Abstract
The bone morphogenic proteins (BMPs) play important roles in vertebrate development. In Xenopus, BMPs act as epidermal inducers and also as negative regulators of neurogenesis. Antagonism of BMP signaling results in neuralization. BMPs signal through the cell-surface receptors and downstream Smad molecules. Upon stimulation with BMP, Smad1, Smad5, and Smad8 are phosphorylated by the activated BMP receptors, form a complex with Smad4, and translocate into the nucleus, where they regulate the expression of BMP target genes. Here, we show that the Ski oncoprotein can block BMP signaling and the expression of BMP-responsive genes in both Xenopus and mammalian cells by directly interacting with and repressing the activity of BMP-specific Smad complexes. This ability to antagonize BMP signaling results in neuralization by Ski in the Xenopus embryo and blocking of osteoblast differentiation of murine W-20-17 cells. Thus, Ski is able to repress the activity of all receptor-associated Smads and may regulate vertebrate development by modulating the signaling activity of transforming growth factor-β family members.
The bone morphogenic protein (BMP) family of proteins regulates many aspects of vertebrate development including mesoderm patterning, neurogenesis, bone formation, and organogenesis (1). In in vitro culture systems, BMP2 not only stimulates maturation of the osteoblast progenitor cells, it also induces osteoblast differentiation of nonosteoblast cells, such as the pluripotent mesenchymal progenitor cell line, C2C12 (2), or the bone marrow stromal cell line, W-20-17 (3), as assessed by induction of alkaline phosphatase activity and expression of osteocalcin. In Xenopus, BMPs act as epidermal inducers and as negative regulators of neurogenesis (4).
BMPs exert their effects through types I and II cell-surface serine/threonine kinase receptors (BMPRI and BMPRII). Binding of BMP to its receptor complex results in the activation of BMPRI, which in turn phosphorylates Smad1, Smad5, and Smad8 molecules. Upon phosphorylation, these BMP-specific Smads form a complex with the co-Smad, Smad4, and translocate into the nucleus (5, 6). In the nucleus, the Smad1/Smad4 complex can bind with low affinity to the GCCG or CAGA motif in the promoter regions of many BMP-responsive genes (6, 7). They can also be recruited to the promoters of BMP-responsive genes by high affinity cofactors such as OAZ, which binds to the promoter of the Xvent2 gene (8). Smad1 was found to complex with p300/CBP and STAT3, and this complex may be involved in the transactivation of the glial fibrillary acidic protein gene, a marker for astrocyte differentiation (9). In addition, Smad1 has been shown to interact with SIP1, the acute myelogenous leukemia protein, and the homeodomain transcription factor Hoxc-8 (6), although the physiological significance of these interactions is not yet clear.
In Xenopus, high levels of BMP induce epidermis, and antagonism of BMP signaling results in neurogenesis (4). Neural specification involves antagonism of the BMP pathway at several key levels and by multiple mechanisms: extracellular antagonists, membrane-bound proteins, and intracellular regulators that prevent the activation of Smad proteins or stimulate the degradation of Smad1. For example, overexpression of a winged-helix transcriptional repressor, BF-2, causes neuralization by inhibition of BMP4 expression (10, 11). Noggin, Chordin, Follistatin, Cerberus, and Xnr3 antagonize BMP signaling either by directly and strongly associating with the BMPs to block access to BMP receptors or by competing with BMPs for receptor binding (Xnr3) (4). Dominant negative BMP receptors or pseudoreceptors such as BAMBI (12) prevent receptor activation and induce neuralization. Blocking the activation of Smad1 or Smad5 by overexpression of the inhibitory Smad6 and Smad7 results in neural specification (5, 6). Finally, Smurf1 induces the ubiquitination and degradation of Smad1 and leads to neuralization (13). Thus, multiple mechanisms blocking the BMP signaling pathway all result in neural tissue specification.
Recently, the Xenopus homologue of a nuclear oncoprotein, Ski, has been shown to induce secondary neural axis formation and neural-specific gene expression in ectoderm explants when injected into the Xenopus embryos (14). However, the mechanism by which XSki neuralizes the ectoderm is not clear. Ski was originally identified as the transforming protein of the avian Sloan–Kettering retrovirus (v-Ski; refs. 15 and 16). The cellular homologue, c-ski, is expressed in all normal adult and embryonic tissues at a low level (17), but the expression is elevated in some human tumor cells (18, 19). The transcript is also increased during differentiation of muscle, central and peripheral nervous systems, and respiratory tissues in the mouse embryo (17, 20) and from very early stage of oogenesis to the late neurula stage in Xenopus (14, 21, 22). Thus, Ski may play a role in both carcinogenesis and embryonic development. Indeed, overexpression of c-Ski induces both oncogenic transformation and terminal differentiation of quail embryo fibroblasts (23). Moreover, mice lacking c-ski displayed defective myogenesis and neural tube formation, resulting in lethality at birth (24).
