.2 | 21.2 ± 1.1 | 819 ± 143 | |
| 80 | 6 | 154.1 ± 4.4 | 0.0 ± 0.0 | 2.1 ± 0.1 | 70.3 ± 9.1 | 19.2 ± 1.0 | 978 ± 126 | |
| 96 | 7 | 148.3 ± 4.3 | 0.0 ± 0.0 | 2.5 ± 0.2 | 71.6 ± 6.0 | 18.2 ± 2.5 | 1,207 ± 56 | |
| 144 | 8 | 137.0 ± 3.7 | 0.0 ± 0.0 | 3.2 ± 0.4 | 72.3 ± 4.5 | 15.9 ± 0.2 | 1,264 ± 91 | |
| RF | 80 | 9 | 148.2 ± 1.7 | 136.5 ± 3.9 | 2.2 ± 0.1 | 71.0 ± 4.1 | 22.2 ± 1.7 | 1,264 ± 91 |
| 96 | 10 | 116.6 ± 3.6 | 0.8 ± 1.4 | 4.2 ± 0.4 | 149.6 ± 15.2 | 17.5 ± 4.4 | 987 ± 366 | |
| 144 | 11 | 24.9 ± 10.8 | 0.0 ± 0.0 | 8.9 ± 1.0 | 210.0 ± 33.7 | 22.7 ± 0.8 | 1,856 ± 134 |
CF, control fermentation (267 mg of N·liter−1); LF, N-limiting fermentation (66 mg of N·liter−1); RF, refed fermentation. Results are expressed as averages ± standard deviations.
Effect of low nitrogen and nitrogen starvation on gene expression.
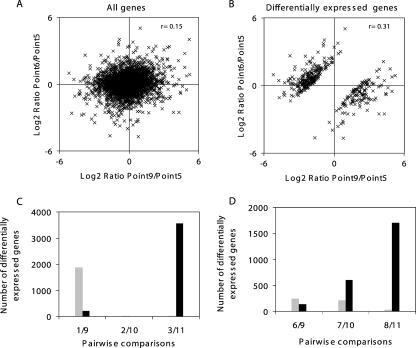
To evaluate how yeast cells adjusted to low nitrogen, time points 2 and 4, corresponding to different experiments and fermentation times but having similar extracellular nitrogen levels, as shown in Table 2, were compared to point 1 as a reference. Scatter plots were constructed for the 5,227 genes that could be scored in both comparisons and for the 1,236 that had significantly altered expression (P < 0.05 according to a z-test) in both comparisons (Fig. 2A and B) (see Table S2 in the supplemental material). A total of 664 genes, mainly associated with protein synthesis (P = 9.5 × 10−37), RNA metabolism (P = 2.3 × 10−12), and nucleic acid metabolism (P = 1.1 × 10−7), were downregulated at both time points. Approximately 46% of genes were upregulated, including those encoding proteins involved in a multiplicity of functions, such as the generation of energy (P = 9.9 × 10−13), carbohydrate metabolism (P = 5.2 × 10−10), oxidoreductase activity (P = 4.5 × 10−9), respiratory chain phosphorylation (P = 1.7 × 10−8), transporter activity (P = 4.6 × 10−7), respiration (P = 2.0 × 10−5), response to oxidative stress (P = 2.4 × 10−5), and oxygen and reactive oxygen species metabolism (P = 4.2 × 10−5). Only seven genes exhibited differential behavior within the two comparisons.
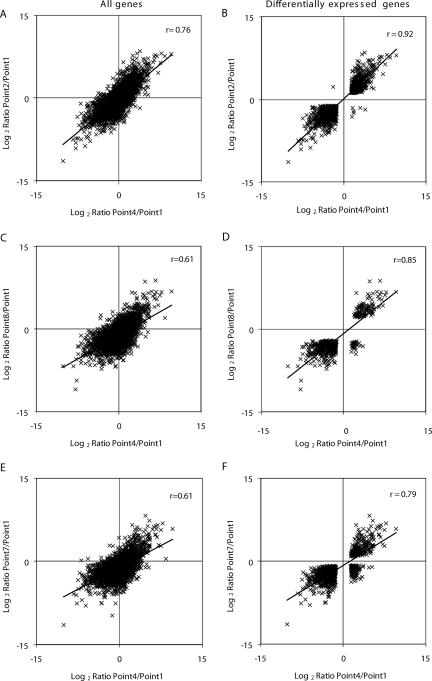
FIG. 2.
