Abstract
One of the primary regulators of Receptor activator of NF-κB ligand (RANKL) is 1,25-dihydoxyvitamin D3 (1,25(OH)2D3). To elucidate the mechanism whereby 1,25(OH)2D3 activates RANKL expression we screened some 300 kb of the RANKL gene locus using a ChIP on chip analysis and identified five potential regulatory regions lying significant distances upstream of the transcription start site (TSS), the farthest over 70 kb from the TSS. A direct ChIP analysis confirmed the presence of the VDR/RXR heterodimer at these sites. The binding of the VDR was associated with histone modification and enhanced entry of RNA polymerase II, indicating an important functional consequence to the localization of these transcription factors in response to 1,25(OH)2D3. The region -76kb upstream from the TSS, termed D5, was capable of mediating VDR-dependent transcriptional output in response to 1,25(OH)2D3 in luciferase assays. The identified VDRE in this region was able to confer dramatic 1,25(OH)2D3 sensitivity to heterologous promoters. This region was highly evolutionarily conserved and functionally active in the human RANKL gene as well. We propose that the RANKL gene is regulated via multiple enhancers that while located at significant distances from the TSS, likely form a chromatin hub centered on the RankL promoter.
Keywords: RankL; ChIP; ChIP/chip; vitamin D receptor; enhancer; VDRE; glucocorticoids; RNA polymerase II recruitment center; osteoblasts; 1,25-dihydroxyvitamin D3
1. Introduction
RankL is the molecule that is now considered to be both necessary and sufficient for osteoclastogenesis in vivo and in vitro. RankL is a TNF-like factor that is produced by stromal cells and osteoblasts as well as a variety of other cell types [1]. This factor actively promotes not only the process of osteoclast differentiation, but is also required for the cell’s bone resorbing activity and for its survival [2]. It is evident that RankL and its receptor Rank are essential in osteoclast formation, which is most strongly supported by the skeletal phenotypes of both RankL- and Rank-null mice, neither of which are capable of producing osteoclasts in vivo [3, 4]. Abnormalities in the expression of RankL are implicated in a variety of bone diseases as extensively reported. RankL is synthesized and expressed on the surface of regulatory cells in response to 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) and other stimulators. Despite earlier studies [5, 6], regulatory sites within the RankL gene that mediate the actions of 1,25(OH)2D3 remain unclear [7, 8].
The absence of bona fide target sites for VDR action within the first 8 kb of the RankL gene, as reported by us and others [7, 8], prompted a more expansive approach to delineate RankL regulatory regions. We therefore used contemporary ChIP/chip analysis in this endeavor. We discovered five sites of action located at upstream regions from the mouse RankL gene TSS, the most distant at approximately 76 kb. This latter region, termed D5, was transcriptionally active in transfection studies and contained a unique VDRE sequence. This region as well as the VDRE sequence itself was highly evolutionarily conserved among species and functionally active in the human RANKL gene as well, suggesting a role of this region in the regulation of this gene by other transcriptional activators. We propose that the RankL gene is regulated via the D5 region through interaction with additional upstream VDR binding sites that create a chromatin loop for transferring pre-initiation complexes (PIC) to the downstream RankL promoter.
2. Materials and methods
2.1. Cell culture
Mouse MC3T3-E1 and ST2 osteoblastic cells were cultured in α-MEM and MEM-α medium, respectively. Primary calvarial osteoblasts (mOBs) were obtained as previously described [9] and cultured in α-MEM. Human osteosarcoma MG63 cells were grown in Dulbeccos’s modified Eagle’s medium (DMEM) supplemented with 1% non-essential amino acids. COS-7 fibroblasts were also cultured in DMEM. Each medium was supplemented with 10% fetal bovine serum obtained from Hyclone (Logan, UT), 100 U/ml penicillin and 100 ug/ml streptomycin.
2.2. Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation assays were performed as previously described [10].
2.3. Tiled oligonucleotide microarray analysis
ChIP/chip analysis was carried out as described by others [11–14]. In brief, DNA was isolated by ChIP methodology followed by ligation mediated PCR as described by Oberley et al. [14]. The resulting ~500 bp amplicons were then labeled with Cy3 or Cy5 dyes using an indirect labeling protocol and mixed in the presence of CoT-1 DNA, denatured and co-hybridized to custom oligonucleotide microarrays (Nimblegen Systems Inc, Madison, WI) as described. The microarrays were washed extensively and scanned using an Axon 4000B scanner at the appropriate wavelengths.
