Abstract
Although it has been widely used as a feed supplement to reduce manure phosphorus pollution of swine and poultry, Aspergillus niger PhyA phytase is unable to withstand heat inactivation during feed pelleting. Crystal structure comparisons with its close homolog, the thermostable Aspergillus fumigatus phytase (Afp), suggest associations of thermostability with several key residues (E35, S42, R168, and R248) that form a hydrogen bond network in the E35-to-S42 region and ionic interactions between R168 and D161 and between R248 and D244. In this study, loss-of-function mutations (E35A, R168A, and R248A) were introduced singularly or in combination into seven mutants of Afp. All seven mutants displayed decreases in thermostability, with the highest loss (25% [P < 0.05]) in the triple mutant (E35A R168A R248A). Subsequently, a set of corresponding substitutions were introduced into nine mutants of PhyA to strengthen the hydrogen bonding and ionic interactions. While four mutants showed improved thermostability, the best response came from the quadruple mutant (A58E P65S Q191R T271R), which retained 20% greater (P < 0.05) activity after being heated at 80°C for 10 min and had a 7°C higher melting temperature than that of wild-type PhyA. This study demonstrates the functional importance of the hydrogen bond network and ionic interaction in supporting the high thermostability of Afp and the feasibility of adopting these structural units to improve the thermostability of a homologous PhyA phytase.
Phytase catalyzes the hydrolysis of phytate (myo-inositol hexakisphosphate), the major storage form of phosphorus in plant seeds (20), to phosphate and myo-inositol. The enzyme has been effectively used as an animal feed supplement to improve the bioavailability of phytate phosphorus to simple-stomached swine and poultry (4, 13). Because temperatures in the practical feed pelleting process can reach as high as 70 to 90°C (18), phytase enzymes with sufficiently high thermal stability are desirable but rare among the naturally occurring sources (14). Therefore, different strategies have been used to enhance the thermal stability of phytase (22).
Aspergillus fumigatus phytase (Afp) (19) is a well-known heat-resilient phytase; it retains 90% of its initial activity after being heated at 100°C for 20 min (21). In comparison, Aspergillus niger PhyA phytase (30) displays much less heat resistance, despite a higher specific activity and a better pH profile than those of Afp (17, 29, 33, 34). More intriguingly, these two phytases have 66% sequence homology and very similar overall crystal structures (9, 15, 35). Both enzymes contain a small α domain and a large α/β domain. The small α domain is composed of a long α helix and seven short α helices, and the large α/β domain contains a six-stranded β sheet surrounded by two long α helices at one side and several short α helices at the other side. Nevertheless, detailed structure comparisons between these two enzymes indicate that three amino acid residues in Afp—E35, R168, and R248—may be critical in maintaining its heat resilience (35). Specifically, E35 is predicted to be involved in a hydrogen bond network in the E35-to-S42 region, and R168 and R248 are predicted to interact with D161 and D244, respectively, to form multiple salt bridges (35). Based on crystal structure and sequence alignment, we identified three amino acid residues in PhyA—A58, Q191, T271—that correspond to E35, R168, and R248, respectively, in Afp. As these three residues in PhyA are not predicted to form any hydrogen bonding in the crystal structure of the enzyme (9, 28), they may be associated with the lower thermostability of PhyA. These structural predications require experimental verification. Therefore, we constructed a series of Afp and PhyA mutants by site-directed mutagenesis to (i) determine the individual and combined contributions of E35, R168, and R248, involved hydrogen bonding and ionic interactions, to the thermal stability of Afp, and (ii) improve the thermal stability of PhyA by substituting residues for those in the corresponding positions in Afp with the presumed ability to mediate putative hydrogen bonding and ionic interactions.
MATERIALS AND METHODS
Media and reagents.
The bacterial and yeast strains, plasmids, and primers used in this study are listed in Table 1. Escherichia coli DH5α was cultured at 37°C in LB medium. Pichia pastoris X33 was cultured at 30°C in either yeast extract-peptone-dextrose (YPD) medium or BMGY/BMMY medium (Invitrogen, San Diego, CA). Zeocin (Invitrogen, San Diego, CA) was added at 100 μg ml−1 to YPD or BMGY/BMMY medium for yeast and at 25 μg ml−1 to LB medium for E. coli. Restriction enzymes were obtained from Promega (Madison, WI). Oligonucleotides were synthesized at MWG-Biotech (High Point, NC). Phytic acid (inositol hexaphosphoric acid, dodecasodium salt from rice [P-3138]), ammonium molybdate tetrahydrate (A-7302), and l-ascorbic acid (A-0278) were purchased from Sigma (St. Louis, MO). Sulfuric acid (A300-212) and trichloroacetic acid (A322) were purchased from Fisher (Pittsburgh, PA). Automatic DNA sequencing was performed at Cornell Biotechnology Center.
TABLE 1.