Log2 ratio scatter plots comparing expression profiles from different sampling points. The log2 ratios of expression for all genes scored in both comparisons (A, C, and E) and for all genes with statistically significant (P < 0.05) changes in expression, according to ArrayStat, in both comparisons (B, D, and F) were plotted against one another.
To assess if the above yeast cell response to low nitrogen is maintained during the late stage of the N-limiting fermentation, the changes in the pairwise comparisons between time points 4 and 1 and either 7 and 1 or 8 and 1 were analyzed. The scatter plots constructed for the 5,284 genes that could be scored in both comparisons (time points 4 and 1 versus 8 and 1) and for 628 genes with significantly altered expression (P < 0.05) (Fig. 2C and D; see Table S2 in the supplemental material) revealed that, despite the variation in expression levels (supported by a lower correlation coefficient, r = 0.61), the transcriptional response triggered by low nitrogen was sustained during the late stages. The genes that displayed lower expression levels during the N-limiting fermentation (time points 4 and 1 versus 8 and 1) were approximately the same that were found at the early yeast cell response (time points 4 and 1 versus 2 and 1). Only in the case of 32 genes did the relative expression specifically decrease after a long-term nitrogen starvation (Fig. 2D, lower right quadrant) and these were mainly associated with cell wall (P = 9.3 × 10−6) functions. The difference in the numbers of genes with a statistically significant (P < 0.05) change obtained in the pairwise comparisons for time points 4 and 1 versus 7 and 1 did not alter the general trends pointed out above for time points 4 and 1 versus 8 and 1, as the overrepresented functional categories were largely the same (Fig. 2E and F; see Table S2 in the supplemental material).
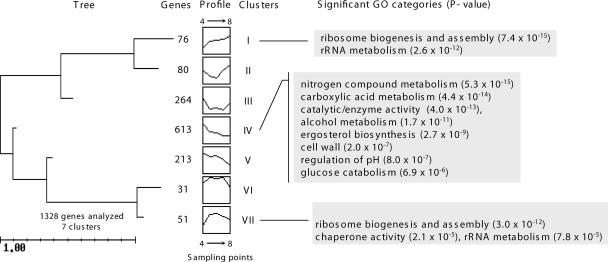
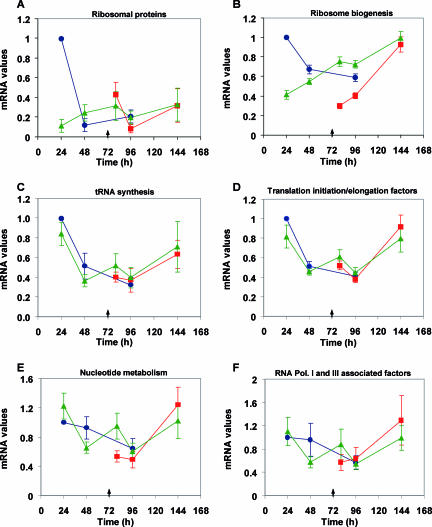
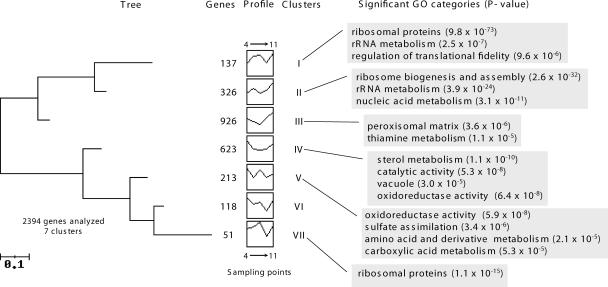
Global analyses of samples from N-limiting fermentation (time points 4, 5, 6, 7, and 8) revealed that 1,328 genes, approximately 23% of the yeast transcriptome analyzed, were significantly changed (P < 0.05). Genes were grouped according to their expression profiles, enabling the definition of seven clusters of genes (Fig. 3) (see Fig. S3 in the supplemental material). From these results, 826 genes (clusters III, IV, V), linked to metabolism, the cell wall, and regulation of pH, were downregulated. The nitrogen catabolite repression (NCR) genes were included in those clusters. The upregulated genes at time point 8 were included in clusters I, II, and VII. Genes encoding proteins with functions in ribosome structure or biogenesis as well as genes involved in rRNA metabolism are included in these clusters, suggesting they had an important role in cell survival under nitrogen starvation conditions. As yeast cells are experiencing nitrogen starvation, genes encoding cytoplasmatic ribosomal proteins and those that compose the RiBi regulon (15), including those coding for ribosome biogenesis and subunits of RNA polymerase I and III, enzymes involved in nucleotide metabolism, tRNA synthetases, and translation factors, were considered for further analysis (Fig. 4A to F). Surprisingly, but in agreement with data shown in Fig. 3, genes related to cytoplasmic ribosomal proteins and, especially, to ribosome biogenesis were induced during the N-limiting fermentation, contrary to what was observed with the control fermentation, where the decrease in expression levels coincided with cell growth arrest. It is possible that ribosomes are present in modified forms for optimal translational activity, similar to that occurring in the stationary phase (29), and that they therefore cannot be altered to a form that is more tolerant of limiting nitrogen without resynthesis. Genes related to tRNA synthesis and nucleotide metabolism and those encoding subunits of RNA polymerases I and III showed similar behavior in the control and the N-limiting fermentations. It is interesting to note the clear difference found between ribosome biogenesis genes and the other groups of genes belonging to the RiBi regulon.
