Abstract
The Listeria monocytogenes genome contains genes encoding several internalins and internalin-like proteins. As L. monocytogenes is present in many environments and can infect numerous, diverse host species, the environmental temperature was hypothesized to be a signal that might affect internalin gene transcription. A subgenomic microarray was used to investigate temperature-dependent transcription of 24 members of the internalin gene family in L. monocytogenes 10403S. The levels of internalin gene transcripts for cells grown at 37°C were compared to the levels of transcripts for cells grown at 16, 30, and 42°C using competitive microarray hybridization, and the results were confirmed by performing quantitative reverse transcriptase PCR for 14 internalin genes. Based on these studies, the internalin genes can be grouped into the following five temperature-dependent categories: (i) four σB-dependent internalin genes (inlC2, inlD, lmo0331, and lmo0610) with the highest levels of transcripts at 16°C and generally the lowest levels of transcripts at 37°C; (ii) three partially PrfA-dependent internalin genes (inlA, inlB, and inlC) with the lowest levels of transcripts at 16°C and the highest levels of transcripts at 37 and 42°C; (iii) four genes (inlG, inlJ, lmo0514, and lmo1290) with the lowest levels of transcripts at 16°C and the highest levels of transcripts at 30 and/or 37°C; (iv) one gene (lmo0327) with the highest levels of transcripts at 16°C and low levels of transcripts at higher temperatures; and (v) 12 internalin genes with no differences in the levels of transcripts at the temperatures used in this study. The temperature-dependent transcription patterns suggest that the relative importance of different internalins varies by environment, which may provide insight into the specific functions of these proteins.
The genus Listeria includes both human and animal pathogens (L. monocytogenes and L. ivanovii), as well as apparently nonpathogenic species (L. innocua, L. seeligeri, L. grayi, and L. welshimeri), which either have not been linked or only rarely have been linked to human or animal infections (11, 40, 54, 63). L. monocytogenes and L. ivanovii are capable of invading a number of different eukaryotic cell types and can subsequently multiply and spread intracellularly (46). In addition to being a human pathogen, L. monocytogenes also has been shown to infect over 40 species of wild and domesticated animals (25). On the other hand, L. ivanovii appears to have a narrower host range; while it predominantly causes abortions in sheep and cattle, it also has been responsible for a small number of opportunistic human infections (43). The pathogenic and nonpathogenic Listeria spp. appear to be able to survive well outside animal hosts and have been isolated from a diverse range of environments (43, 46); the possible exception is L. ivanovii, which appears to have been isolated only rarely from environmental sources (56). Listeria spp. are thus often considered saprophytes.
Interestingly, the genomes of both pathogenic and nonpathogenic Listeria spp. contain genes encoding several internalins and internalin-like proteins (12, 13, 19, 20, 28, 51). The internalins comprise a family of proteins characterized by the presence of repeat motifs of regularly spaced leucine or isoleucine residues (45). The leucine-rich repeat motif, which is broadly present in many groups of proteins, is thought to provide a versatile structural framework for protein-protein interactions (37, 38). The Listeria internalins are either surface bound (through an LPXTG binding motif or, less commonly, through a GW domain) or secreted (7). Comparative analyses of four L. monocytogenes genomes resulted in identification of between 24 and 29 genetic loci encoding internalin-like proteins in the strains sequenced (51). Genomic microarray characterization of 113 Listeria isolates representing all six species showed that internalin-like genes are present in all members of this genus and revealed the presence of at least four L. seeligeri genes with homology to L. monocytogenes internalin genes (13). A total of 14 genes encoding internalin-like proteins have been identified to date in L. ivanovii (12, 19, 20). By comparison, analyses of the L. innocua and L. welshimeri genome sequences revealed the presence of 19 and 8 genes encoding internalin-like proteins, respectively (28).
While a number of internalin genes have been characterized in L. monocytogenes, specific functions have been identified for only a few internalin proteins. InlA and InlB have been clearly demonstrated to be important for host cell invasion and virulence (14, 22). InlA interacts with the adhesion molecule E-cadherin (48), while InlB interacts with at least three cell surface molecules, including the hepatocyte growth factor receptor Met (58), gC1qR (5), and glycosaminoglycans (31). InlA, which by itself triggers inefficient invasion of Caco-2 cells, appears to require the support of InlB, InlC, and InlGHE (3). A null mutation in L. monocytogenes inlJ significantly attenuates virulence in intravenously infected mice or in orally inoculated mice expressing human E-cadherin (55). Lmo0327 appears to be an enzyme with murein-hydrolyzing activity. A null mutation in lmo0327 did not result in reduced adhesion or invasion of human embryo intestinal epithelial INT407 cells (52). Similarly, a null mutation in inlI (lmo0333) did not result in reduced invasion of tissue culture cells or reduced virulence in a mouse model (55). In L. ivanovii, 12 genes encoding internalin-like proteins are present in a single virulence gene island, while two internalin genes (i-inlC and i-inlD) are located elsewhere in the chromosome (12, 19, 20). Characterization of null mutations in two L. ivanovii internalin-like protein genes (i-inlE and i-inlF) indicated that these genes contribute to virulence in a mouse model (12, 20). The functions of the remaining internalin-like protein genes in L. ivanovii remain to be elucidated. Similarly, the functions of the internalin-like protein genes in the nonpathogenic Listeria spp. also remain to be determined.