Strains, plasmids, and synthetic oligonucleotide primers
| Item | Descriptiona | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | α complementation | Stratagene |
| P. pastoris X33 | Protein expression host | Invitrogen |
| Plasmids | ||
| pPICZα | ColE1 ori, ZeoR; for integration in P. pastoris | Invitrogen |
| pGAPZα | ColE1 ori, ZeoR; for integration in P. pastoris | Invitrogen |
| pPICZα-afp | afp fragment cloned into the EcoRI and XbaI sites of pPICZα | This study |
| pGAPZα-phyA | phyA fragment cloned between the EcoRI and XbaI sites of pGAPZα | This study |
| pafp E35A | E35A substitution of Afp in pPICZα-afp | This study |
| pafpR168A | R168A substitution of Afp in pPICZα-afp | This study |
| pafpR248A | R248A substitution of Afp in pPICZα-afp | This study |
| pafpE35A/R168A | E35A R168A substitution of Afp in pPICZα-afp | This study |
| pafpE35A/R248A | E35A R248A substitution of Afp in pPICZα-afp | This study |
| pafpR168A/R248A | R168A R248A substitution of Afp in pPICZα-afp | This study |
| pafpE35A/R168A/R248A | E35A R168A R248A substitution of Afp in pPICZα-afp | This study |
| pphyAA58E | A58E substitution of PhyA in pGAPZα-phyA | This study |
| pphyAQ191R | Q191R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAT271R | T271R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAA58E/Q191R | A58E Q191R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAA58E/T271R | A58E T271R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAQ191R/T271R | Q191R T271R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAA58E/Q191R/T271R | A58E Q191R T271R substitution of PhyA in pGAPZα-phyA | This study |
| pphyAA58E/P65S | A58E P65S substitution of PhyA in pGAPZα-phyA | This study |
| pphyAA58E/P65S/Q191R/T271R | A58E P65S Q191R T271R substitution of PhyA in pGAPZα-phyA | This study |
| Oligonucleotide primers | ||
| afp-F2 | 5′-GGATTTCGATGTTGCTGTTTTG-3′ | |
| afp-M1(E35A) | 5′-CAGCTCGTCCGCGAGCGAAAAG-3′ | |
| afp-M2(R168A) | 5′-GCGACGAACGCCGCCGCTCCG-3′ | |
| afp-M3(R248A) | 5′-ACGGTAGCGGCCACCAGCGAC-3′ | |
| afp-R2 | 5′-GTCGAGTTAGTGCTGGTGTGGT-3′ | |
| phyA-F | 5′-CGGAATTCCTGGCAGTCCCCGCCT-3′ | |
| phyA-A58E | 5′-ACCGATTCGTTTTCCAGAGAGAAGA-3′ | |
| phyA-P65S | 5′-GCGGGCACCTCAGAGGAGATGACC-3′ | |
| phyA-Q191R | 5′-TTGGGCGACGATCGGCCGGGCTGGG-3′ | |
| phyA-T271R | 5′-ACACCATCTCCAGAAGCACCGTCGA-3′ | |
| phyA-R | 5′-GCTCTAGACTAAGCAAAACACTCC-3′ |
Underlined primer sequence represents the mutated codon for the target amino acid residue.
Rational design of mutations.
To assess the contribution of E35, R168, and R248, involved hydrogen bonding and ionic interactions, to the thermostability of Afp, we altered each of the three residues to alanine so that these residues no longer participated in the predicted interactions. A total of seven mutants were constructed, including three single mutants (E35A, R168A, and R248A), three double mutants (E35A R168A, R168A R248A, and E35A R248A), and one triple mutant (E35A R168A R248A) (Table 1).
To test whether the thermostability of PhyA could be improved by adopting the putative hydrogen bonding and ionic interactions in Afp, we replaced residues A58, P65, Q191, and T271 in PhyA with residues at the corresponding positions from Afp (E35, S42, R168, and R248, respectively) (10, 35). In addition, the P65S mutation in PhyA was designed to form a hydrogen bonding network with E58, as in the case of Afp between E35 and S42 (35). In all, nine mutants were constructed: three single mutants (A58E, Q191R, and T271R), four double mutants (A58E Q191R, A58E T271R, Q191R T271R, and A58E P65S), one triple mutant (A58E Q191R T271R), and one quadruple mutant (A58E P65S Q191R T271R) (Table 1).
Site-directed mutagenesis.
Site-directed mutagenesis of the A. fumigatus afp and the A. niger phyA genes was conducted with megaprimer PCR mutagenesis. Briefly, DNA fragments containing the desired point mutations were produced in two sequential PCRs. The first reaction used a 22- to 25-nucleotide primer containing a single nucleotide alteration (Table 1) and a 22- to 25-nucleotide reverse primer. The resulting PCR products were size fractionated with a 1.5% agarose gel and purified with the QIAquick gel extraction kit (QIAGEN, Valencia, CA). These PCR fragments were then used as megaprimers in a second PCR, along with a forward primer. The second PCR products were then digested at each end by two restriction enzymes and introduced into the wild-type (WT) template to replace the corresponding gene fragments. The multiple mutations were generated by splicing the template containing the single mutations after restriction enzyme digestion and combining the mutations. Plasmids containing the mutations were verified by automated DNA sequencing at Cornell Biotechnology Center.