FIG. 3.
Tree obtained after clustering the significantly changed genes during the nitrogen-limiting fermentation. On the x axis are plotted the time points 4, 5, 6, 7, and 8. On the y axis are plotted the average normalized mRNA levels. Only highly significant GO categories, according to the FuncAssociate tool, are shown.
FIG. 4.
Expression profiles of ribosomal proteins (A) and RiBi regulon genes: ribosome biogenesis (B), tRNA synthesis (C), translation initiation and elongation factors (D), nucleotide metabolism (E), and RNA polymerases I- and III-associated factors (Pol. I and III) (F). Expression profiles of gene expression ratio results from mean values of the genes associated to each category compared to the reference stage (sampling point 1). •, control fermentation (267 mg of N·liter−1); ▴, N-limiting fermentation (66 mg of N·liter−1); and ▪, refed fermentation (66 + 200 mg of N·liter−1). The standard error of the mean is shown with a bar, and the arrow indicates the time of nitrogen addition.
Effect of nitrogen refeeding on gene expression.
To assess how starved cells responded to nitrogen addition, pairwise comparisons between time points 9 versus 5 and 6 versus 5 were combined (Fig. 5A and B). The analysis revealed profound alterations caused by nitrogen addition, corroborated by the low correlations found between the two ratios. The effect of nitrogen addition was more clearly seen in the subsequent pairwise comparisons of time points 7 versus 10 and 8 versus 11 (Fig. 5C). In the first pairwise comparison, the expression of 814 genes was significantly different (P < 0.05). Of those, 215 genes were less expressed at point 10 and were surprisingly connected to ribosomal proteins (P = 1.1 × 10−10), the small nucleolar ribonucleoprotein complex (P = 3.8 × 10−10), the nucleolus (P = 1.4 × 10−9), and RNA processing and metabolism (P = 1.4 × 10−5). The 598 genes with higher expression levels at time point 10 than at point 7 were included in several functional categories, such as cell wall functions (P = 9.0 × 10−11), glucose catabolism (P = 3.7 × 10−6), protein folding (P = 6.1 × 10−6), energy pathways (P = 1.1 × 10−5), alcohol metabolism (P = 2.6 × 10−5), and glycolysis (P = 7.0 × 10−5). The comparison between time points 8 and 11 indicated that the number of highly expressed genes at time point 11 strongly increased (1,705 genes) (Fig. 5C). Functional analysis revealed that the majority of the gene ontology (GO) categories described previously were enriched and others, such as response to stimulus (P = 5.0 × 10−8), response to stress (P = 1.2 × 10−7), ion transport (P = 2.3 × 10−6), and transcription regulator activity (P = 4.4 × 10−5), as well as genes associated with fermentation (P = 6.0 × 10−5), were also overrepresented. The results obtained from these comparisons, regarding the expression of ribosomal protein and stress genes, suggested that the refed fermentation yeast cells were progressively detecting the stressful conditions, while at time points 7 and 8 only minor changes occurred.
FIG. 5.
Log2 ratio scatter plots comparing expression profiles from time point 6 and time point 9 relative to time point 5 for all genes (A) and for those significantly changed (P < 0.05) between time points 6 and 9 (B). Graphical representation of the number of genes showing significant (P < 0.05) changes in pairwise comparisons between sampling points of the refed fermentation with those of the (C) control (time points 1 and 9, 2 and 10, and 3 and 11) and of the (D) N-limiting fermentation (6 and 9, 7 and 10, 8 and 11). Gray and black bars represent the number of under- and overexpressed genes at the refed fermentation sampling points (9, 10, and 11), respectively.