The presence of genes encoding internalin-like proteins in all members of the genus Listeria suggests that these proteins have diverse functions in different hosts and/or environments, which are likely not limited to virulence-associated roles. We thus hypothesized that temperature may be an important factor in regulating transcription of these genes and that a better understanding of temperature-dependent transcription of the genes encoding internalins and internalin-like proteins may help identify hosts and/or environments where different internalins facilitate L. monocytogenes survival and multiplication. This hypothesis is supported by the previous observation that the level and activity of L. monocytogenes positive regulatory factor A (PrfA), a key regulator of virulence gene expression, are both affected by temperature (30, 39). We used microarrays to compare the levels of transcripts of (i) 24 genes encoding internalin-like proteins identified in L. monocytogenes 10403S, (ii) 24 housekeeping genes, and (iii) five control genes (prfA, sigB, clpE, plcA, and opuCA) in L. monocytogenes grown at 37°C to the levels of transcripts in cells grown at 16, 30, and 42°C. The temperature-dependent levels of transcripts of selected internalin genes and control genes were also confirmed by quantitative reverse transcriptase PCR (qRT-PCR). We selected conditions that reflect temperatures that may be encountered by L. monocytogenes in the environment and in cold-blooded animals (16 and 30°C) or warm-blooded hosts (37 and 42°C, as body temperatures in birds can be as high as 42°C). The levels of transcripts for sigB, which encodes the stress-responsive sigma factor σB, the σB-dependent gene opuCA, prfA, which encodes PrfA, and the PrfA-dependent gene plcA were evaluated as σB- and PrfA-dependent regulation of at least some L. monocytogenes internalins has been demonstrated previously (18, 32, 34, 36, 41, 57).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. monocytogenes 10403S (serotype 1/2a) was used throughout this study (4). Before each experiment, this strain was grown in brain heart infusion broth (BHI) at 37°C with shaking (200 rpm) to an optical density at 600 nm (OD600) of 0.4 and then diluted 1:100 into fresh BHI and grown again to an OD600 of 0.4 to increase the proportion of cells present in the log phase.
To measure the growth of L. monocytogenes at different temperatures, log-phase L. monocytogenes 10403S cells (OD600, 0.4) were diluted 1:100 into BHI at 16, 30, 37, or 42°C. The cultures were incubated at these temperatures with shaking (200 rpm), and the OD600 was determined at regular intervals. Growth curves were constructed for each temperature by using the average OD600 values at different times postinoculation, as determined in three independent experiments.
For RNA isolation, a log-phase culture (OD600, 0.4) of L. monocytogenes 10403S was diluted 1:100 in BHI at 16, 30, 37, or 42°C and subsequently incubated with shaking (200 rpm) at the same temperature until the early stationary phase, defined as 3 h after the OD600 reached 1.0 for cells grown at 30, 37, and 42°C. For cells grown at 16°C, the early stationary phase was defined, based on growth curve data, as 6 h after the OD600 reached 1.0. When cells reached the stationary phase, RNA was stabilized by addition of 2 volumes of RNAprotect (QIAGEN, Valencia, CA) to the bacterial culture. Bacterial cells were then harvested by centrifugation and stored at −80°C for no more than 24 h before RNA isolation.
RNA collection and purification.
RNA was isolated from frozen bacterial pellets with an RNeasy kit (QIAGEN) by using the manufacturer's protocol for enzymatic and mechanical disruption, except that cells were sonicated twice on ice with three 30-s bursts at 18 to 21 W using a Sonicator 3000 (Misonix, Farmingdale, NY). Contaminating DNA was degraded using Turbo DNase according to the manufacturer's instructions (Ambion Inc., Austin, TX). The total nucleic acid concentration and purity were estimated using A260/A280 values obtained with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Microarray construction and validation.
Construction and validation of a subgenomic microarray targeting the 24 internalin-like protein genes present in L. monocytogenes 10403S have been described elsewhere (P. McGann, S. Raengpradub, R. Ivanek, M. Wiedmann, and K. J. Boor, submitted for publication). Briefly, two 70-mer oligonucleotides were designed for each of the 24 internalin genes, 24 housekeeping genes, and five control genes (prfA, plcA, sigB, opuCA, and clpE) using ArrayOligoSelector (http://arrayoligosel.sourceforge.net/) (see Table S1 in the supplemental material for probe sequences and characteristics). The oligonucleotides were synthesized by Operon Biotechnologies Inc. (Huntsville, AL). To assist in background correction, 70-mer oligonucleotides targeting five yeast genes encoding the mating pheromone α-factors (mfα1, mfα2), mating type α-factor pheromone receptor (ste3), actin (act1), and GTP-binding protein involved in the regulation of the cyclic AMP pathway (ras1) were used as negative controls (67). To assist in signal normalization, eight twofold serial dilutions of 10403S chromosomal DNA (from 200 to 0.8 ng/μl) were prepared in printing buffer and also spotted on the array. All oligonucleotides and controls were spotted in duplicate on UltraGAPS slides (Corning, Corning, NY) using a custom-built XYZ microarrayer. Printing quality was checked by hybridization with Cy3-labeled random sequence nonamers and subsequent scanning using a GenePix 4000b scanner (Axon Instruments Inc., Foster City, CA). Printed microarrays were fixed with a UV cross-linker at 300 mJ for 1 min and stored in a desiccator at room temperature.
Probe labeling and microarray hybridization.
cDNA was synthesized from 10 μg of purified total RNA and labeled with Alexa Fluor dyes using the SuperScript Plus indirect cDNA labeling system (Invitrogen Inc., Carlsbad, CA). Before hybridization, microarray slides were treated for 10 min in a methyl pyrrolidone solution containing 0.18 M succinic anhydride and 44 mM sodium borate (pH 8.0) and then washed in fresh methyl pyrrolidone. The slides were washed successively (1 min each) in (i) H2O, (ii) 0.1% sodium dodecyl sulfate (SDS) (three washes), (iii) water (five washes), and (iv) 95% ethanol (one wash). The slides were dried by centrifugation and immersed for 1 h in blocking solution (0.1% bovine serum albumin, 0.1% SDS, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at 42°C. After blocking, the slides were washed (1 min each) in H2O (five washes) and 95% ethanol (two washes) and then dried by centrifugation. Labeled cDNA of L. monocytogenes grown at the two temperatures to be compared was resuspended in 50 μl of hybridization buffer (5× SSC, 0.1% SDS, 0.1 mM dithiothreitol, 0.5× formamide, 600 μg/ml salmon sperm DNA), denatured at 95°C for 5 min, and then applied to the slides using mSeries LifterSlips (Erie Scientific, Portsmouth, NH). Hybridization was performed overnight at 42°C in a water bath. After hybridization, the slides were washed in 2× SSC-0.1% SDS (5 min at 42°C) and then at room temperature in 2× SSC-0.1% SDS (5 min), 2× SSC (5 min), 0.2× SSC (5 min), and 0.05× SSC (1 min); this was followed by a final rinse in H2O. The slides were dried by centrifugation and scanned using a GenePix 4000b scanner. The images acquired were analyzed using GenePix Pro 6.0 (Molecular Devices Corp., Sunnyvale, CA). For each growth condition described above, three independent sets of RNA isolation procedures were performed on different days to provide distinct biological replicates, and each set of RNA samples was used to perform competitive hybridization microarray experiments. For each set of biological replicates, labeled cDNA from cells cultured at 37°C was hybridized to labeled cDNA from cells cultured at 16, 30, or 42°C.