We and others have recently identified Ski and the closely related SnoN as important negative regulators of transforming growth factor-β (TGFβ) signaling. Ski and SnoN interact with Smad2, Smad3, and Smad4 on a TGFβ-responsive promoter element and repress their ability to activate TGFβ target genes (25–28). Thus, the elevated level of Ski protein in human cancer cells can potentially inactivate the tumor-suppressive actions of the Smads, and this may account, at least partially, for the transforming activity of Ski. The mechanism of Ski action during embryonic development, however, remains undefined. Because the BMP-specific Smads share considerable homology with the TGFβ Smads and because Smad4 acts as a co-Smad in both pathways, we asked whether Ski may neuralize in the Xenopus embryo by antagonizing BMP signaling. Specifically, we examined whether Ski could interact with the BMP Smads and repress their ability to activate BMP target genes.
Materials and Methods
Cells and Plasmids.
The human 293T cells and the murine W-20-17 cells (3) were maintained in DMEM supplemented with 10% FBS. The human hepatoma Hep3B cell line (American Type Culture Collection) was maintained in MEM with 10% FBS. The human BMP2 was purchased from R & D Systems. The AB2 chimera is described in Eimon and Harland (29).
Plasmids containing the Smad proteins and Ski have been described (26). The constitutively active type I BMP and TGFβ receptors [Alk3(Q233D) and Alk5(T204D)] were generated by PCR. For expression in Xenopus embryos, human wild-type and mutant ski cDNAs were subcloned into pCS2 (11).
Embryo Culture and Dissection.
Eggs from Xenopus laevis females were fertilized
in vitro and cultured as described (11). Embryos were staged
and cultured in 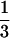 × Marc's Ringer's solution (0.1 M
NaCl/2.0 mM KCl/1 mM MgSO4/2 mM
CaCl2/1 mM EDTA/5 mM Hepes, pH 7.8). Animal
caps were explanted at stages 8–9 and cultured in 75% normal
amphibian media with 0.5 mg/ml gentamycin (11).
× Marc's Ringer's solution (0.1 M
NaCl/2.0 mM KCl/1 mM MgSO4/2 mM
CaCl2/1 mM EDTA/5 mM Hepes, pH 7.8). Animal
caps were explanted at stages 8–9 and cultured in 75% normal
amphibian media with 0.5 mg/ml gentamycin (11).
Microinjection.
Synthetic capped mRNA was prepared and purified on a Sephadex column. A total of 10 nl of RNA was injected per embryo as described in Vize et al. (30). For expression in the ectoderm, embryos were injected at the one-cell stage at the animal pole. For ski mRNA expression in ventral mesoderm, embryos were injected at the four-cell stage into two ventral blastomeres at the marginal zone with two 250-pg injections.
Reverse Transcriptase (RT)-PCR.
RNA was isolated from explants by incubating lysed tissue in a proteinase K buffer (11) for 30 min at 42°C. After treatment with DNase I to remove genomic DNA, RT-PCR analysis was carried out as described (31). All experiments included reactions for EF1α to assess RNA recovery. Primer sets and PCR conditions for Xbra, EF1α, gsc, noggin, XAG, muscle actin, and neural cell adhesion molecule (NCAM) have been described (31–34).
In Situ Hybridization.
In situ hybridization was carried out as described (11). The bound probe was detected with antidigoxigenin antibody conjugated to alkaline phosphatase and the substrate BM Purple (Roche Molecular Biochemicals). After staining and fixation, pigment was removed by bleaching. Specimens were cleared in benzyl benzoate/benzyl alcohol (2:1).
Transfection and Transcriptional Reporter Assay.
293T cells were transiently transfected by using the calcium phosphate protocol (35). Hep3B cells and W-20-17 cells were transfected with Lipofectamine (GIBCO).
For the transcriptional reporter assay, 15xGCCG-Luc (7) was cotransfected together with cDNAs encoding various Smad proteins or active type I receptors. Luciferase activity was measured 48 h after transfection. A β-galactosidase-expressing construct was cotransfected in all samples to normalize for transfection efficiency.
Glutathione S-Transferase (GST) Pull-Down.
A recombinant Ski fragment (residue 2–605) was expressed in and purified from Escherichia coli as a GST-fusion protein (26). Recombinant Smad1 and Smad4 were similarly purified as GST-fusion proteins followed by thrombin cleavage. For binding assays, 1.5 μg of GST-Ski immobilized on glutathione Sepharose were blocked at 4°C first with bacterial cell lysates followed by 0.2% BSA in GST binding buffer (20 mM Hepes/10% glycerol/0.2 mM EDTA/1 mM DTT/100 mM NaCl/0.5% Nonidet P-40/0.25 mM KCl). The GST-Ski was then incubated with 2 μg of purified recombinant Smad1 or Smad4 for 1 h in the same buffer. After extensive washing, the Ski-bound Smads were eluted by glutathione and detected by immunoblotting with anti-Smad1 (T-20, Santa Cruz Biotechnology) or anti-Smad4 (C-20, Santa Cruz Biotechnology) antibodies.
Immunoprecipitation and Immunoblotting.
Flag- or hemagglutinin (HA)-tagged Smad and Ski proteins were isolated from transfected 293T cells by immunoprecipitation with anti-Flag agarose (M2, Sigma) or anti-HA beads followed by elution with Flag or HA peptides and analyzed by Western blotting as described (27).
Alkaline Phosphatase Assay.