Custom oligonucleotide arrays were synthesized by Nimblegen Systems, Inc (Madison, WI). The microarray probes consisted of maskless-array, in situ-synthesized 50-mer oligonucleotides at 2 bp intervals representing a screen of over 300 kb of the mouse RANKL gene locus from 200 kb upstream of the gene’s TSS to 100 kb downstream of the final 3′ non-coding exon. The tiled arrays were synthesized in duplicate in both the forward as well as reverse directions, providing 4 independent measurements at each site within the gene. In addition, each analysis was carried out using two independently-derived ChIP DNA samples. After sample co-hybridization, the logarithmic enrichment ratio of Cy5 to Cy3 hybridization intensities (log2 ) were plotted as a function of chromosome nucleotide position. While all the peaks representative of enhanced VDR or RXR binding either in the presence of 1,25(OH)2D3 or as compared to input DNA are presented as raw data, a peak finding algorithm was utilized to score the relative level of binding in the five regions identified [15].
2.4. Plasmids
The mouse RankL D5 (-76045/-74973) and human RankL D5 (-96903/-95805) regions were amplified from genomic DNA and cloned into the HindIII/BamHI sites of the TK-luc vector. The mRL-VDRE (-75620/-75590) containing several overhanging 5′ and 3′ nucleotides were synthesized, annealed and similarly cloned into the HindIII/BamHI sites of pTK-luc. All plasmid constructs were sequenced to verify successful cloning.
2.5. Transfection assays
MC3T3-E1 and/or ST2 cells were seeded into 24-well plates at appropriate densities and cultured in α-MEM or MEM-α medium containing 10% FBS. Cells were transfected 24 hrs later with Lipofectamine PLUS in serum and antibiotic-free medium. Individual wells were transfected with 250 ng of a luciferase reporter vector, 50 ng of pCH110-βgal and 50 ng of pcDNA-hVDR (unless otherwise indicated). After transfection, the cells were cultured for 24 hr in a medium supplemented with 20% FBS and treated with vehicle, 1,25(OH)2D3, Dex or combination of both. Cells were then harvested and the lysates assayed for luciferase and β-galactosidase activities as previously described [16]. Luciferase activity was normalized to β-galactosidase activity in all cases.
3. Results and discussion
Earlier studies indicated that 1,25(OH)2D3 induces RankL expression in a variety of osteoblast-like cells including the mouse ST2 cell line [6]. In order to find VDRE sequences responsible for induction of this gene, we used ChIP-chip analysis to scan the entire mouse RankL gene locus for VDR binding sites. ST2 cells were first treated for 6 hr with either vehicle or 1,25(OH)2D3 and then subjected to a standard chromatin immunoprecipitation (ChIP) using antibodies to VDR, RXR or a non-specific IgG. Fig. 1 documents the hybridization signals generated across this tiled array for the following comparisons: 1) IgG in the presence and absence of hormone, 2) VDR in the absence or presence of hormone. As highlighted in the figure, no preferential enrichment of RankL DNA was evident when a hormone/vehicle comparison was made using DNA derived from immunoprecipitations with control IgG. In contrast, however, five regions of DNA enrichment were observed when similar comparisons were made using ChIP DNA derived from anti-VDR immunoprecipitation. Interestingly, all of these potential sites of VDR binding were located at significant distances upstream of the RankL TSS: -16 kb (mRLD1), -22 kb (mRLD2), -60 kb (mRLD3), -69 kb (mRLD4) and -76 kb (mRLD5); no sites were observed within or downstream of the RankL gene itself. Importantly, VDR binding coincided with RXR biding (data not shown). Each of these binding sites is intergenic in view of the location of AkapII, the annotated gene lying approximately 250 kb upstream. While all of these peaks were obvious to visual inspection, a peak finding analysis of these data [14] indicated that binding of the VDR and RXR to the mRLD5 region was 2 to 5 fold more robust than that observed for mRLD1-mRLD4; mRLD4 generated the weakest signal. These data suggest the locations of at least five potential VDR/RXR binding sites situated at large distances upstream of the RankL gene. Direct ChIP analysis of immunoprecipitated DNA derived from 1,25(OH)2D3-treated ST2 cells confirmed the presence of VDR and RXR at the mRLD1-mRLD5 regions in Fig. 2 but not at intervening sites within the RankL upstream region (data not shown).
Figure 1. ChIP/chip analysis reveals five VDR/RXR interacting regions at significant distances upstream of the mRankL gene TSS.