To evaluate whether yeast cells returned to a state similar to that of the control fermentation, the following pairwise comparisons were performed: time points 9 versus 1, 10 versus 2, and 11 versus 3. (Fig. 5D). It was found that 2,069 genes were significantly (P < 0.05) differentially expressed despite the good correlation between comparison 1 versus 9 (r = 0.80). Of those, the 205 genes with higher expression at time point 9 were included in functional categories related to generation of precursor metabolites and energy pathways (in particular, electron transport activity), carbohydrate metabolism, and energy reserve metabolism. This result could be related to the different growth stage found at time points 9 and 1. Conversely, most of the genes (1,864 genes) with lower expression at time point 9 were related to ribosome biogenesis and assembly, RNA metabolism and RNA processing, protein biosynthesis, and amino acid metabolism. Accordingly, the genomic expression profile at time point 9 seemed to be quite similar to that observed at time point 4, which is corroborated by the highly correlated (r = 0.81) expression ratios between both conditions. These data point out that, 8 h after nitrogen addition, cells have not completely returned to the situation found in the control fermentation. The comparison between time points 2 and 10 showed that only 21 genes were significantly (P < 0.05) changed, suggesting that only 24 h after nitrogen addition, cells returned to a state similar to the control conditions. However, a comparison of time points 3 to 11 showed 3,574 genes that were differentially expressed, probably due to differences in yeast growth stages.
The expression of 2,394 genes, corresponding to nearly 40% of the yeast transcriptome, was significantly changed over the time course of refed fermentation. The genome-wide profiles of these genes as well as the significant GO categories are shown in Fig. 6 (see Fig. S4 in the supplemental material). Genes whose expression increased until nitrogen addition were included in clusters I, II, and VII. Genes with earlier responses to nitrogen addition appeared in clusters II, V, VI, and VII, while those with a later response to refeeding belonged to cluster III and IV. Some of the NCR-sensitive genes, such as DAL82, GAT1, GDH3, GLN3, MEP3, PUT4, URA10, and UGA4, were included in cluster III, while 22 of 56 genes reported to be NCR-responsive genes (5, 10, 13) were allocated to cluster IV.
FIG. 6.
Tree obtained after clustering the significantly changed genes during the refed fermentation. On the x axis are plotted the time points 4, 5, 9, 10, and 11. On the y axis are plotted the average normalized mRNA levels. Only highly significant GO categories, according to the FuncAssociate tool, are shown.
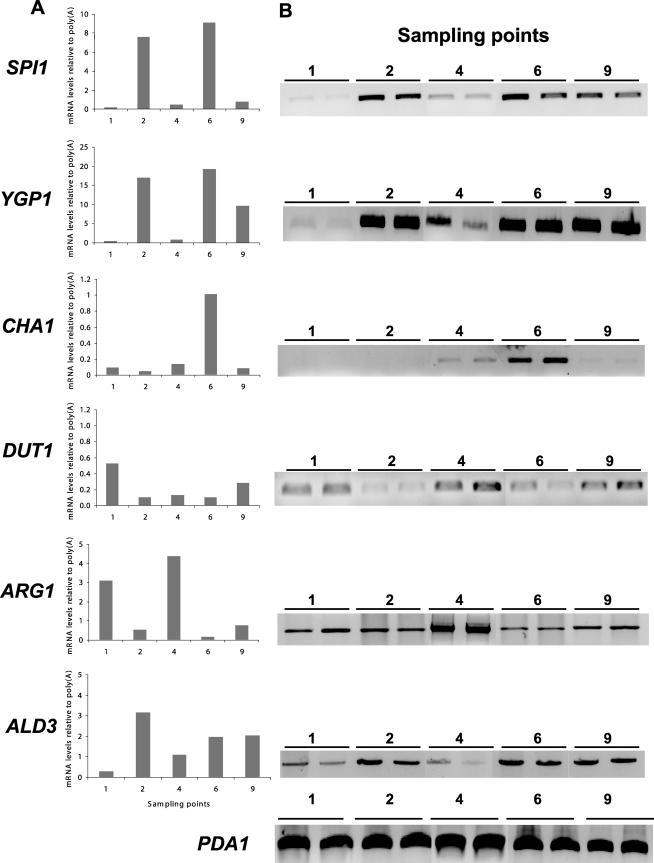
Validation of the macroarray data by RT-PCR.