W-20-17 cells were seeded overnight in a well in a 96-well cluster and then transfected with or without c-ski. After 24 h, the cells were treated with 250 ng/ml BMP2 for an additional 40 h. The alkaline phosphatase activity was measured by using p-nitrophenyl phosphate (Sigma) as a substrate as described (3). Enzymatic activities were determined by measuring absorption at 405 nm in a model 550 microplate reader (Bio-Rad) and expressed as p-nitrophenol produced in nmol/min per mg protein.
Results
Human Ski Neuralizes Ectoderm in Xenopus Embryos.
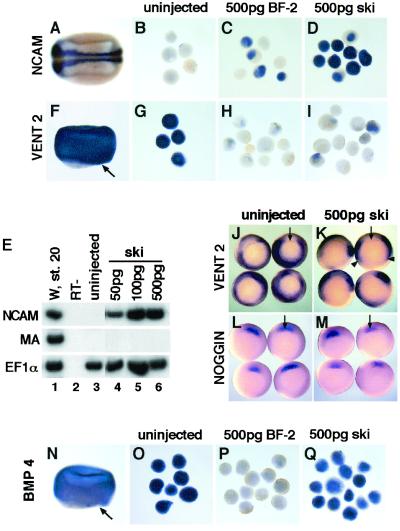
Animal cap assays were used to characterize the biological activities of human c-Ski in Xenopus laevis embryos. One-celled embryos were injected in the animal pole with RNA encoding either human Ski or BF-2. Explants were taken at the blastula stage and cultured until control embryos expressed marker genes of interest. Explants were then harvested and analyzed for gene expression by in situ hybridization and RT-PCR. Ectodermal explants from uninjected embryos developed into epidermis and did not express the general neural marker, NCAM (Fig. 1 B and E, lane 3). As reported (11), injection of BF-2 resulted in a dramatic increase in NCAM expression (Fig. 1C). Human ski RNA at both high (500 pg) and low (50 pg) levels neuralized the explants and induced NCAM expression (Fig. 1 D and E). This neuralization occurs independent of mesoderm because neither brachyury (Xbra) nor muscle actin, markers of general and paraxial mesoderm respectively, was expressed (Figs. 1E and 2A). In addition, embryos expressing human Ski displayed phenotypes identical to those injected with the Xenopus gene (data not shown). Given the high degree of similarity between the human and the Xenopus Ski (85%), it is not surprising that human Ski also neuralizes ectodermal tissue.
Figure 1.
(A–E) Human ski neuralizes Xenopus ectoderm. Explants were cultured until stage 20 for analysis of NCAM expression. (A) Embryos express NCAM only in the central nervous system. Anterior is to the left and dorsal is to the top. (B) Uninjected explants. (C) Explants expressing 500 pg of BF-2 mRNA. (D) Explants expressing 500 pg of human ski mRNA. (E) RT-PCR analysis. Lanes 1 and 2, whole embryo control (stage 20) followed by the RT minus negative control; lane 3, uninjected explants; lanes 4–6, explants injected with increasing amounts of ski mRNA. The elongation factor 1 alpha (EF1α) primers were used to determine RNA recovery. Primers for muscle actin were used to assess mesoderm development. W, whole embryo; RT−, no RT. (F–I) Ski inhibits Xvent2 expression in ectoderm. Ectodermal explants were cultured until the midgastrula stage (stage 10.5). (F) In the embryo, Xvent2 is expressed in the animal cap and the marginal zone (dorsal lip is indicated by the arrow). (G) Uninjected explants. (H) Explants expressing 500 pg of BF-2 mRNA. (I) Explants expressing 500 pg of ski mRNA. (J–M) Ski inhibits Xvent2 expression in the embryo. (J) Control embryos (ventral view) showing Xvent2 expression in the marginal zone (stage 10.5). (K) Embryos injected with 500 pg of ski mRNA; arrowheads point to where Xvent2 expression is inhibited. (L) Control embryos probed for noggin expression. (M) Embryos injected with 500 pg of ski mRNA probed for noggin expression. (N–Q) Ski does not inhibit BMP4 expression in the ectoderm. Explants were cultured until the midgastrula stage (stage 10.5). (N) In the embryo, BMP4 is expressed strongly in the ectoderm and the marginal zone (dorsal lip is indicated by the arrow). (O) Uninjected ectodermal explants. (P) Explants expressing 500 pg of BF-2. (Q) Explants expressing 500 pg of ski.
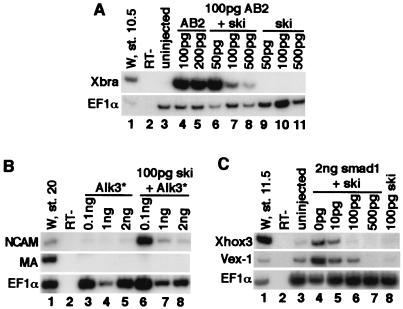
Figure 2.