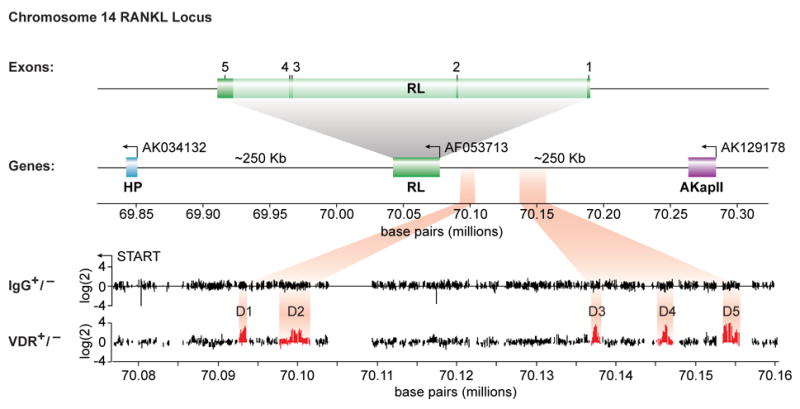
Schematic diagram of the mouse RankL gene with its 6 exons and its position relative to adjacent downstream (AK034132) and upstream AK129178 genes on chromosome 14. The reverse arrow indicates the direction of transcription on the reverse strand. The nucleotide base pairs indicate nucleotide location on chromosome 14 (Dec 2004 Assembly). Individual data tracks representing the enrichment ratio of Cy5 to Cy3 hybridization intensity (log(2)) for IgG plus or minus hormone or VDR plus or minus hormone. The nucleotide base pairs on the x axis indicate position on chromosome 14.
Figure 2. 1,25(OH)2D3 induces selective VDR and RXR binding to the mRLD1-mRLD5 regions of the mRankL gene.
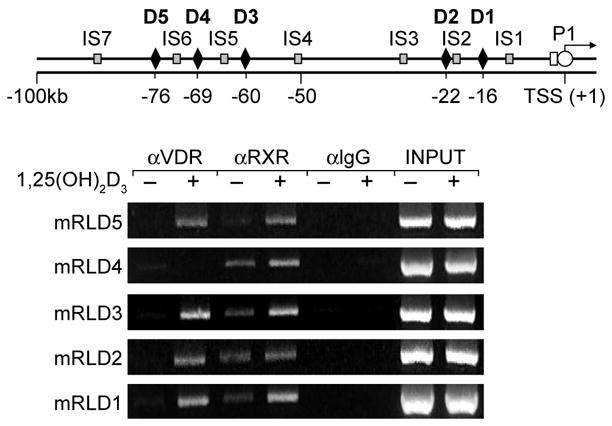
ST2 cells were treated with either vehicle or 1,25(OH)2D3 (10−7 M) and then subjected to ChIP analysis using antibodies to VDR, RXR(pan) or control IgG. Upper panel, Location of primer sets used to detect RankL DNA fragments. Lower panel, Direct ChIP analysis at the RankL locus.
We next amplified each of the five regions (whose coordinates were defined by both conservation and the tiling array data, and are documented in Fig. 3a), and cloned these fragments into a TK promoter-luciferase reporter vector. We assessed transcriptional activity of each cloned fragment in response to 1,25(OH)2D3, DEX or a combination of the two ligands. Strikingly, the results depicted in Fig. 3b reveal that mRLD5 mediated a strong, dose-dependent sensitivity to 1,25(OH)2D3 that was potentiated by DEX. DEX did not function on its own, rather it appeared to potentiate the activity of 1,25(OH)2D3 when used in combination suggesting a potentially additive or synergistic effect on RankL expression. Surprisingly, the regions comprising mRLD1-mRLD4 were unable to convey 1,25(OH)2D3 response to the TK promoter, although an unusual suppression of activity by the individual hormones was noted with mRLD4 (data not shown). These experimental results suggest that the mRLD5 region is likely an important, if not central, component that mediates the regulation of RankL gene expression by 1,25(OH)2D3 and perhaps the GC’s. The significant VDR/RXR binding noted at mRLD1-mRLD4, however, supports these regions as potential regulators of RankL gene expression.
Figure 3. The mRLD5 region of the mRankL gene mediates transcriptional induction by 1,25(OH)2D3 and DEX.