Protein expression.
Pichia pastoris X33 was used as an expression host. The A. fumigatus phytase gene and seven mutants were cloned in pPICZα vector and transformed into P. pastoris X33 by electrophoresis with an ECM 600 Electro Cell Manipulator (BTX Instrument Division, Gentronics, Inc., San Diego, CA). Individual transformants were grown in BMGY medium for 1 day before being transferred to BMMY inducible medium. Cells were grown at 28.5°C for up to 144 h with aeration (220 rpm). Methanol was added as an inducer first after 24 h of culturing at a final concentration of 0.5% and then after 48, 72, 96, 120, and 144 h of culturing, to maintain a constant concentration of methanol. At each time point, 1 ml of culture was collected and centrifuged at 12,000 × g for 30 min. Supernatants were saved at −20°C for analysis later. The WT phyA gene and the nine mutants were cloned into pGAPZα vector and transformed into P. pastoris. Individual transformants were grown in YPD expression medium with aeration (220 rpm) at 30°C for 48 to 72 h until the optical density at 600 nm (OD600) reached 20.
Purification of the expressed phytases.
Cultures of WT and mutant Afp transformants were centrifuged at 12,000 × g for 30 min to remove cell debris. After the supernatants were concentrated approximately 10-fold by an Amicon centrifugal filter device (Centriplus YM-30 [molecular weight cutoff, 30,000]; Fisher, Pittsburgh, PA), the expressed enzymes were purified by nickel-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography (QIAGEN). Appropriate amounts of Ni-NTA resin (10 μl of resin for 50 to 100 μg of His6-tagged protein) were added to the supernatant and mixed gently for 30 min. The resin was precipitated by centrifugation for 10 s at 15,000 × g and then washed twice with a washing buffer (50 mM NaH2PO4-300 mM NaCl-20 mM imidazole, adjusting the pH to 8.0 with NaOH). Proteins were eluted by incubating the resin with the elution buffer (50 mM NaH2PO4-300 mM NaCl-250 mM imidazole, adjusting the pH to 8.0) three times.
After cultures of WT and mutant PhyA transformants were centrifuged at 12,000 × g for 30 min to remove cell debris, the supernatants were concentrated approximately 20-fold by ultrafiltration. The concentrate was subjected to DEAE cation-exchange chromatography (Sigma). The DEAE column was balanced with 1,000 ml of 10 mM Tris-HCl buffer (pH 7.4). The proteins were eluted with 300 ml of an elution buffer (10 mM Tris-HCl, pH 7.4) with a linear gradient of NaCl from 0 to 0.3 M. The flowthrough fractions were collected by an automatic fraction collector. The fraction profiles of OD280 and phytase activity were checked to determine the desired protein peaks. The peak fractions were pooled and concentrated to less than 2 ml by an Amicon centrifugal filter device and then loaded onto a Sephadex100 sizing column (Sigma) previously equilibrated with 10 mM Tris-HCl buffer (pH 7.4) containing 0.15 M NaCl. Peak fractions were used for further characterization.
Biochemical characterization of the expressed phytases.
Phytase activity was assayed as previously described (7). The pH activity profile of phytase was determined at 37°C with two different buffers, 0.2 M glycine-HCl buffer for pH 2.0 to 3.0 and 0.2 M citrate buffer for pH 3.5 to 6.5 (6, 7). The optimal temperature of the phytase was tested with 0.2 M citrate buffer at pH 5.5. The thermal stability of the phytase was tested with both the culture supernatants and purified phytase proteins. For supernatants, the samples were diluted with 0.2 M citrate buffer (pH 5.5) to 0.2 U of phytase activity per ml. For purified proteins, the samples were diluted with 0.2 M citrate buffer (pH 5.5) to 10 μg of phytase protein per ml. Concentration of the purified proteins was determined by the Lowry assay (16). The diluted samples were incubated for 10 min at each of the following temperatures: 50, 60, 70, 80, 90, and 100°C. Immediately after heat treatment, the samples were placed on ice for 30 min (6, 7). Phytase activity was measured at 37°C and pH 5.5, as described above.
Differential scanning calorimetry.
Melting temperatures (Tm) of the purified WT and mutant A58E P65S Q191R T271R PhyA proteins were determined with a differential scanning calorimeter (model Q10; TA Instruments, New Castle, DE) equipped with a refrigerated cooling system and Thermal Advantage for Q Series software. The purified phytases were concentrated to 50 mg/ml by freeze-drying with a Jouan RC1010 speed vacuum in 50 mM Tris-HCl buffer, pH 7.4. After 8 mg of each protein sample was sealed in stainless steel cells, equilibrated at 10°C followed by isothermaling for 2 min, the proteins were scanned from 30°C to 100°C at a heating rate of 2°C per min. Data were collected at a rate of 0.1 s per point.
Kinetic parameters Km and Vmax.