Microarray data analysis.
Analyses of microarray data were performed in R (http://www.R-project.org) with Bioconductor (26) using the LIMMA software (65). To correct for background noise, the background intensity was subtracted from the foreground intensity for each spot (69). For normalization within arrays, the data were weighted for the housekeeping genes and genomic DNA controls and were normalized using a print-tip loess (59). The initial data analysis indicated that housekeeping genes were differentially expressed as a function of temperature, with five and two genes having higher and lower levels of transcripts, respectively, at 16 than at 37°C (Table 1). Therefore, the initial normalization was conducted by weighting only for the genomic DNA controls. Differentially expressed housekeeping genes identified with data normalized using only genomic DNA were removed from subsequent normalization procedures and designated “test” genes for all analyses. Finally, between-array normalization was performed by application of scale normalization to scale the log ratios to the same median absolute deviation across arrays. Two independent probes for each gene, each printed in duplicate, were included on a single array, and two arrays were printed per slide. To provide the most comprehensive and robust data analyses, the two independent probes for each gene were initially analyzed separately. In addition, the two arrays on each slide were treated as independent blocks for statistical examination and were analyzed accordingly. A linear model was fitted to the normalized log ratios, and four B-statistics, t-statistics, and P values were generated for each gene, corresponding to the two profiles from each array block and the two independent probes. Differential gene expression was reported only if the average fold difference between two temperatures was greater than 2 for both probes and if the difference in transcript levels was significant (P < 0.05) for both probes; this approach was previously validated by our group and was shown to reduce false-positive results (McGann et al., submitted).
TABLE 1.
Relative fold differences in levels of transcripts of housekeeping genes and clpE as determined by microarray analysis
| Gene locus (common name)a | Relative fold differences in transcript levels for cells grown atb:
|
|||||
|---|---|---|---|---|---|---|
| 16 and 37°C
|
30 and 37°C
|
42 and 37°C
|
||||
| Probe 1 | Probe 2 | Probe 1 | Probe 2 | Probe 1 | Probe 2 | |
| lmo0490 (aroE) | 2.3 | 1.5 | ||||
| lmo0257 (rpoB) | −4.2 | −3.8 | −2.9 | −2.2 | ||
| lmo1064 (corA) | 1.4 | 1.6 | ||||
| lmo1093 (nadE) | 1.5 | 1.5 | ||||
| lmo1327 (rbfA) | 1.9 | 1.8 | ||||
| lmo1403 (mutS) | −2.3 | −2.3 | ||||
| lmo2459 (gap) | 10.9 | 33.6 | ||||
| lmo0997 (clpE) | −1.8 | −1.6 | 20.9 | 22.6 | ||
Gene names are based on the annotation of the L. monocytogenes EGD-e genome available from the NCBI (accession number AL591824; http://www.ncbi.nlm.nih.gov).
The relative difference in transcript levels based on the microarray data was calculated using Bioconductor for R by dividing the average spot intensity for L. monocytogenes 10403S at the first temperature listed by the average spot intensity at the second temperature. For each gene the fold difference is shown for each of the two probes targeting the gene (probe 1 and probe 2); for each probe the value is the average of the fold differences for the probe on the upper and lower arrays. The microarray data represent the data for three separate slides, each hybridized with cDNA from an independent replicate RNA isolation. A negative value indicates that the level of expression at the first temperature listed was lower than the level of expression at the second temperature. Relative differences in transcript levels are shown only if the difference was significant (P < 0.05).
qRT-PCR.
qRT-PCR using TaqMan primers and probes was performed to confirm the temperature-dependent levels of transcripts for selected internalin and control genes. All primers and probes have been described previously and are shown in Table S2 in the supplemental material. qRT-PCR was performed in 25-μl reaction mixtures essentially as previously described (10, 34), except that we used iTaq Supermix with Rox (Bio-Rad, Hercules, CA). All qRT-PCR experiments were performed in triplicate with the three independent RNA isolates used for the microarray expression analyses. All data were log transformed to obtain a normal distribution, and the log-transformed levels of transcripts were normalized to the geometric means for the rpoB and gap housekeeping genes as previously described (10, 34). One-way analysis of variance and Tukey's honestly significant difference test were used to investigate differences in the levels of transcripts obtained at the four temperatures for a given gene. All statistical analyses were performed with S-Plus 6.2 (Insightful Corp, Seattle, WA).
Microarray data accession number.
Raw data and microarray files in the MIAME format have been deposited in the Gene Expression Omnibus (GEO) database (1, 17) under accession number GSE6471.
RESULTS
Expression of seven housekeeping genes and the clpE heat shock gene changes in response to the external temperature.