The differential expression of genes obtained by macroarray analysis was confirmed by RT-PCR using the same RNA from the original macroarray experiments. Six genes with different expression profiles throughout the experiment (YPG1, ALD3, DUT1, SPI1, CHA1, and ARG1) were selected for validation by RT-PCR. As can be seen in Fig. 7, the data obtained from RT-PCR experiments (panel B) was essentially consistent with the results from the macroarray analyses (panel A). SPI1 and YGP1 genes, originally identified as being induced upon entry into stationary phase, displayed the highest mRNA levels after nitrogen depletion, corresponding to time points 2 and 6. The CHA1 gene, which encodes an l-serine (and l-threonine) deaminase, also showed the maximal expression at time point 6. In contrast, for DUT1 encoding a dUTPase, which catalyzes the hydrolysis of dUTP to dUMP and PPi, the lowest levels were found at time points 2 and 6, where nitrogen was limiting. In the case of ARG1 (encoding arginosuccinate synthetase), the highest mRNA values both in macroarrays and in RT-PCRs were found at time point 4 and the lowest at time point 6. Finally, ALD3, whose expression is known to be induced by stress and repressed by glucose, showed the highest expression at time points 2, 6, and 9.
FIG. 7.
RT-PCR analysis of selected genes. (A) Data obtained by macroarray analyses; (B) image of an agarose gel corresponding to samples treated as described in Materials and Methods. Two independent samples were used for the RT-PCR validation. In both panels, the results are normalized with those obtained with the PDA1 gene.
DISCUSSION
In the present study, array technology was used to evaluate gene expression profiles during three different fermentations, in order to assess at a genome-wide level how the yeast strain responded to the progressive nitrogen depletion and how it exited from such a metabolic state once nitrogen was added. These experiments were done using batch cultures, under conditions mimicking the enological environment; to our knowledge, these are the first studies done that take a genome-wide approach integrating the different situations of nitrogen supply. As previously shown (12, 15) yeast genome plasticity enables yeast cells to respond to different stress conditions with global changes at the transcriptome level. Accordingly, in the eleven samples analyzed, combining low and/or high concentrations of glucose, nitrogen, and ethanol and taken at different stages in each experiment, a total of 4,116 genes (roughly 70% of transcriptome) changed their mRNA levels in at least one condition across the experiments. The decrease in the number of genes differentially expressed during sluggish fermentation at time points 1 versus 4 (1,735 genes), 2 versus 5 (186 genes), and 3 versus 7 (210 genes) and the increase observed in that number after refeeding at time points 6 versus 9 (379 genes), 7 versus 10 (814 genes), and 8 versus 11 (1,734 genes) indicate that yeast cell responses are coordinated as nitrogen deficiency is sustained, and these results clearly show a role for nitrogen availability in the subsequent effects on the gene expression profile.
From the analysis of the transcriptome of yeast cells challenged by nitrogen deficiency, different situations can be highlighted. The early yeast cell response to low nitrogen availability (time points 2 and 4 compared to point 1) was characterized by the upregulation of genes involved in energy, carbohydrate metabolism, respiration, transport activity, and response to oxidative stress and a large number of genes with no predicted biological role. In contrast, genes involved in protein synthesis, as well as RNA metabolism, transcription, translation, and nucleotide and nucleobase metabolism, were downregulated, resembling the stereotypical changes known as the ESR (15) or common environmental response (12).
As the N-limiting fermentation progressed, the majority of genes involved in catabolism decreased in expression, including those involved in carbohydrate and nitrogen metabolism, suggesting that some metabolic pathways, namely glycolysis and fermentation, slowed down to enable cell survival, leading to sluggish fermentation. However, the oxidative metabolism of glucose seemed to be fully operational, according to the elevated mRNA levels of mitochondrial associated genes, despite the high amounts of glucose present. Previous reports suggested that when yeast cells sense intracellular nitrogen limitation, either by its absence from the medium or by ammonium transport inactivation, protein synthesis stops and the glucose transport begins to be irreversibly inactivated (9, 11, 22). This disabling of glucose transport could lead ultimately to an alleviation of the well-known Crabtree effect, associated with the decrease in fermentation rate and activation of respiratory genes. Accordingly, it has been suggested that the transcription activation of genes involved in the tricarboxylic acid cycle and respiration may be associated with low sugar uptake capacity and/or redox imbalance (20), with an obvious advantage once less sugar is needed to obtain the same amount of ATP (35) needed for cellular maintenance. In the current study, it was found that during the N-limiting fermentation, the expression levels of the glucose transport genes, both the low-affinity (HXT1 and HXT3) and the high-affinity (HXT2, HXT6, HXT7) carriers, were always greater than those seen in control fermentations (results not shown), indicating that the regulation of the glucose transporters is associated with translation or posttranslation levels rather than occurring at the transcriptional level.