Ski antagonizes BMP signaling in Xenopus explants. (A) Ski blocks BMP2 activity. Explants were cultured until stage 10.5 and analyzed for Xbra expression by RT-PCR. Lanes 1 and 2, whole embryo control and no RT (RT−); lane 3, uninjected explants; lanes 4 and 5, explants expressing AB2; lanes 6–8, explants expressing AB2 with increasing amounts of ski mRNA; lanes 9–11, explants expressing ski mRNA only. (B) Ski fails to neuralize in the presence of high levels of the constitutive active BMPRI (Alk3*). Explants were analyzed at stage 20 for NCAM expression. Lanes 3–5, explants expressing AB2 mRNA only; lane 6–8, explants expressing ski and increasing amounts of Alk3* as indicated. Alk3*, (Q233D)Alk3. (C) Ski blocks the activity of Smad1. Lane 3, uninjected sample; lane 4, Smad1 stimulates the expression of the ventral markers, Xvex-1 and Xhox-3; lanes 5–7, explants expressing Smad1 mRNA in combination with increasing amounts of ski mRNA; Lane 8, ski alone.
Ski Neuralizes Xenopus Explants by Antagonizing BMP Signaling.
The observation that Ski neuralizes Xenopus ectoderm strongly suggests that it represses BMP signaling in some way. Consistent with this idea, expression of the BMP-responsive gene, Xvent2, was repressed in explants from embryos injected with Ski (Fig. 1I), like those expressing BF-2 (Fig. 1H), but not in those from uninjected embryos (Fig. 1G). In addition, when Ski was ectopically expressed on the ventral side of the gastrulating embryo, Xvent2 (Fig. 1K) and Xvent1 (data not shown) were also down-regulated in the marginal zone where BMP signaling confers ventral and posterior fates. The cells expressing Ski on the ventral side were not simply being converted to more dorsal mesodermal fates because no ectopic expression of the organizer markers noggin and goosecoid (gsc) was apparent (Fig. 1M and data not shown). In fact, when coinjected with Smad2, Ski blocked expression of these mesoderm markers induced by Smad2 (data not shown).
We next examined the ability of Ski to directly block BMP-induced signaling events. High-level expression of BMP2 in the ectoderm results in mesoderm induction as indicated by the up-regulation of Xbra expression (4). We therefore examined the ability of Ski to block BMP2-induced Xbra expression. To obtain a more active form of BMP2, we used a hybrid activin/BMP2, AB2, where the N terminus of prepro-activin (a well-processed TGFβ family member) was fused to the BMP2 C terminus (29). Ectodermal explants from embryos injected with AB2 expressed a high level of Xbra (Fig. 2A, lanes 4 and 5). Coinjections of AB2 with increasing amounts of Ski resulted in a striking decrease in Xbra expression (Fig. 2A, lanes 6–8). Thus, in Xenopus ectoderm, Ski acts as a potent inhibitor of BMP2 activity.
Antagonism of BMP signaling could occur at multiple levels of BMP expression and signaling. In contrast to BF-2, which blocked BMP4 expression (Fig. 1P; ref. 11), Ski did not inhibit BMP signaling at the level of BMP mRNA expression because explants from embryos injected with ski expressed BMP-4 mRNA at the gastrula stage (Fig. 1Q). Furthermore, the blocking of BMP target gene expression by Ski cannot be rescued by addition of exogenous BMP (Fig. 2A), again confirming that Ski does not directly block BMP expression.
Factors from the Spemann organizer, such as Noggin and Chordin, directly associate with BMPs to block access to BMP receptors. However, the expression of noggin, chordin, and gsc was not up-regulated in ectodermal explants from embryos injected with ski (Fig. 1M and data not shown), suggesting that Ski did not inhibit BMP signaling at the level of ligand binding.
We next examined the ability of Ski to neuralize in the presence of increasing levels of active BMPRI. If Ski could still neuralize to the same degree even in the presence of high amounts of active BMPRI, Ski likely functions independent of blocking of BMP signaling. However, if high levels of active BMPRI overcome the ability of Ski to neuralize, then Ski likely acts by abrogating BMP signaling. Injection of the embryos with low amounts of active BMPRI effectively ventralized embryos (data not shown) but had no effect on Ski-induced NCAM expression (Fig. 2B, lane 6). Increasing amounts of active BMPRI, in contrast, blocked the neuralizing activity of Ski completely (Fig. 2B, lanes 7 and 8). Thus, the ability of Ski to neuralize the ectoderm is not independent of BMP signaling.
When Smad1 is expressed in Xenopus ectoderm, it up-regulates expression of ventral mesoderm markers including Xhox3 (32, 33) and Xvex-1 (34) (Fig. 2C, lane 4). Coexpression of increasing amounts of Ski resulted in a corresponding decrease in Xhox3 and Xvex-1 expression (Fig. 2A, lanes 5–7). Thus, Ski can effectively attenuate the activity of Smad1 and repress the expression of BMP-target genes.
Ski Represses BMP Signaling in Mammalian Cells.
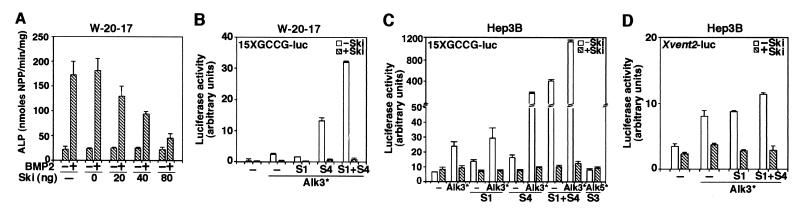
To examine whether Ski also represses BMP signaling in mammalian cells, we used the W-20-17 bone marrow stromal cell line, which up-regulates alkaline phosphatase (ALP) when stimulated with BMP2 (ref. 3; Fig. 3A). Introduction of increasing amounts of ski cDNA resulted in a corresponding decrease in the BMP2-induced ALP activity (Fig. 3A). Similar results were obtained with another BMP-responsive cell line C2C12 (data not shown).