The kinetic parameters Km and Vmax of selected phytases were determined at both pH 3.5 and pH 5.5. Purified samples were diluted with 0.2 M citrate buffer (pH 3.5 or 5.5) to a final concentration of 0.1 U of phytase activity per ml. The phytase activity assay was carried out with phytic acid dodecasodium salts used as the substrate at 13 different concentrations (2.5, 5, 7.5, 10, 25, 50, 75, 100, 250, 500, 750, 1,000, and 2,500 μM). Four parallel series of phytase reactions were carried out with different reaction times: 5, 10, 15, and 20 min. Data were analyzed as following: (i) a plot of phytase activity versus reaction time to calculate initial velocities (μmol/min), (ii) a plot of initial velocities versus substrate concentrations (V0 versus [S]), and (iii) a reciprocal plot of V0 and [S] to make a Lineweaver-Burk plot and calculation of the Km and Vmax, respectively.
Hydrolysis of phytate phosphorus from soybean meal.
The hydrolysis of phytate phosphorus from soybean meal was conducted by incubating WT and mutant (A58E Q191R, A58E Q191R T271R, A58E P65S, and A58E P65S Q191R T271R) PhyA enzymes with soybean meal at ratios of 250, 500, 750, and 1,000 U of phytase per kg of soybean meal. The hydrolysis was carried out at pH 5.5 or 3.5 at 37°C for 1 h. The released inorganic phosphorus was determined as previously described (7).
Statistical analysis.
Data were analyzed by Minitab (release 14; Minitab Inc., State College, PA). The Bonferroni t test was used to compare mean differences. Significance was set at a P value of <0.05.
RESULTS
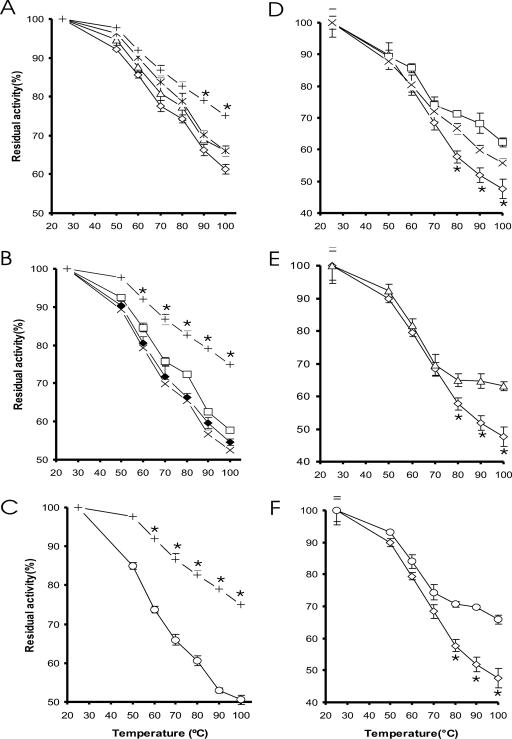
Mutations in residues E35, R168, and R248 of Afp decrease its thermal stability.
Each of the single and combined substitutions of residues E35, R168, and R248 with alanine in Afp resulted in a reduction in enzyme thermostability (Fig. 1A). Among the three single mutants, E35A decreased (P < 0.05) the enzyme thermostability at ≥70°C. The other two single mutants (R168A and R248A) showed decreases (P < 0.05) at ≥90°C. Three double mutants showed lower (P < 0.05) thermostability than that of the single mutants or the WT, at ≥60°C (Fig. 1B). Among the seven mutants, the triple mutant had the lowest thermostability, showing 25% lower residual activity than the WT after heat treatment at 100°C (Fig. 1C). The seven purified Afp mutants and the WT enzyme exhibited similar specific activities (31.8 to 35.1 U/mg protein) and pH activity profiles. All of these expressed Afp phytases also shared a pH optimum of 5.5 and a temperature optimum of 55°C (data not shown).
FIG. 1.
Effects of mutations on the residual activities of the A. fumigatus Afp (A to C) and A. niger PhyA (D to F) phytases after being heated to various temperatures for 10 min. (A) WT (+), E35A (⋄), R168A (▵), and R248A (*). (B) WT (+), E35A R168A (×), E35A R248A(□), and R168A R248A(♦). (C) WT (+) and E35A R168A R248A (○). (D) WT (⋄), A58E Q191R (□), and A58E P65S (×). (E) WT (⋄) and A58E Q191R T271R (▵). (F) WT (⋄) and A58E P65S Q191R T271R (○). An asterisk indicates a difference (P < 0.05) between the residual activities of the respective WT protein and the mutants.
Substitutions A58E, P65S, Q191R, and T271R in PhyA enhance its thermostability.
In the one-temperature (80°C for 10 min) screening assay of PhyA mutants, two double mutants (A58E Q191R and A58E P65S), one triple mutant (A58E Q191R T271R), and one quadruple mutant (A58E P65S Q191R T271R) showed promising improvement in residual activity (data not shown). Thus, these four PhyA mutants were selected for further characterization and, along with the WT, were heated at different temperatures from 50 to 100°C for 10 min. Each of the four mutants showed greater residual activity (P < 0.01) than that of the WT at ≥80°C (Fig. 1D to F). The quadruple mutant (A58E P65S Q191R T271R) showed the highest heat stability, with 20% higher residual activity than that of the WT after being heated at 100°C (Fig. 1F). While the melting temperature of WT was 66.3°C, the thermogram of the quadruple mutant showed transition midpoints at 71.5 and 73.8°C, respectively.