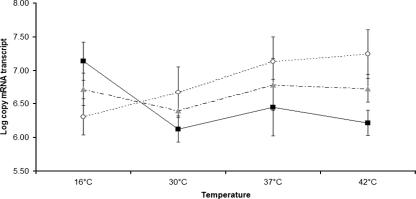
Analyses of the microarray data showed that the levels of transcripts of seven housekeeping genes and the clpE gene encoding a class III heat shock protein (Clp ATPase) were temperature dependent (Table 1). Specifically, the levels of transcripts of five housekeeping genes (aroE, corA, nadE, rbfA, and gap) were higher at 16°C than at 37°C, and the levels of transcripts of two housekeeping genes (rpoB and mutS) were lower at 16°C than at 37°C; the levels rpoB transcripts were also lower at 30°C than at 37°C. clpE was included in the microarray as a control gene predicted to exhibit increased transcription at elevated temperatures. As expected, the levels of the clpE transcripts were higher at 42°C than at 37°C, while the levels of these transcripts were lower at 30°C than at 37°C. The levels of the transcripts of rpoB and gap, which have previously been used by workers in our group to normalize the levels of transcripts of different target genes in qRT-PCR experiments (10, 34), differed significantly for L. monocytogenes grown at some temperatures (Table 1). Therefore, we investigated levels of transcripts of these genes at different temperatures using qRT-PCR. Analyses using Tukey's honestly significant difference test showed that the levels of rpoB transcripts were significantly lower at 16°C than at 42°C, while the levels of gap transcripts were significantly higher at 16°C than at 30 or 42°C. While a significant effect of temperature on the levels of rpoB and gap transcripts was confirmed by analysis of variance, temperature did not have a significant effect on the geometric mean of the levels of the rpoB and gap transcripts (Fig. 1). These data support the conclusion that normalizing the levels of mRNA transcripts to the geometric mean of the levels of the rpoB and gap transcripts, as previously reported by workers in our group (10, 34), is a valid approach for normalizing levels of mRNA transcripts determined by qRT-PCR using RNA from cells grown at different temperatures. Therefore, in all qRT-PCR experiments, the levels of transcripts for each target gene were normalized using the geometric mean of the levels of the rpoB and gap transcripts.
FIG. 1.
Line plots of pooled, log-transformed absolute levels of transcripts for gap (▪) and rpoB (○) and the geometric mean for rpoB and gap transcript levels ( ). The levels of transcripts were determined by qRT-PCR for L. monocytogenes 10403S grown to the early stationary phase at 16, 30, 37, or 42°C. The error bars indicate one standard deviation based on three independent replicates.
). The levels of transcripts were determined by qRT-PCR for L. monocytogenes 10403S grown to the early stationary phase at 16, 30, 37, or 42°C. The error bars indicate one standard deviation based on three independent replicates.
Temperature-dependent levels σB-dependent inlC2, inlD, lmo0331, and lmo0610 transcripts are similar to the levels of σB-dependent opuCA transcripts.
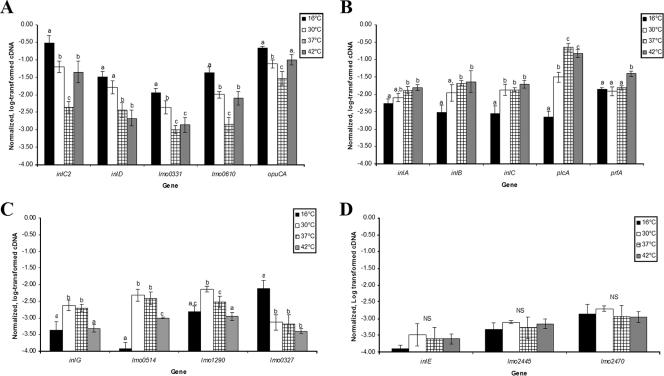
Previous reports have shown that inlC2, inlD, lmo0331, and lmo0610 are at least partially σB dependent, and putative σB-dependent promoters have been identified upstream of the coding regions of these genes (32; McGann et al., submitted). Microarray data showed that the levels of inlC2, inlD, lmo0331, and lmo0610 transcripts, as well as the levels of opuCA and sigB transcripts, were higher at both 16 and 30°C than at 37°C (Table 2). In addition, the levels of inlC2, lmo0610, opuCA, and sigB transcripts were also higher at 42°C than at 37°C, while the levels of inlD and lmo0331 transcripts were not different at 37 and 42°C. qRT-PCR confirmed the microarray results (Fig. 2A) and revealed that inlC2, inlD, lmo0331, lmo0610, and opuCA had very similar temperature-dependent transcription patterns; the highest levels of transcripts were observed in cells grown at 16°C, and the lowest levels of transcripts were observed in cells grown at 37°C (the one exception was inlD, whose transcript levels were lowest at 42°C).
TABLE 2.
Relative fold differences in levels of transcripts of internalin and internalin-like protein genes as determined by microarray analyses
| Gene locus (common name)a | Relative fold differences in transcript levels for cells grown atb:
|
|||||
|---|---|---|---|---|---|---|
| 16 and 37°C
|
30 and 37°C
|
42 and 37°C
|
||||
| Probe 1 | Probe 2 | Probe 1 | Probe 2 | Probe 1 | Probe 2 | |
| Category I | ||||||
| NAc (inlC2) | 7.4 | 9.5 | 4.3 | 4.3 | 3.1 | 3.7 |
| NA (inlD) | 6.7 | 7.4 | 3.1 | 2.6 | ||
| lmo0331 | 9.3 | 8.2 | 2.4 | 2.9 | ||
| lmo0610 | 17.3 | 12.3 | 2.6 | 3.0 | 3.6 | 3.9 |
| lmo1428 (opuCA) | 4.3 | 4.4 | 2.2 | 2.3 | 2.5 | 2.4 |
| lmo0895 (sigB) | 4.9 | 5.7 | 2.6 | 3.0 | 2.5 | 2.4 |
| Category II | ||||||
| lmo0433 (inlA) | −3.4 | −3.8 | ||||
| lmo0434 (inlB) | −4.7 | −5.6 | ||||
| lmo1786 (inlC) | −4.3 | −3.6 | ||||
| lmo0201 (plcA) | −16.5 | −23.3 | −11.0 | −3.8 | ||
| lmo0200 (prfA) | 3.2 | 4.8 | ||||
| Category III | ||||||
| lmo0262 (inlG) | −6.7 | −4.1 | −2.7 | −2.9 | ||
| lmo0514 | −2.8 | −3.7 | −2.6 | −2.7 | ||
| lmo2821 (inlJ) | −3.3 | −2.8 | 2.9 | 3.2 | ||
| lmo1290 | −4.2 | −3.3 | ||||
| lmo0327 | 6.1 | 6.4 | ||||
Gene names are based on the annotation of the L. monocytogenes EGD-e genome available from the NCBI (accession number AL591824; http://www.ncbi.nlm.nih.gov). The only exceptions are inlC2 and inlD, which are not present in the EGD-e genome.