The low mRNA levels of ribosomal protein genes at the early time point of the N-limiting fermentation were not directly associated with entrance into stationary phase, as happened in the control fermentation; since the cells were still at the exponential growth phase, this may be a mechanism of yeast cell adaptation to nitrogen limitation. This response is probably related to the already low nitrogen levels at time point 4; although cells are still actively growing, gene expression was anticipating the growth arrest that took place later on, as supported by a similar result for ribosome biogenesis genes (Fig. 4B). A reduction in yeast ribosomal protein mRNA levels that was not synchronized with a detectable change in growth rate or cell number was also observed after rapamycin treatment (27). A continuous decrease in mRNA levels of ribosomal protein genes over the time course of the sluggish fermentation would be expected. Nevertheless, the mRNA increase that followed suggests that the expression of ribosomal protein genes has a continuous role in cell survival under nitrogen starvation conditions as previously observed under other stress conditions, such as low temperature (30) or high salinity (41). The adaptation to the low-nitrogen circumstances could also explain the smaller decrease in poly(A) relative to total RNA, in spite of growth rate decay. It should be noted that genes belonging to the RiBi regulon (21, 38) that are tightly coregulated (15, 19, 21, 38) behaved distinctively in this study. Whereas ribosome biogenesis genes (Fig. 4B) showed a marked increase in expression during N starvation (points 4 to 8), the other genes included in the regulon (those encoding tRNA synthetases, translation factors, nucleotide metabolism, and RNA polymerases I and III) showed a different behavior with a decrease in either rich or poor nitrogen medium. To our knowledge, this is the first time that such a different response has been seen, which suggests the existence of an unknown regulatory mechanism apart from that already described for this regulon (reviewed in reference 21). Further studies will be necessary to clarify this aspect.
With the addition of a nitrogen source to nitrogen-starved yeast cells, in the presence of high glucose levels it would be expected that activation of the fermentable-growth medium pathway characterized by enhanced ribosomal protein synthesis and repression of stress response element-controlled genes would occur (34). However, these effects did not appear to be triggered under the conditions used in this study. In fact, genes involved in growth-related functions, such as those encoding ribosomal proteins and those involved in RNA processing and metabolism, were repressed. Nevertheless, nitrogen addition clearly enables the yeast strain to overcome the previous nitrogen starvation stress and restart alcoholic fermentation. Many genes involved in glycolysis, thiamine metabolism, and energy pathways were upregulated, which is consistent with a high fermentative activity.
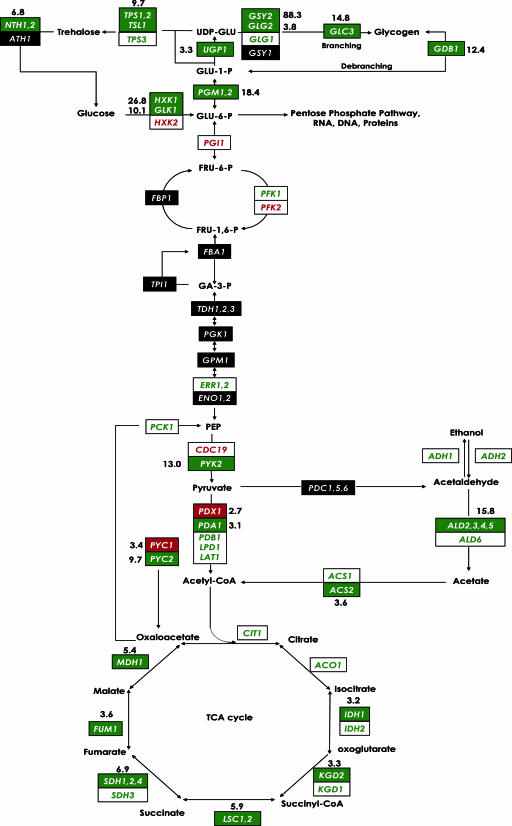
As a final remark, the results obtained within the present work have enabled a detailed evaluation of the most prevalent features in the yeast transcriptome and have provided new insights into changes in gene expression of S. cerevisiae challenged by nitrogen deficiency during an alcoholic fermentation at high glucose concentrations. The experimental conditions were different from those previously selected (3, 24, 28, 37, 42), and this made it possible to identify novel expression responses. First, it should be stressed that despite a high glucose concentration, yeast cells responded to a low-nitrogen challenge by inducing a great number of genes involved in carbon and energy metabolism, as summarized in Fig. 8. Second, the NCR-sensitive genes, which normally become derepressed with a poor nitrogen source or during nitrogen starvation in a target of rapamycin (TOR) protein response to nitrogen limitation (4, 5), are among the genes highly expressed at the onset of nitrogen-limiting fermentation; during the later stages, expression was strongly reduced. Third, genes encoding proteins with functions in ribosome structure or biogenesis as well as genes involved in rRNA metabolism were specifically induced during nitrogen-limiting fermentation and were repressed after nitrogen addition. Globally, the results provide a broad and integrated view of the gene expression changes that may occur under conditions mimicking those in the enological environment. The large amount of data made available from this study will be used to develop more efficient strategies and methods for the prediction and cure of problematic fermentations due to nitrogen deficiency.