Substitutions A58E and P65S in PhyA improve its binding affinity toward sodium phytate.
Steady-state kinetic measurements revealed that the apparent Km of the WT was 167.5 μM sodium phytate (Table 2). The apparent Km of the PhyA mutants A58E P65S and A58E P65S Q191R T271R decreased by approximately 22% and 36%, respectively, compared with that of WT. In contrast, the WT and four PhyA mutants had similar Vmax and specific activity. Likewise, these four mutants showed no shift in pH activity profile, optimal pH (2.5 and 5.5), or optimal temperature (55°C) from that of the WT (data not shown).
TABLE 2.
Specific activities and kinetic parameters of WT and selected mutant PhyA phytases at pH 5.5
| PhyA phytase | Sp act (U/mg) (mean ± SD) | Vmax (μM/min ·mg) | Km (μM) |
|---|---|---|---|
| WT PhyA | 80.9 ± 1.1 | 135.1 | 167.5 |
| PhyA(A58E Q191R) | 82.2 ± 1.2 | 128.2 | 154.0 |
| PhyA(A58E Q191R T271R) | 83.8 ± 4.1 | 169.5 | 162.3 |
| PhyA(A58E P65S) | 84.2 ± 3.4 | 109.9 | 130.4 |
| PhyA(A58E P65S Q191R T271R) | 83.2 ± 2.1 | 117.6 | 106.4 |
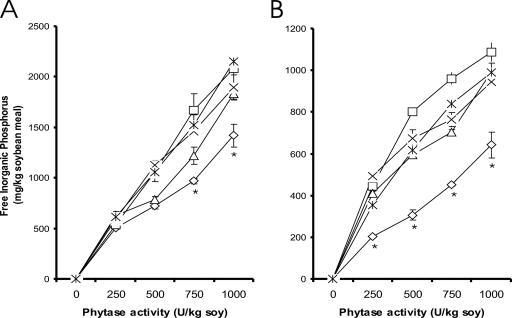
Substitutions A58E, P65S, Q191R, and T271R in PhyA elevate hydrolysis of phytate-phosphorus from soybean meal.
At pH 5.5, mutants A58E Q191R, A58E Q191R T271R, A58E P65S, and A58E P65S Q191R T271R released greater (P < 0.01) amounts of inorganic phosphate from phytate in soybean meal than those by the WT at 750 and 1,000 U per kg of soybean meal (Fig. 2A). At pH 3.5, such differences became significant (P < 0.01) at all tested ratios of phytase and soybean meal (Fig. 2B).
FIG. 2.
Effects of mutations in A. niger PhyA on hydrolysis of phytate-phosphorus from soybean meal at concentrations of 250, 500, 750, and 1,000 U/kg of soybean meal at 37°C in 0.2 M citrate buffer for 1 h at pH 5.5 (A) and pH 3.5 (B). Represented are WT (⋄), A58E Q191R (□), A58E Q191R T271R (▵), A58E P65S (×), and A58E P65S Q191R T271R (*). An asterisk indicates a difference (P < 0.05) between each mutant and the WT.
DISCUSSION
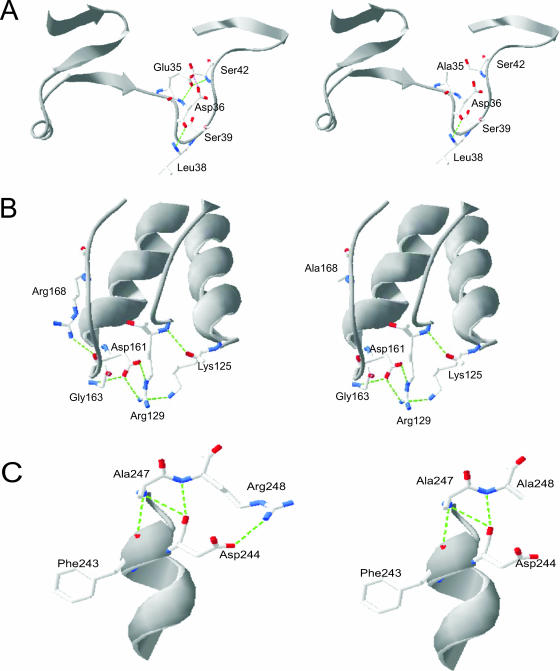
Based on the crystal structure of Afp (35), two strong hydrogen bonds with distances of 2.54 and 2.95 Å can be formed between E35 and S42 (Fig. 3A). Because these bonds may help maintain the hydrogen bond network in the E35-to-S42 region and thereby contribute to enzyme stability, disruption of the hydrogen bonding by the substitution in the E35A mutant explains its thermal stability reduction compared with the WT. The residues R168 and R248 in the WT (35) can interact with D161 and D244, respectively, through salt bridges to form a C-terminal capping box structure that stabilizes the helical conformation of oligopeptides and hydrogen bonds with distances from 2.7 to 2.9 Å (Fig. 3B and C). As expected, the replacement of Arg with Ala in the mutant R168A disrupted these structures, lowering the enzyme thermostability (Fig. 3B). Similarly, the same substitution in mutant R248A (Fig. 3C) and three other mutants—E35A R248A, R168A R248A, and E35A R168A R248A—interrupted the hydrogen bond between the side chain of R248 and the carboxylate group of D244, causing decreased thermostability in all of these mutants. Overall, the three residues (E35, R168, and R248) bear similar contributions to the thermostability of Afp. It is noteworthy that the deleterious effect of each single mutation was additive, as the triple mutant showed the lowest thermostability.