Figure 3.
Ski represses BMP signaling in mammalian cells. (A) Ski inhibits BMP-induced ALP in W-20-17 cells. W-20-17 cells plated in a 96-well cluster were transfected with increasing amounts of c-ski as indicated. Twenty-four hours after transfection, the cells were treated with 250 ng/ml BMP2 for 40 h. ALP was measured by an Elisa reader and expressed as p-nitrophenol produced in nmol/min per mg protein. Ski represses BMP-induced transcriptional activation in W-20-17 (B) and Hep3B (C and D) cells. W-20-17 and Hep3B cells were cotransfected with 0.75 μg of 15xGCCG-Luc (B and C) or Xvent2-Luc (D), 0.5 μg of constitutive active type I receptors (Alk3* or Alk5*), 0.75 μg of ski, and 0.15 μg of Smads. Luciferase activity was measured 48 h later. Alk3*, (Q233D)Alk3; Alk5*, (T204D)Alk5; S1, Smad1; S4, Smad4.
To examine whether Ski represses BMP-induced transcriptional activation in mammalian cells and whether the repression is Smad-dependent, we transfected the W-20-17 (Fig. 3B), Hep3B (Fig. 3C), and C2C12 cells (data not shown) with a BMP-specific 15xGCCG-Luc reporter construct (7). Activation of BMP signaling by cotransfection of a constitutively active BMPRI and the Smad proteins markedly increased the luciferase activity (Fig. 3 B and C). Cotransfection of ski cDNA strongly repressed this activation (Fig. 3 B and C). As a negative control, cotransfection of the constitutively active type I TGFβ receptor (Alk5*) and Smad3 did not activate 15xGCCG-luc, confirming that the observed effect was specific to BMP signaling. The closely related SnoN did not interact with Smad1 and Smad5 and did not repress the transactivation of 15xGCCG-luc (data not shown). Similar results were obtained with the natural BMP-responsive promoter Xvent2-Luc (Fig. 3D). Thus, Ski can repress BMP-induced and Smad1/4-dependent transcriptional activation.
Ski Interacts with Smad1 and Smad5.
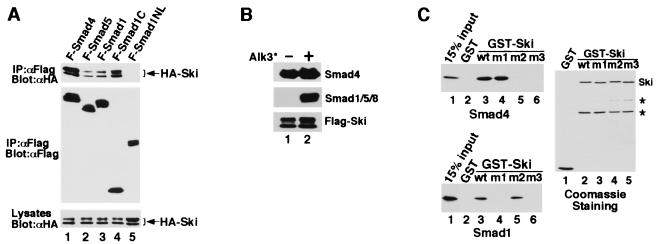
Our results show that c-Ski antagonizes BMP signaling in both Xenopus and mammalian cells by repressing the BMP-specific Smads. Because Ski associates with Smad2, Smad3, and Smad4 through the MH2 domains (25, 26, 28), we asked whether Ski also interacts with Smad1 and Smad5. In 293T cells cotransfected with the Flag-tagged Smads and HA-Ski, HA-Ski was found to associate with the full-length and the MH2 domains but not the MH1 domains of Smad1 and Smad5 (Fig. 4A and data not shown). This interaction is direct because recombinant Smad1 and a Ski fragment purified from E. coli associated with each other readily (Fig. 4C). More importantly, Ski was found to associate with endogenous Smad1/5 and Smad4 in BMP-responsive W-20-17 (Fig. 4B) and C2C12 cells (data not shown). The Ski/Smad1/5 interaction was induced by the constitutively active BMPRI, whereas the Ski/Smad4 association occurred independent of receptor activation (Fig. 4B). Because Smad1 is localized in the cytoplasm and only translocates into the nucleus after activation by the BMP receptor, its interaction with Ski depends on receptor activation. On the other hand, because Smad4 is found in both the nucleus and cytoplasm (26, 27), its interaction with Ski is constitutive. The affinity of Ski for Smad1 or Smad5 is lower than that for Smad2/Smad3 (data not shown) or Smad4 (Fig. 4A).
Figure 4.
Ski interacts with Smad1 and Smad5. (A) Flag-tagged full-length or truncated Smad proteins were cotransfected into 293T cells together with HA-tagged Ski. The Smad-bound Ski was isolated by immunoprecipitation with anti-Flag M2 mAb and detected by Western blotting with an anti-HA mAb. Cell lysates were blotted directly as a control for HA-ski expression. (B) Ski associated with endogenous Smad1 and Smad4 in W-20-17 cells. W-20-17 cells were transfected with Flag-Ski together with or without the constitutively active type I BMP receptor. Ski-associated Smad complex was isolated by immunoprecipitation with anti-Flag agarose and detected by immunoblotting with an anti-Smad1 (T-20) or anti-Smad4 (C-20) antibody. (C) Ski binds to Smad1 directly in vitro. GST-Ski immobilized on glutathione-Sepharose was incubated with purified recombinant Smad1 or Smad4 as described in Materials and Methods. The Ski-bound Smads were eluted by glutathione and detected by immunoblotting with anti-Smad1 or anti-Smad4. GST alone was used as negative control. The amounts of wild-type or mutant GST-Ski used in the binding assay were shown by Coomassie staining. * indicates nonspecific background bands.