The relative difference in transcript levels based on the microarray data was calculated using Bioconductor for R by dividing the average spot intensity for L. monocytogenes 10403S at the first temperature listed by the average spot intensity at the second temperature. For each gene the fold difference is shown for each of the two probes targeting the gene (probe 1 and probe 2); for each probe the value is the average of the fold differences for the probe on the upper and lower arrays. The microarray data represent the data for three separate slides, each hybridized with cDNA from an independent replicate RNA isolation. A negative value indicates that the level of expression at the first temperature listed was lower than the level of expression at the second temperature. Relative differences in transcript levels are shown only if the difference was significant (P < 0.05).
NA, not applicable.
FIG. 2.
Normalized, log-transformed levels of transcripts for category I genes (A), category II genes (B), selected category III genes (C), and selected category IV genes (D). For details, see Table 2. Levels of transcripts were determined by qRT-PCR for L. monocytogenes 10403S grown to the early stationary phase at 16, 30, 37, or 42°C. The data are average log-transformed levels of transcripts (normalized to the geometric mean for rpoB and gap) for three independent replicates (i.e., three RNA isolations performed on different days); the error bars indicate one standard deviation. For each gene, Tukey's multiple-comparison procedure was used to determine whether the levels of transcripts of the gene differed at the four temperatures; different letters above bars indicate that the levels of transcripts were significantly different (P < 0.05), while identical letters above bars indicate that the levels of transcripts were not significantly different. NS, levels of transcripts at the different temperatures were not significantly different.
Levels of PrfA-dependent inlA, inlB, and inlC transcripts are temperature dependent, like the levels of PrfA-dependent plcA transcripts.
Previous studies have shown that inlA, inlB, and inlC are partially PrfA dependent (18, 41), and PrfA binding domains (a “PrfA box”) have been identified upstream of the inlAB operon (16) and inlC (18). In addition, transcription of inlAB is also σB dependent, and a σB-dependent promoter has been identified upstream of this operon (32). Microarray data showed that the levels of inlA, inlB, inlC, and plcA transcripts were significantly lower at 16°C than at 37°C (Table 2), and the differences were more pronounced for plcA (16. 5- to 23.3-fold) (Table 2) than for inlA, inlB, and inlC (3.4- to 5.6-fold). While the levels of plcA transcripts were also lower at 30°C than at 37°C, the levels of inlA, inlB, and inlC transcripts were not significantly different at 30 and 37°C. These microarray results were confirmed by qRT-PCR (Fig. 2B), which showed that the temperature-dependent transcription patterns for inlA, inlB, inlC, and plcA were similar. For all four genes, the levels of transcripts at 16°C were significantly lower than the levels of transcripts in cells grown at 37 or 42°C. The levels of plcA transcripts were also significantly lower in cells grown at 30°C than in cells grown at 37 or 42°C, consistent with the microarray data. The levels of prfA transcripts were similar in cells grown at 16, 30, and 37°C, but they were significantly higher at 42°C (Fig. 2B), also consistent with the microarray data (Table 2).
Levels of inlG, inlJ, lmo0514, and lmo1290 transcripts are low at 16°C and highest at 30 and/or 37°C.
Microarray data showed that the levels of inlG, inlJ, and lmo0514 transcripts were significantly lower in cells grown at 16°C than in cells grown at 37°C (Table 2). The levels of both inlG and lmo0514 transcripts were significantly lower in cells grown at 42°C than in cells grown at 37°C. In the microarray experiments, the levels of lmo1290 transcripts were also significantly lower in cells grown at 42°C than in cells grown at 37°C. qRT-PCR confirmed that inlG, lmo0514, and lmo1290 had similar temperature-dependent transcription patterns, with low levels of transcripts at 16°C and the highest levels of transcripts at 30 and/or 37°C.
Levels of lmo0327 transcripts are highest at 16°C and low at higher temperatures.
Both microarray (Table 2) and qRT-PCR data (Fig. 2C) showed that lmo0327 had a temperature-dependent transcription profile that was unique among the genes included on the subgenomic microarray; the levels of transcripts were significantly higher at 16°C than at 30, 37, and 42°C (Fig. 2C), and the levels of transcripts at 30, 37, and 42°C were not significantly different from each other.
Twelve internalin-like protein genes showed no evidence of temperature-dependent transcription levels.
The microarray data revealed that temperature did not have a significant effect on the levels of transcripts of 12 internalin-like protein genes at the temperatures tested (see Table S3 in the supplemental material). qRT-PCR analysis of three genes, lmo0264 (inlE), lmo2445, and lmo2470 (Fig. 2D), confirmed that temperature did not have a significant effect on the levels of transcripts of these genes. qRT-PCR also showed that the levels of transcripts of these three genes were consistently low and generally lower than the levels of transcripts found for category I, II, and III genes (Fig. 2).
DISCUSSION
As genes encoding internalin and internalin-like proteins are present in both pathogenic and nonpathogenic Listeria spp. (13), in addition to the recognized role that some of these proteins play in virulence, it is likely that other internalin-like proteins have physiological functions that are not linked to virulence. This hypothesis is supported by the observation that the internalin Lmo0327 has been characterized as an enzyme with murein-hydrolyzing activity, which appears to be involved in cell separation and turnover of murein but not in adhesion to or entry into eukaryotic cells (52). We hypothesized that the environmental temperature might be an important environmental cue affecting expression of members of the internalin gene family. A comprehensive understanding of temperature-dependent transcription of the genes encoding internalins and internalin-like proteins may help identify host and/or environmental conditions in which specific internalins facilitate L. monocytogenes survival, multiplication, or other critical functions. Microarray and qRT-PCR-based characterization of internalin gene transcription showed that internalin genes can be separated into five broad categories based on temperature-dependent transcription patterns and also showed that transcription of a number of L. monocytogenes housekeeping genes is significantly affected by temperature.