FIG. 3.
Structural prediction of the residual interactions in A. fumigatus phytase (Afp) before and after substitutions at E35, R168, and R248. Green dotted lines represent the hydrogen bond interactions: predicted changes in hydrogen bond interactions caused by the substitution E35A (A); predicted changes in ionic interactions caused by the substitution R168A (B); and predicted changes in ionic interactions caused by the substitution R248A (C).
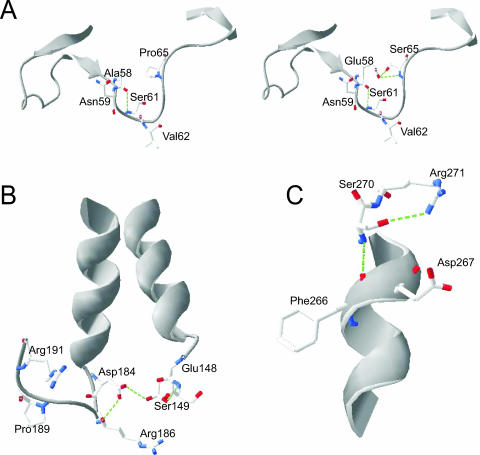
The loss of thermostability in Afp by disruption of selected hydrogen bonds and salt bridges prompted us to take the opposite approach in PhyA in hopes of improving its heat stability. In fact, the double mutation in the mutant A58E P65S introduced two new hydrogen bonds with predicted distances of 3.01 Å and 3.66 Å, respectively (5) (Fig. 4A). Though hydrogen bonds with such distances are rather weak, the actual distances for these bonds between the two amino acids (E58 and S65) located in the loop region may be as close as 1.2 to 2.5Å through the rotamer change of their side chains. The importance of these hydrogen bonds and the need for the mutation P65S to form such bonds are clearly illustrated by the fact that only the double mutant A58E P65S, and not the single mutant A58E, showed a detectable increase in thermostability over the WT after being heated at 80°C for 10 min.
FIG. 4.
Structural rationale of the designated mutations in A. niger PhyA. Green dotted lines represent the hydrogen bond interactions. (A) A comparison of hydrogen bonding between the WT (left) and the double mutant A58E P65S (right) shows that two new hydrogen bonds with distances of 3.01 and 3.66 Å, respectively, were introduced between substitutions E58 and S65 (right). (B) The single mutation Q191R removes a repulsive ionic interaction in the loop region, and R191 can interact with D184 through a salt bridge to stabilize the structure. (C) The single mutation T271R can render the residue interacting with D267 through a salt bridge and also introduces a new hydrogen bond between R271 and S270 with a distance of 2.71 Å.
Mutations Q191R (Fig. 4B) and T271R (Fig. 4C) in PhyA created new salt bridges between D184 and D267, respectively. Also, the former mutation (Q191R) removed repulsive ionic interactions between Q191 and D184 in the loop region (Fig. 4B), and the latter mutation (T271R) introduced a rather strong new hydrogen bond between R271 and S270, with a distance of 2.71 Å (Fig. 4C). These modifications certainly help to stabilize PhyA. However, mutations Q191R and T271R exerted much less positive impacts on PhyA thermostability than did mutation A58E P65S. This is probably because A58 and P65 are located in the loop region, while Q191 and T271 are both located in the more rigid α helix regions. As the secondary structure change in the loop region directly affects the tertiary structure of the protein (2), mutations there could alter thermostability of proteins to a larger extent than those in the α helix regions.
A number of studies have shown that electrostatic and hydrogen bonding interactions in thermophilic proteins are responsible for much of the increased stability over their mesophilic counterparts (1, 8, 12, 23, 26, 27, 31, 32). The hydrogen bonding appears to be the most important factor for thermal stability in 16 families of proteins (32). The number of ion pairs is also found to be associated with thermal stability, but the correlation is not as strong as with hydrogen bonding (32). Because the temperature-induced denaturing of proteins is affected by these noncovalent interactions (24), the enhanced thermotolerance, including the 7°C increase in melting temperature, in mutant A58E P65S Q191R T271R could be attributed to the introduced hydrogen bonding and salt bridges by the four amino acid substitutions. The two peaks seen in the thermogram of the mutant may represent independent folding of the two protein domains, the α domain and the α/β domain (11).