Interaction Between Ski and Smad1/Smad4 Is Required for Repression of BMP Signaling.
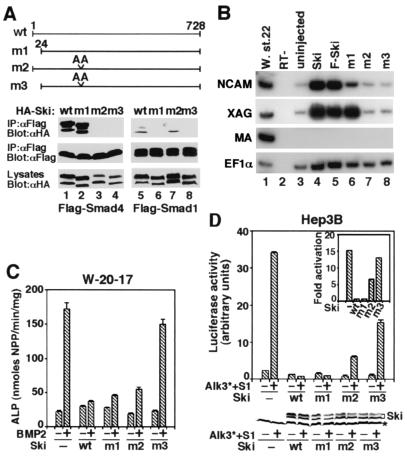
To examine whether the interaction between Ski and the Smads is necessary for the repression of BMP signaling, we used mutant Ski proteins that no longer interact with the Smads. We had previously narrowed the Smad4 binding site in Ski to a region between residues 203 and 239 (26). By comparison with the closely related SnoN and by mutagenesis analyses (data not shown), residues 222 and 223 in Ski were found to be required for binding to Smad4. Mutation of these two residues to Ala (mutant m2) abolished interaction with Smad4 but not with Smad1 (Figs. 4C and 5A). Similarly, the first 24 residues of Ski were required for binding to Smad1 but not with Smad4 (Fig. 4C and 5A). Mutation of both regions resulted in a Ski mutant (m3) that associated with neither Smad1 nor Smad4 (Figs. 4C and 5A).
Figure 5.
Interactions between Ski and the Smads are required for repression of BMP signaling by Ski. (A) Interaction between mutant Ski proteins and the Smads. (Upper) Schematic drawings of the Ski mutants are shown. (Lower) HA-tagged, wild-type, or mutant Ski proteins were cotransfected into 293T cells together with Flag-tagged Smad1 or Smad4 as indicated. The Smad-bound Ski proteins were isolated by immunoprecipitation with anti-Flag M2 mAb and detected by Western blotting with an anti-HA mAb. Cell lysates were blotted directly as a control for HA-Ski expression. (B) Mutant analysis in Xenopus ectoderm. Explants were harvested at stage 22 and assayed for NCAM, XAG, and muscle actin expression. Lanes 1 and 2, whole embryo and no RT (RT−) control; lane 3, uninjected sample; lanes 4 and 5, explants expressing 500 pg of wild-type ski mRNA; lanes 6–8, explants expressing 500 pg of mRNA of ski mutants m1, m2, or m3. (C) To test the ability of the Ski mutants to repress BMP-induced ALP activity, various ski mutants were transfected into W-20-17 cells. ALP activity was assayed as described in Materials and Methods. (D). Hep3B cells were cotransfected with 0.75 μg of 15xGCCG-Luc, 0.5 μg of constitutive active BMPRI (Alk3*), 0.75 μg of HA-tagged, wild-type, or mutant ski, and 0.15 μg of Smads. Luciferase activity was measured 48 h later. (Lower) The levels of Ski proteins present in the transfected cells were detected by Western blotting with the anti-HA antibody. * indicates a nonspecific background band.
In the Xenopus animal cap assay, whereas the wild-type Ski and m1 mutant induced NCAM expression (Fig. 5B, lanes 4 and 5), expression of m2 (unable to bind Smad4) or m3 (unable to bind Smad4 or Smad1) was inactive and resulted in no NCAM expression (Fig. 5B, lanes 7 and 8). The explants expressing m2 or m3 maintained the epidermis fate (data not shown) and did not become a cement gland because the expression of a cement gland marker, XAG, was not induced (Fig. 5B, lanes 7 and 8). In W-20-17 cells, m1 and, to a slightly lesser extent, m2 repressed BMP-induced ALP activity, similar to the wild-type Ski (Fig. 5C). In contrast, the m3 mutant could not block the induction of ALP activity (Fig. 5C). Because Smad1 and Smad4 form a heteromeric complex, these results suggest that Ski can be recruited to the Smad1/Smad4 complex through interaction with either Smad1 or Smad4. Only when both interaction sites are disrupted is the ability of Ski to repress BMP signaling abolished. Similar results were observed in the transcriptional activation assay (Fig. 5D). When expressed at similar levels (Fig. 5D Lower), both the m1 and m2 mutants, but not the m3 mutant, repressed transactivation of the 15xGCCG-luc reporter construct. Thus, the Ski mutants behave similarly when they are expressed in either mammalian or Xenopus cells.
Taken together, these results suggest that interaction of Ski with the BMP-specific Smad1/Smad4 complex is critical for repression of BMP-induced signals both in mammalian cells and in neural fate determination in Xenopus embryos.