Levels of transcripts of some housekeeping genes are affected by temperature.
While housekeeping genes are commonly used as controls and for normalization in quantitative gene expression studies (49), we clearly showed that the levels of transcripts of a number of L. monocytogenes housekeeping genes are affected by temperature, which is consistent with the results of other studies that have also shown that the levels of housekeeping gene transcripts in L. monocytogenes (9, 10, 60) and other bacteria (62) can be affected by growth conditions. Interestingly, for the majority of the L. monocytogenes housekeeping genes that showed temperature-dependent transcription in our study, differential transcription occurred at 16°C. While our results specifically showed that the levels of rpoB and gap transcripts in L. monocytogenes are affected by temperature (particularly 16°C), the geometric mean of the levels of transcripts of these two genes was not affected by temperature, supporting the validity of using the geometric mean of the levels of rpoB and gap transcripts for normalization of the levels of gene transcripts determined for L. monocytogenes cultured under different environmental conditions, including in the presence of NaCl and charcoal (10, 32). These results are consistent with the findings of Vandesompele et al. (62), who demonstrated that measurement of the levels of transcripts of a single housekeeping gene can be inadequate for normalizing the levels of transcripts of target genes in cells grown under very different physiological conditions. Use of a consistent, statistically validated approach with data for multiple housekeeping genes (e.g., both rpoB and gap) can facilitate reliable comparisons of levels of transcripts across studies (e.g., the levels of transcripts reported here and the levels of transcripts in intracellular L. monocytogenes reported by Kazmierczak et al. [34]).
Levels of transcripts of the σB-dependent internalin genes inlC2, inlD, lmo0331, and lmo0610 generally are highest at 16°C and lowest at 37°C.
The levels of the transcripts of inlC2, inlD, lmo0331, and lmo0610, as well as the σB-dependent opuCA genes, were highest at 16°C and lowest at 37°C (the only exception was inlD, whose transcript levels were lower at 42°C than at 37°C). These findings are consistent with previous reports that have shown that there is increased transcription of σB-dependent genes at low temperatures in L. monocytogenes (2, 42) and in Bacillus subtilis (6, 68), which is closely related to Listeria. In B. subtilis, σB is essential for low-temperature adaptation, and the growth of a sigB null mutant is impaired at 15°C (6). The overall expression patterns of inlC2, inlD, lmo0331, lmo0610, and opuCA are also consistent with the well-documented role of σB as a stress-responsive alternative sigma factor in L. monocytogenes (2, 21, 60, 64, 66) and other gram-positive species (33), and σB-dependent gene expression is generally lowest at the optimal growth temperature for L. monocytogenes (37°C) and higher at temperatures that may impose physiological stress on the organism (temperatures lower and higher than 37°C).
In addition to regulating the expression of genes involved in the global stress response, σB plays a critical role during the gastrointestinal stage of listeriosis in the guinea pig (24). The ability of a L. monocytogenes sigB null mutant to invade human intestinal epithelial cells also is impaired (23, 35, 36). Thus, it is tempting to speculate that the σB-dependent internalins (i.e., the internalins encoded by inlC2, inlD, lmo0331, and lmo0610) contribute to gastrointestinal invasion and pathogenesis in L. monocytogenes, which is consistent with evidence that the inlGHE operon (which includes inlH, a fusion of inlC2 and inlD) contributes to gastrointestinal virulence in mice (53), even though clear roles for inlC2 and inlD in virulence have not been defined yet. Upregulation of these genes at temperatures lower than mammalian body temperatures may allow enhanced expression of these internalins, which may help prime L. monocytogenes for infection, which is consistent with the finding that certain stress conditions that induce σB activity enhance L. monocytogenes invasiveness for human intestinal epithelial cells (23). Interestingly, while inlC2 and inlD appear to be specific to L. monocytogenes, consistent with their proposed role in virulence, homologues of lmo0331 and lmo0610 have been found in L. innocua and L. welshimeri (only lmo0610) (27, 28), possibly indicating a physiological function(s) other than virulence for these internalins.
Levels of the transcripts of the PrfA-dependent inlA, inlB, and inlC internalin genes are lowest at 16°C.
Our data showed that the levels of the transcripts of the PrfA-dependent inlA, inlB, and inlC genes (18, 41), as well as plcA, were lowest in cells grown at 16°C. These findings are consistent with the well-documented temperature-dependent activation of PrfA, a pleiotropic transcriptional activator of virulence genes in L. monocytogenes (8, 47). The levels of the PrfA protein and the activity are both affected by temperature (30, 39). Translation of prfA mRNA is temperature dependent, apparently as a result of the secondary structure of the untranslated mRNA preceding prfA, which masks the transcript's ribosome binding region at temperatures below 37°C (30). PrfA activity (as measured by expression of PrfA-dependent genes) is also higher at 37°C than at 30°C and lower temperatures (39). Our data provide further evidence that there is posttranscriptional regulation of PrfA activity, as the levels of prfA transcripts were similar at 16, 30, and 37°C, while PrfA activity appeared to increase at 30 and 37°C (as indicated by the levels of PrfA-dependent plcA transcripts).