The increased melting temperature of mutant A58E P65S Q191R T271R will help the enzyme to resist heat inactivation during feed pelleting (18). It is also promising that the point mutations in PhyA for better thermal stability did not produce negative impacts on its catalytic properties (25). The four thermostable PhyA mutants had similar specific activities, but higher efficiency in hydrolysis of phytate phosphorus from soybean meal at both pH 3.5 and 5.5 at 37°C, than did the WT. This is an especially desired feature for phytase, since soybean meal, a commonly used animal feed ingredient, is the major source of dietary phytate for swine and poultry (3). Neither Afp nor PhyA mutants displayed shifts in pH activity profiles, optimal pH, or optimal temperature compared to the respective WT enzymes. The structural basis for the thermostability of these two phytases is seemingly fairly independent of that for other properties of the enzymes (10).
In summary, our research has provided experimental evidence for the predicted hydrogen bonding or salt bridge as the main structural basis for the superior thermal stability of Afp to PhyA (35). More excitingly, the introduction of unique tertiary structures from Afp to PhyA by rational design resulted in significant improvements in the thermal stability and melting temperature of PhyA. Contrary to the conventional view (25), the gain of thermostability in PhyA at high temperatures did not compromise its function at the (low) body temperature of animals but enhanced its efficiency in hydrolysis of phytate from soybean meal. These combined improvements render the mutant one step closer to being an “ideal phytase” (14).
Acknowledgments
This research was supported in part by a Cornell Biotechnology Program grant (to X.G.L.).
We thank David B. Wilson and Q. Hao for helpful suggestions and Syed S. H. Rizvi and Philipina A. Marcelo for technical assistance with differential scanning calorimetry.
Footnotes
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Acharya, P., E. Rajakumara, R. Sankaranarayanan, and N. M. Rao. 2004. Structural basis of selection and thermostability of laboratory evolved Bacillus subtilis lipase. J. Mol. Biol. 341:1271-1281. [DOI] [PubMed] [Google Scholar]
- 2.Bogin, O., I. Levin, Y. Hacham, S. Tel-Or, M. Peretz, F. Frolow, and Y. Burstein. 2002. Structural basis for the enhanced thermal stability of alcohol dehydrogenase mutants from the mesophilic bacterium Clostridium beijerinckii: contribution of salt bridging. Protein Sci. 11:2561-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cromwell, G. L. 1980. Biological availability of phosphorus in feedstuffs for swine. Feedstuffs 52:14-16. [Google Scholar]
- 4.Gentile, J. M., K. R. Roneker, S. E. Crowe, W. G. Pond, and X. G. Lei. 2003. Effectiveness of an experimental consensus phytase in improving dietary phytate-phosphorus utilization by weanling pigs. J. Anim. Sci. 81:2751-2757. [DOI] [PubMed] [Google Scholar]
- 5.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 6.Han, Y., and X. G. Lei. 1999. Role of glycosylation in the functional expression of an Aspergillus niger phytase (PhyA) in Pichia pastoris. Arch. Biochem. Biophys. 364:83-90. [DOI] [PubMed] [Google Scholar]
- 7.Han, Y., D. B. Wilson, and X. G. Lei. 1999. Expression of an Aspergillus niger phytase gene (phyA) in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 65:1915-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karshikoff, A., and R. Ladenstein. 1998. Proteins from thermophilic and mesophilic organisms essentially do not differ in packing. Protein Eng. 11:867-872. [DOI] [PubMed] [Google Scholar]
- 9.Kostrewa, D., F. Gruninger-Leitch, A. D'Arcy, C. Broger, D. Mitchell, and A. P. G. M. van Loon. 1997. Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution. Nat. Struct. Biol. 4:85-190. [DOI] [PubMed] [Google Scholar]
- 10.Kostrewa, D., M. Wyss, A. D'Arcy, and A. P. G. M. van Loon. 1999. Crystal structure of Aspergillus niger pH2.5 acid phosphatase at 2.4Å resolution. J. Mol. Biol. 288:965-974. [DOI] [PubMed] [Google Scholar]
- 11.Kundu, S., M. Sundd, and M. V. Jagannadham. 2002. Alcohol and temperature induced conformational transitions in ervatamin B: sequential unfolding of domains. J. Biochem. Mol. Biol. 35:155-164. [DOI] [PubMed] [Google Scholar]
- 12.Ladenstein, R., and G. Antranikian. 1998. Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv. Biochem. Eng. Biotechnol. 61:37-85. [DOI] [PubMed] [Google Scholar]
- 13.Lei, X. G., P. K. Ku, E. R. Miller, D. E. Ullrey, and M. T. Yokoyama. 1993. Supplemental microbial phytase improves bioavailability of dietary zinc to weanling pigs. J. Nutr. 123:1117-1123. [DOI] [PubMed] [Google Scholar]