Discussion
Ski is a unique oncoprotein in that it induces not only oncogenic transformation but also terminal differentiation of quail embryo fibroblasts (23). We have previously reported that Ski interacts with Smad2, Smad3, and Smad4 and represses their ability to activate TGFβ target genes (25, 26, 28). In this report, we show that Ski can also antagonize BMP signaling in both Xenopus and mammalian cells. The repression by Ski is not the indirect result of activating Spemann organizer genes that block BMP signaling, but instead is due to a direct interaction with the Smad1/5 and Smad4 complexes to repress transcriptional activation of BMP target genes. Thus, Ski can interact with and repress the activity of all R-Smads and the co-Smad, Smad4, and is therefore a general repressor of BMP, activin, and TGFβ signaling. The ability of Ski to inactivate the tumor suppressor pathway of TGFβ may contribute to the transforming activity of Ski, and its ability to antagonize BMP and activin signaling may play a role in the regulation of vertebrate development. This differs from its closely related family member, SnoN, which does not interact with the BMP-specific Smad1/5/8 and does not repress the activation of BMP-responsive promoters in mammalian cells (data not shown).
Ski Represses BMP Signaling in Both Xenopus and Mammalian Cells.
Ski plays an important role in vertebrate development. Mice lacking c-ski displayed defective myogenesis and neural tube formation that result in lethality at birth (24). In Xenopus, XSki protein is maternal and is maintained at a high level until the late neurula stage (14). Injection of ski RNA into the Xenopus embryos induces secondary neural axis formation and neural-specific gene expression in ectodermal explants (14). However, the mechanism by which Ski regulates differentiation was not clear. In this report, we showed that in Xenopus, Ski neuralizes by attenuating BMP signaling and the expression of BMP early responsive genes such as Xvent-1 and Xvent2. This direct inactivation of the Smads by Ski provides a mechanism for specifying neural cells. In addition to Smad1, Ski also inhibits mesoderm induction mediated by Smad2 (data not shown). This is probably independent of the neuralizing activity of Ski because inhibition of Smad2 signals alone does not result in neuralization. Because the relative affinity of Ski for Smad2 is higher than that for Smad1 (data not shown), the ability of Ski to repress dorsal mesoderm should be at least equal to, if not greater than, its ability to neuralize.
Mechanism of Repression of BMP Signaling by Ski.
Ski could repress BMP-Smad function by several mechanisms. Ski may disrupt the heteromeric complex of Smad1 and Smad4 through binding to the MH2 domains. Second, Ski may inhibit Smad1 by up-regulating the expression of the inhibitory Smads, Smad6 or Smad7. Finally, Ski may actively recruit the nuclear corepressor N-CoR and its associated histone deacetylase complex to the Smads (36). We found that the ability of Smad1 to hetero-oligomerize with Smad4 was not affected by coexpression of Ski (data not shown), although at this point, we cannot exclude the possibility that the Smad1/Smad4 complex in the presence of Ski adopts a new conformation and may be inactive. Northern blotting analysis showed that overexpression of Ski did not up-regulate Smad6 or Smad7 expression (data not shown). Thus, Ski most likely represses Smad1/Smad4 function through recruitment of a transcriptional repressor complex. Indeed, we have shown previously that Ski could recruit a nuclear corepressor N-CoR to Smad4 and Smad2 (26).
Our result that Ski interacts with the BMP-Smads and represses BMP signaling differs from those by Akiyoshi et al. (25). In that report, interaction between Ski and the full-length Smad1/5 was not detected by yeast two-hybrid and coimmunoprecipitation, and Ski also failed to repress transcription from the pTlx-luc in P19 cells. Because Ski interacts with the MH2 domain much better than with the full-length Smad1, and because this interaction requires activation of BMP receptors, the inability to detect Ski–Smad1 interaction in yeast may be due to the lack of receptor activation and low stoichiometry of interaction with the full-length Smad1. It is not clear why the Ski–Smad1 interaction was not detected in transfected COS-7 cells. The pTlx-luc reporter responds to BMP activation only in P19 cells but not in other BMP-responsive cells (7). Furthermore, the BMP-responsive region in the Tlx promoter does not share sequence similarity with the Xvent2 promoter or 15xGCCG-Luc, and the Smad1 cofactor, OAZ, does not activate transcription from pTlx-Luc (8). Thus, this promoter may represent a special subset of BMP-responsive genes, and Ski does not affect the activation of these genes. This is also consistent with the observation that Tlx is expressed at around embryonic day 6.5 (E6.5) in the mouse embryo before the elevation of ski expression (E8.5 to E9.5) (17, 24, 37). Our results that demonstrate the inhibitory effect of Ski on BMP signaling rely not only on reporter gene expression but also on the physiological response of Xenopus embryos or cultured mammalian cells.
In summary, we have shown that Ski is a general corepressor of BMP, activin, and TGFβ Smads. By repressing the ability of these Smad proteins to activate downstream target genes, Ski may participate in the regulation of cell proliferation and differentiation.
Acknowledgments
We thank Drs. K. Miyazono for 15xGCCG-Luc, C. Niehrs for Xvent2-Luc, N. Nomura for c-ski cDNA, and J. Massague for Smad1 cDNA. We also thank A. Hemmati-Brivanlou for Smad5 cDNA, C. Kintner for pTN1, J. Smith for bmp4, P. Wilson for AB2, and W. Smith for X-Alk3*. We are grateful to S. Stroschein for critically reading the manuscript. This work was supported by an American Cancer Society grant, a March of Dimes research grant, and the Human Frontier Science Program (to K.L.); by National Institutes of Health Grant GM49346 (to R.H.); and by predoctoral grants from the National Science Foundation and Genentech (to F.M.). W.W. is a Ph.D. candidate at the University of Science and Technology of China and is completing his thesis research at the Lawrence Berkeley National Laboratory.