While the levels of plcA transcripts were significantly lower at 30°C than at 37°C, the levels of inlA, inlB, and inlC transcripts were similar at 30 and 37°C. These data support the hypothesis that there are additional, PrfA-independent mechanisms that contribute to transcription of these three genes, as previously proposed (41, 44), which is consistent with the observation that the inlAB operon is also regulated by σB (in addition to PrfA) and the observation that inlC transcription can be initiated from overlapping PrfA-dependent and -independent promoters (44). Regulation of inlA, inlB, and inlC by multiple PrfA-dependent and -independent regulators appears to allow levels of transcription of these genes over a wider temperature range that are higher than the levels of transcription of the genes that are solely PrfA dependent (as observed for plcA). This feature may be important for genes that are critical for the early (gastrointestinal) stage of L. monocytogenes infection (e.g., inlA [22]) to ensure baseline transcription at temperatures lower than mammalian body temperatures (prior to ingestion) in order to prime cells for rapid invasion following ingestion and during subsequent gastric passage. Overall, the temperature-dependent transcription profiles for inlA, inlB, and inlC (i.e., low levels of transcripts at 16°C and high levels of transcripts at 37°C) are consistent for genes with confirmed roles in mammalian virulence (14, 18, 22, 41). inlA, inlB, and inlC have not been identified in any nonpathogenic Listeria spp. (13, 28), further supporting the hypothesis that they are virulence genes, as well as the hypothesis that elucidation of temperature-dependent expression profiles can help define putative virulence genes. Thus, studies of temperature-dependent regulation of internalin expression in other Listeria spp. may provide further insight into the evolution of Listeria internalins and their regulation.
σB- and PrfA-independent internalins have multiple and distinct temperature-dependent transcription profiles.
Temperature-dependent transcription profiles of 17 σB- and PrfA-independent genes that encode internalins and internalin-like proteins identified three groups of genes, including (i) four genes (inlG, inlJ, lmo0514, and lmo1290) whose levels of transcripts are lowest at 16°C and highest at 30 and/or 37°C, (ii) one gene (lmo0327) whose levels of transcripts are highest at 16°C and low at higher temperatures, and (iii) 12 genes whose levels of transcripts did not differ at the temperatures used in this study. As the levels of inlG, inlJ, lmo0514, and lmo1290 transcripts are lowest at 16°C and highest at mammalian body temperatures, these genes may be important for virulence and/or interactions with mammalian hosts. Some experimental evidence supports the conclusion that inlG and inlJ contribute to virulence (3, 53, 55). inlG is not present in all L. monocytogenes strains and is not present in other Listeria spp. (27, 61). While no clear attenuation of virulence has been described for an inlG null mutant, inlGHE appears to be required for InlA-dependent invasion of Caco-2 cells in the absence of InlB (3) and to contribute to virulence in a mouse model (53). inlJ has recently been shown to play a role in virulence, as an inlJ null mutant was significantly attenuated for virulence after intravenous infection of mice or oral inoculation of mice expressing human E-cadherin (55). Similar to inlG, no homolog of the inlJ gene has been described in Listeria species other than L. monocytogenes to date (13). lmo0514 and lmo1290 have not been characterized previously, but based on our expression data, these genes are hypothesized to contribute to interactions of L. monocytogenes with mammalian hosts. The sizes of the two proteins encoded by these genes are very similar (605 and 598 amino acids, respectively), and both have structural features indicating the presence of two PKD repeat units (7). Because the PKD domain contains an immunoglobulin-like fold, these structures are proposed to form ligand-binding sites in cell surface proteins. PKD1 has been shown to colocalize to form a complex with E-cadherin and α-, β- and γ-catenins (29), further supporting the hypothesis that these genes have roles in binding to mammalian cell surface molecules.
The levels of lmo0327 transcripts were highest at 16°C and low at higher temperatures, suggesting that this gene may not contribute to mammalian virulence. This is consistent with the observation that lmo0327 encodes a surface protein that is covalently linked to murein and has murein-hydrolyzing activity, which appears to play a role in cell separation and murein turnover (52). Our data for transcript levels are also consistent with the observation that a Δlmo0327 strain had a significantly lower growth rate than the parent strain at 25°C but not at 30 and 37°C (52).
The 12 internalin-like protein genes that did not show any apparent effect of temperature on transcription included inlE and inlF. Our finding that the levels of transcripts of these genes were low is consistent with the results of a study in which the workers failed to detect InlE by Western blotting in cells cultured in bacterial medium at 37°C (15) and with the results of qRT-PCR studies which showed that the levels of inlE transcripts were not detectable at 37°C (P. McGann, M. Wiedmann, and K. J. Boor, submitted for publication). Our findings may indicate that these genes are transcribed only under very specific environmental conditions or that they may be silent in L. monocytogenes. For inlF, these hypotheses are supported by the fact that an inlF null mutant did not show any apparent phenotype when it was tested in tissue culture assays (15). While an inlGHE null mutant strain exhibited reduced oral virulence in mice, possibly indicating that inlE has a role in virulence, no specific phenotype associated with an inlE null mutation has been reported to date. While further studies of these genes are required to understand their functions, we predict that they are not likely candidates for virulence or virulence-associated genes.
Conclusions.
This study provided further evidence that internalins and internalin-like proteins in L. monocytogenes and other Listeria spp. have a broad spectrum of functions, including facilitating host-pathogen interactions and other physiological processes not linked to virulence. The conclusion that Listeria internalins are functionally diverse is supported by the diverse temperature-dependent gene regulation patterns observed for genes encoding members of the internalin family, including genes that are expressed most highly at temperatures considerably below mammalian body temperatures, along with other genes that are expressed most highly at mammalian body temperatures. Additional support for the conclusion that the internalins are functionally diverse includes (i) the highly variable distribution of internalins among Listeria spp., including the internalins that are strain, species, and genus specific (13, 27, 28, 61), (ii) the transcriptional regulation by diverse transcription factors, including PrfA and σB (16, 18, 32, 34, 36, 41, 50), and (iii) the diverse functions of the internalins characterized to date, including a number of internalins without clearly defined functions (3, 13-15, 18, 22, 52, 53, 55). Further studies of the Listeria internalins are likely to provide broad insight into the evolution of gene families that contribute to virulence and other physiological functions in environmentally transmitted pathogens.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grant RO1-AI052151-01A1 to K.J.B.