FIG. 8.
Modifications in gene expression by comparisons of results from the earlier time points of the N-limiting versus control fermentations (time points 4 and 1). The yeast genes encoding the enzymes that catalyze each step in this metabolic circuit are identified by the names in the boxes. Green boxes with white letters identify genes whose expression is significantly higher at time point 4 of the N-limiting condition. White boxes with green letters identify genes whose expression is higher, but not significantly so, at that time point. Red boxes with white letters identify genes whose expression is significantly lower at time point 4 of the N-limiting condition. White boxes with red letters identify genes whose expression is lower, but not significantly so, at that time point. The magnitude of induction or repression is indicated only for significantly changed genes. For multimeric enzyme complexes, the indicated change (n-fold) represents an average for all the genes listed in the box. Black boxes and white letters indicate genes for which expression information was not available in one of the conditions. CoA, coenzyme A.
Supplementary Material
Acknowledgments
A. Mendes-Ferreira was supported in part by a Fundação Calouste Gulbenkian Grant during her stay at the Departamento de Bioquimica y Biologia Molecular of Universitat de València, València, Spain. Work in the Pérez-Ortín laboratory was funded by project BMC2003-07072-C03-02 from Ministerio de Educación y Ciencia to J.E.P.-O. and Grupos03/096 from Generalitat Valènciana to Vicente Tordera. This work has been also supported by grants AGL2002-01109 from the Spanish Ministerio de Ciencia y Tecnología, AGL2005-00508 from the Spanish Ministerio de Educación y Ciencia, and GRUPOS03/012 from the Generalitat Valenciana.
We thank R. N. Bennett for English revision of the manuscript.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alberola, T. M., J. García-Martínez, O. Antúnez, L. Viladevall, A. Barceló, J. Arino, and J. E. Pérez Ortín. 2004. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7:199-206. [PubMed] [Google Scholar]
- 2.Alexandre, H., and C. Charpentier. 1998. Biochemical aspects of stuck and sluggish fermentation in grape must. J. Ind. Microbiol. Biotech. 20:20-27. [Google Scholar]
- 3.Backhus, L. E., J. DeRisi, P. O. Brown, and L. F. Bisson. 2001. Functional genomic analysis of a commercial wine strain of Saccharomyces cerevisiae under differing nitrogen conditions. FEMS Yeast Res. 1:111-125. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, P. G., J. H. Choi, J. Carvalho, T. F. Chan, W. Ai, and X. F. Zheng. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, W. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 6.Bisson, L. F. 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 50:107-119. [Google Scholar]
- 7.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 8.Brauer, M. J., A. J. Saldanha, K. Dolinski, and D. Botstein. 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16:2503-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busturia, A., and R. Lagunas. 1986. Catabolite inactivation of the glucose transport system in Saccharomyces cerevisiae. J. Gen. Microbiol. 132:379-385. [DOI] [PubMed] [Google Scholar]
- 10.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso, H., and C. Leão. 1992. Sequential inactivation of ammonium and glucose transport in Saccharomyces cerevisiae during fermentation. FEMS Microb. Lett. 73:155-159. [DOI] [PubMed] [Google Scholar]
- 12.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, K. H., A. B. Pinchak, and T. G. Cooper. 1999. Genome-wide transcriptional analysis in S. cerevisiae by mini-array membrane hybridization. Yeast 15:703-713. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Martinez, J., Aranda, A., and J. E. Perez-Ortin. 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15:303-313. [DOI] [PubMed] [Google Scholar]
- 15.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henschke, P. A., and V. Jiranek. 1993. Yeasts—metabolism of nitrogen compounds, p. 77-164. In G. H. Fleet (ed.), Wine microbiology and biotechnology. Harwood Academic, Chur, Switzerland.