Abbreviations
- BMP
bone morphogenic protein
- TGFβ
transforming growth factor-β
- ALP
alkaline phosphatase
- HA
hemagglutinin
- GST
glutathione S-transferase
- RT
reverse transcriptase
- NCAM
neural cell adhesion molecule
References
- 1.Hogan B L. Curr Opin Genet Dev. 1996;6:432–438. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 2.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney J M, Fujisawasehara A, Suda T. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thies R S, Bauduy M, Ashton B A, Kurtzberg L, Wozney J M, Rosen V. Endocrinology. 1992;130:1318–1324. doi: 10.1210/endo.130.3.1311236. [DOI] [PubMed] [Google Scholar]
- 4.Harland R M. Curr Opin Genet Dev. 2000;10:357–362. doi: 10.1016/s0959-437x(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 5.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 6.Massague J, Chen Y G. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 7.Kusanagi K, Inoue H, Ishidou Y, Mishima H K, Kawabata M, Miyazono K. Mol Biol Cell. 2000;11:555–565. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 10.Baker J C, Beddington R S, Harland R M. Genes Dev. 1999;13:3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariani F V, Harland R M. Development (Cambridge, UK) 1998;125:5019–5031. doi: 10.1242/dev.125.24.5019. [DOI] [PubMed] [Google Scholar]
- 12.Onichtchouk D, Chen Y G, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Nature (London) 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Kavsak P, Abdollah S, Wrana J L, Thomsen G H. Nature (London) 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 14.Amaravadi L S, Neff A W, Sleeman J P, Smith R C. Dev Biol. 1997;192:392–404. doi: 10.1006/dbio.1997.8780. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Turck C M, Teumer J K, Stavnezer E. J Virol. 1986;57:1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavnezer E, Brodeur D, Brennan L A. Mol Cell Biol. 1989;9:4038–4045. doi: 10.1128/mcb.9.9.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons G E, Micales B K, Herr M J, Horrigan S K, Namciu S, Shardy D, Stavnezer E. Dev Dyn. 1994;201:354–365. doi: 10.1002/aja.1002010407. [DOI] [PubMed] [Google Scholar]
- 18.Fumagalli S, Doneda L, Nomura N, Larizza L. Melanoma Res. 1993;3:23–27. doi: 10.1097/00008390-199304000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namciu S, Lyons G E, Micales B K, Heyman H C, Colmenares C, Stavnezer E. Dev Dyn. 1995;204:291–300. doi: 10.1002/aja.1002040307. [DOI] [PubMed] [Google Scholar]
- 21.Ludolph D C, Neff A W, Parker M A, Mescher A L, Smith R C, Malacinski G M. Biochim Biophys Acta. 1995;1260:102–104. doi: 10.1016/0167-4781(94)00194-8. [DOI] [PubMed] [Google Scholar]
- 22.Sleeman J P, Laskey R A. Oncogene. 1993;8:67–77. [PubMed] [Google Scholar]
- 23.Colmenares C, Stavnezer E. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 24.Berk M, Desai S Y, Heyman H C, Colmenares C. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 26.Luo K, Stroschein S L, Wang W, Chen D, Martens E, Zhou S, Zhou Q. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroschein S L, Wang W, Zhou S, Zhou Q, Luo K. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Liu X, Eaton E N, Lane W S, Lodish H F, Weinberg R A. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- 29.Eimon P M, Harland R M. Dev Biol. 1999;216:29–40. doi: 10.1006/dbio.1999.9496. [DOI] [PubMed] [Google Scholar]
- 30.Vize P D, Melton D A, Hemmati-Brivanlou A, Harland R M. Methods Cell Biol. 1991;36:367–387. doi: 10.1016/s0091-679x(08)60288-5. [DOI] [PubMed] [Google Scholar]
- 31.Wilson P A, Melton D A. Curr Biol. 1994;4:676–686. doi: 10.1016/s0960-9822(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 32.Altaba A R, Melton D A. Development (Cambridge, UK) 1989;106:173–183. doi: 10.1242/dev.106.1.173. [DOI] [PubMed] [Google Scholar]
- 33.Altaba A R, Melton D A. Nature (London) 1989;341:33–38. doi: 10.1038/341033a0. [DOI] [PubMed] [Google Scholar]
- 34.Shapira E, Marom K, Yelin R, Levy A, Fainsod A. Mech Dev. 1999;86:99–111. doi: 10.1016/s0925-4773(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 35.Luo K, Lodish H F. EMBO J. 1996;15:4485–4496. [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura T, Khan M M, Kaul S C, Dona H-D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang S J, Hoodless P A, Lu Z, Breitman M L, McInnes R R, Wrana J L, Buchwald M. Development (Cambridge, UK) 1998;125:1877–1887. doi: 10.1242/dev.125.10.1877. [DOI] [PubMed] [Google Scholar]