Footnotes
Published ahead of print on 2 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barrett, T., T. O. Suzek, D. B. Troup, S. E. Wilhite, W. C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash, W. Fujibuchi, and R. Edgar. 2005. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 33:D562-D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 5.Braun, L., B. Ghebrehiwet, and P. Cossart. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 19:1458-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigulla, M., T. Hoffmann, A. Krisp, A. Volker, E. Bremer, and U. Volker. 2003. Chill induction of the σB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturongakul, S., and K. J. Boor. 2006. σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72:5197-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummins, A. J., A. K. Fielding, and J. McLauchlin. 1994. Listeria ivanovii infection in a patient with AIDS. J. Infect. 28:89-91. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez-Bernal, G., S. Muller-Altrock, B. Gonzalez-Zorn, M. Scortti, P. Herrmann, H. J. Monzo, L. Lacharme, J. Kreft, and J. A. Vazquez-Boland. 2006. A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol. Microbiol. 59:415-432. [DOI] [PubMed] [Google Scholar]
- 13.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 15.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dramsi, S., C. Kocks, C. Forestier, and P. Cossart. 1993. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator PrfA. Mol. Microbiol. 9:931-941. [DOI] [PubMed] [Google Scholar]
- 17.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 19.Engelbrecht, F., C. Dickneite, R. Lampidis, M. Gotz, U. DasGupta, and W. Goebel. 1998. Sequence comparison of the chromosomal regions encompassing the internalin C genes (inlC) of Listeria monocytogenes and L. ivanovii. Mol. Gen. Genet. 257:186-197. [DOI] [PubMed] [Google Scholar]
- 20.Engelbrecht, F., G. Dominguez-Bernal, J. Hess, C. Dickneite, L. Greiffenberg, R. Lampidis, D. Raffelsbauer, J. J. Daniels, J. Kreft, S. H. Kaufmann, J. A. Vazquez-Boland, and W. Goebel. 1998. A novel PrfA-regulated chromosomal locus, which is specific for Listeria ivanovii, encodes two small, secreted internalins and contributes to virulence in mice. Mol. Microbiol. 30:405-417. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 23.Garner, M. R., K. E. James, M. C. Callahan, M. Wiedmann, and K. J. Boor. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 72:5384-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gellin, B. G., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313-1320. [PubMed] [Google Scholar]
- 26.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 28.Hain, T., C. Steinweg, and T. Chakraborty. 2006. Comparative and functional genomics of Listeria spp. J. Biotechnol. 126:37-51. [DOI] [PubMed] [Google Scholar]
- 29.Huan, Y., and J. van Adelsberg. 1999. Polycystin-1, the PKD1 gene product, is in a complex containing E-cadherin and the catenins. J. Clin. Investig. 104:1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 31.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 32.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2006. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827-1838. [DOI] [PubMed] [Google Scholar]
- 35.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, H., H. Marquis, and K. J. Boor. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobe, B., and J. Deisenhofer. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415-421. [DOI] [PubMed] [Google Scholar]
- 38.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 39.Leimeister-Wachter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lessing, M. P., G. D. Curtis, and I. C. Bowler. 1994. Listeria ivanovii infection. J. Infect. 29:230-231. [DOI] [PubMed] [Google Scholar]
- 41.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Low, J. C., and W. Donachie. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153:9-29. [DOI] [PubMed] [Google Scholar]
- 44.Luo, Q., M. Rauch, A. K. Marr, S. Muller-Altrock, and W. Goebel. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39-52. [DOI] [PubMed] [Google Scholar]
- 45.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 2000. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl. Acad. Sci. USA 97:8784-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLauchlin, J. 1997. Animal and human listeriosis: a shared problem? Vet. J. 153:3-5. [DOI] [PubMed] [Google Scholar]
- 47.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vazquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 48.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 49.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 50.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popowska, M., and Z. Markiewicz. 2006. Characterization of Listeria monocytogenes protein Lmo0327 with murein hydrolase activity. Arch. Microbiol. 186:69-86. [DOI] [PubMed] [Google Scholar]
- 53.Raffelsbauer, D., A. Bubert, F. Engelbrecht, J. Scheinpflug, A. Simm, J. Hess, S. H. Kaufmann, and W. Goebel. 1998. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Gen. Genet. 260:144-158. [DOI] [PubMed] [Google Scholar]
- 54.Rocourt, J., H. Hof, A. Schrettenbrunner, R. Malinverni, and J. Bille. 1986. Acute purulent Listeria seeligeri meningitis in an immunocompetent adult. Schweiz. Med. Wochenschr. 116:248-251. [PubMed] [Google Scholar]
- 55.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauders, B. 2005. Molecular epidemiology, diversity, and distribution, and ecology of Listeria. Ph.D. thesis. Cornell University, Ithaca, NY.
- 57.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InlB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 59.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 60.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 61.Tsai, Y. H., R. H. Orsi, K. K. Nightingale, and M. Wiedmann. 2006. Listeria monocytogenes internalins are highly diverse and evolved by recombination and positive selection. Infect. Genet. Evol. 6:378-389. [DOI] [PubMed] [Google Scholar]
- 62.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 63.Walker, J. K., J. H. Morgan, J. McLauchlin, K. A. Grant, and J. A. Shallcross. 1994. Listeria innocua isolated from a case of ovine meningoencephalitis. Vet. Microbiol. 42:245-253. [DOI] [PubMed] [Google Scholar]
- 64.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. P. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wettenhall, J. M., and G. K. Smyth. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20:3705-3706. [DOI] [PubMed] [Google Scholar]
- 66.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamanaka, K., J. Araki, M. Takano, and J. Sekiguchi. 1997. Characterization of Bacillus subtilis mutants resistant to cold shock-induced autolysis. FEMS Microbiol. Lett. 150:269-275. [DOI] [PubMed] [Google Scholar]
- 69.Yang, M. C., Q. G. Ruan, J. J. Yang, S. Eckenrode, S. Wu, R. A. McIndoe, and J. X. She. 2001. A statistical method for flagging weak spots improves normalization and ratio estimates in microarrays. Physiol. Genomics 7:45-53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.