- 17.Herrero, J., A. Valencia, and J. Dopazo. 2001. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17:126-136. [DOI] [PubMed] [Google Scholar]
- 18.Herrero, J., F. Al-Shahrour, R. Diaz-Uriarte, A. Mateos, J. M. Vaquerizas, J. Santoyo, and J. Dopazo. 2003. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 31:3461-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y. S., J. M. Laplaza, and T. M. Jeffries. 2004. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl. Environ. Microbiol. 70:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagunas, R., C. Domínguez, A. Busturia, and M. J. Sáez. 1982. Mechanisms of appearance of the Pasteur effect in Saccharomyces cerevisiae: inactivation of the sugar transport systems. J. Bacteriol. 152:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manginot, C., J. L. Roustan, and J. M. Sablayrolles. 1998. Nitrogen demand of different yeast strains during alcoholic fermentation. Importance of the stationary phase. Enzyme Microbiol. Technol. 23:511-517. [Google Scholar]
- 24.Marks, V. D., G. K. van der Merwe, and H. J. J. van Vuuren. 2003. Transcriptional profiling of wine yeast in fermenting grape juice: regulatory effect of diammonium phosphate. FEMS Yeast Res. 3:269-287. [DOI] [PubMed] [Google Scholar]
- 25.Mendes-Ferreira, A., A. Mendes-Faia, and C. Leão. 2004. Growth and fermentation patterns of Saccharomyces cerevisiae under different ammonium concentrations and its implications in winemaking industry. J. Appl. Microbiol. 97:540-545. [DOI] [PubMed] [Google Scholar]
- 26.Parrou, J. L., B. Enjalbert, L. Plourde, A. Bauche, B. Gonzalez, and J. Francois. 1999. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast 15:191-203. [DOI] [PubMed] [Google Scholar]
- 27.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossignol, T., L. Dulau, A. Julien, and B. Blondin. 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 16:1369-1385. [DOI] [PubMed] [Google Scholar]
- 29.Saenz-Robles, M. T., M. Remacha, M. D. Vilella, S. Zinker, and J. P. G. Ballesta. 1990. The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta 1050:51-55. [DOI] [PubMed] [Google Scholar]
- 30.Sahara, T., T. Goda, and S. Ohgiya. 2002. Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J. Biol. Chem. 277:50015-50021. [DOI] [PubMed] [Google Scholar]
- 31.Saldanha, A. J., M. J. Brauer, and D. Botstein. 2004. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 15:4089-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmon, J. M. 1989. Effect of sugar transportation inactivation in Saccharomyces cerevisiae on sluggish and stuck enological fermentations. Appl. Environ. Microbiol. 55:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 34.Thevelein, J. M., and J. H. De Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 35.Thomsson, E., L. Gustafsson, and C. Larsson. 2005. Starvation response of Saccharomyces cerevisiae grown in anaerobic nitrogen- or carbon-limited chemostat cultures. Appl. Environ. Microbiol. 71:3007-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trabalzini, L., A. Paffetti, A. Scaloni, F. Talamo, E. Ferro, G. Coratza, L. Bovalini, P. Lusini, P. Martelli, and A. Santucci. 2003. Proteomic response to physiological fermentation stresses in a wild-type wine strain of Saccharomyces cerevisiae. Biochem. J. 370:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varela, C., J. Cárdenas, F. Melo, and E. Agosin. 2005. Quantitative analysis of wine yeast gene expression profiles under winemaking conditions. Yeast 22:369-383. [DOI] [PubMed] [Google Scholar]
- 38.Wade, C., K. A. Shea, R. V. Jensen, and M. A. McAlear. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21:8638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wenzel, T. J., M. A. Luttik, J. A. van den Berg, and H. Y. de Steensma. 1993. Regulation of the PDA1 gene encoding the E1 alpha subunit of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae. Eur. J. Biochem. 218:405-411. [DOI] [PubMed] [Google Scholar]
- 40.Wu, J., N. Zhang, A. Hayes, K. Panoutsopoulou, and S. G. Oliver. 2004. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA 101:3148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yale, J., and H. J. Bohnert. 2001. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276:15996-16007. [DOI] [PubMed] [Google Scholar]
- 42.Zuzuarregui, A., L. Monteoliva, C. Gil, and M. del Olmo. 2006. Transcriptomic and proteomic approach for understanding the molecular basis of adaptation of Saccharomyces cerevisiae to wine fermentation. Appl. Environ. Microbiol. 72:836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.