- 14.Lei, X. G., and C. H. Stahl. 2001. Biotechnological development of effective phytases for mineral nutrition and environmental protection. Appl. Microbiol. Biotechnol. 57:474-481. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Q., Q. Huang, X. G. Lei, and Q. Hao. 2004. Crystallographic snapshots of Aspergillus fumigatus phytase, revealing its enzymatic dynamics. Structure 12:1575-1583. [DOI] [PubMed] [Google Scholar]
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 17.Mullaney, E. J., C. B. Daly, T. Kim, J. M. Porres, X. G. Lei, K. Sethumadhavan, and A. H. J. Ullah. 2002. Site-directed mutagenesis of Aspergillus niger NRRL 3135 phytase at residue 300 to enhance catalysis at pH 4.0. Biochem. Biophys. Res. Commun. 297:1016-1020. [DOI] [PubMed] [Google Scholar]
- 18.Mullaney, E. J., C. B. Daly, and A. H. J. Ullah. 2000. Advances in phytase research. Adv. Appl. Microbiol. 47:157-199. [DOI] [PubMed] [Google Scholar]
- 19.Pasamontes, L., M. Haiker, M. Wyss, M. Tessier, and A. van Loon. 1997. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 63:1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy, N. R., S. K. Sathe, and D. K. Salukhe. 1982. Phytates in legumes and cereals. Adv. Food Res. 28:1-92. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez, E., E. J. Mullaney, and X. G. Lei. 2000. Expression of the Aspergillus fumigatus phytase gene in Pichia pastoris and characterization of the recombinant enzyme. Biochem. Biophys. Res. Commun. 268:373-378. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, E., Z. A. Wood, P. A. Karplus, and X. G. Lei. 2000. Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch. Biochem. Biophys. 382:105-112. [DOI] [PubMed] [Google Scholar]
- 23.Scandurra, R., V. Consalvi, R. Chiaraluce, L. Politi, and P. C. Engel. 1998. Protein thermostability in extremophiles. Biochimie 80:933-941. [DOI] [PubMed] [Google Scholar]
- 24.Scopes, R. K. 1994. Protein purification: principles and practice. Springer-Verlag, New York, NY.
- 25.Shoichet, B., W. Baase, R. Kuroki, and B. Matthews. 1995. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA 92:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szilagyi, A., and P. Zavodszky. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8:493-504. [DOI] [PubMed] [Google Scholar]
- 27.Tigerstrom, A., F. Schwarz, G. Karlsson, M. Okvist, C. Alvarez-Rua, D. Meader, F. T. Robb, and L. Sjolin. 2004. Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin. Biochemistry 43:12563-12574. [DOI] [PubMed] [Google Scholar]
- 28.Tomschy, A., M. Wyss, D. Kostrewa, K. Vogel, M. Tessier, S. Hofer, H. Burgin, A. Kronenberger, R. Remy, A. P. G. M. van Loon, and L. Pasamontes. 2000. Active site residue 297 of Aspergillus niger phytase critically affects the catalytic properties. FEBS Lett. 472:169-172. [DOI] [PubMed] [Google Scholar]
- 29.Ullah, A. H. J., K. Sethumadhavan, X. G. Lei, and E. J. Mullaney. 2000. Biochemical characterization of cloned Aspergillus fumigatus phytase (PhyA). Biochem. Biophys. Res. Commun. 275:279-285. [DOI] [PubMed] [Google Scholar]
- 30.van Hartingsveldt, W., C. M. J. van Zeijl, G. M. Harteveld, R. J. Gouka, M. E. G. Suukerbuyk, R. G. M. Luiten, P. A. van Paridon, G. C. M. Selten, A. E. Veenstra, R. F. M. van Gorcom, and A. M. J. J. van den Hondel. 1993. Cloning, characterization and overexpression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene 127:87-94. [DOI] [PubMed] [Google Scholar]
- 31.Vogt, G., and P. Argos. 1997. Protein thermal stability: hydrogen bonds or internal packing? Folding Design 2:S40-S46. [DOI] [PubMed] [Google Scholar]
- 32.Vogt, G., S. Woell, and P. Argos. 1997. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 269:631-643. [DOI] [PubMed] [Google Scholar]
- 33.Wyss, M., L. Pasamontes, A. Friedlein, R. Remy, M. Tessier, A. Kronenberger, A. Middendorf, M. Lehmann, L. Schnoebelen, U. Rothlisberger, E. Kusznir, G. Wahl, F. Muller, H.-W. Lahm, K. Vogel, and A. P. G. M. van Loon. 1999. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl. Environ. Microbiol. 65:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyss, M., L. Pasamontes, R. Remy, J. Kohler, E. Kusznir, M. Gadient, F. Muller, and A. P. G. M. van Loon. 1998. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl. Environ. Microbiol. 64:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang, T., Q. Liu, A. M. Deacon, M. Koshy, I. A. Kriksunov, X. G. Lei, Q. Hao, and D. J. Thiel. 2004. Crystal structure of a heat-resilient phytase from Aspergillus fumigatus, carrying a phosphorylated histidine. J. Mol. Biol. 339:437-445. [DOI] [PubMed] [Google Scholar]