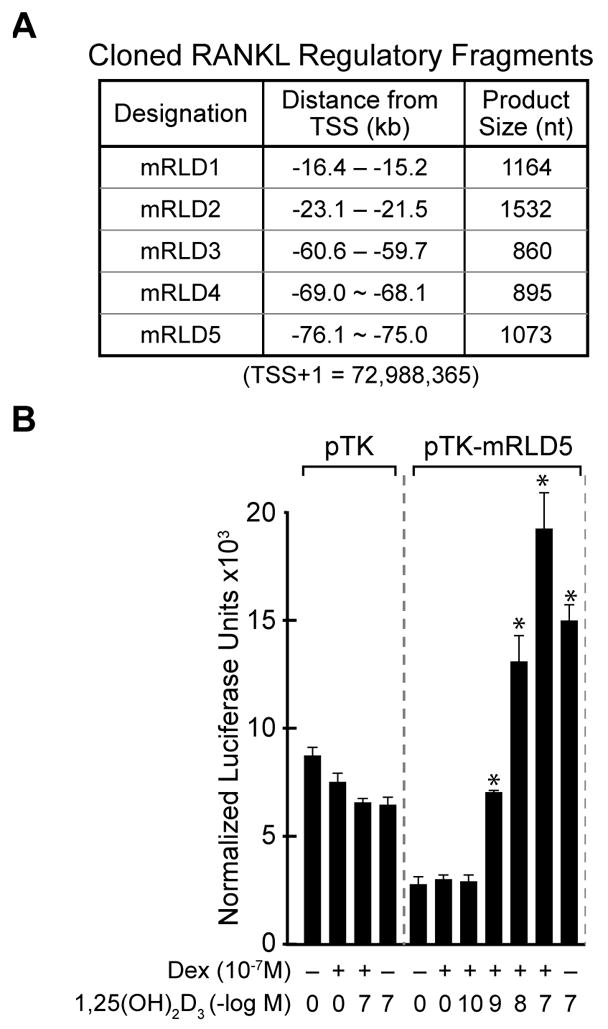
A, Features of the cloned mRLD1-mRLD5 regions of the mRankL gene, including the size of the fragments cloned for evaluation, the distance from the RankL TSS and the boundaries of the fragments on chromosome 14 (Feb 2006 Assembly). B, Basal and hormone-inducible activity of mRLD1-mRLD5 in ST2 cells. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng) or TK-mRLD5. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M) or DEX (10−7 M) plus increasing concentrations of 1,25(OH)2D3 (10−10 – 10−7 M) and evaluated after 24 hr for both luciferase and β-gal activity as described in Materials and Methods. Each point represents the normalized RLU average ± SEM for a triplicate set of transfections.
In order to map VDRE sequence in D5 region, we created a series of 5′ and 3 deletion constructs of D5 region (data not shown), and subsequently found two highly evolutionarily conserved VDRE-like sequences separated by a single base pair. This element is located at -75620 to -75590 upstream of the mouse RankL transcription start site as seen in Fig. 4a. A single copy of the mRL-VDRE was strongly induced by 1,25(OH)2D3 in a dose-dependent fashion when cloned into pTK-luc plasmid vector (Fig. 4b). We also confirmed that this VDRE sequence was capable of binding to VDR and RXR by bandshift assay using purified VDR and RXR as well as nuclear extract from ST2 cells (data not shown). Further analysis of the mRL-VDRE indicated that the introduction of a 3 base pair change in any one of the four mRL-VDRE half-sites fully compromised response to 1,25(OH)2D3 (data not shown).
Figure 4. Mapping the mRL-VDRE.
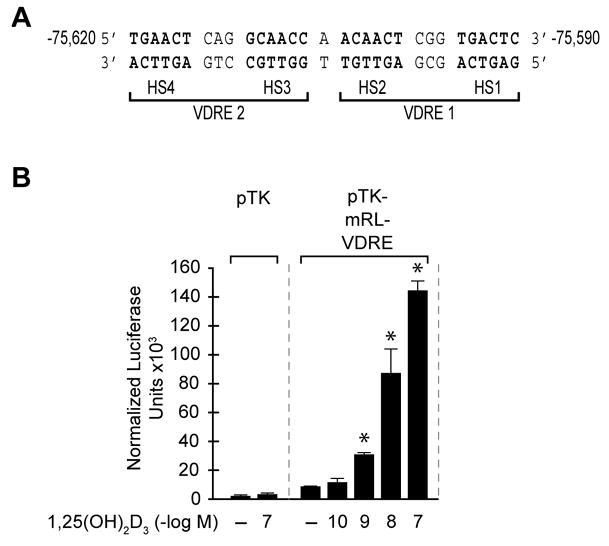
A, DNA sequence of a putative mRL-VDRE revealed by in silico analysis (CONSITE (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite). The nucleotide numbering represent the boundaries of the mRL-VDRE relative to the RankL TSS (Feb 2006 Assembly). Nucleotide bases above the arrows represent the triplet alterations introduced by site-directed mutagenesis into each of the half-sites (HS1–HS4) of the two VDREs (VDRE1 and VDRE2) that constitute the mRL-VDRE. B, Transcriptional activity of the mRL-VDRE or its two component VDREs in ST2 cells. ST2 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng) and either the pTK control vector (250 ng) or pTK-mRL-VDRE. Cells were treated with either vehicle or increasing concentrations of 1,25(OH)2D3 (10−10–10−7 M) and evaluated after 24 hr for both luciferase and β-gal activity.
The mRLD5 region of the mouse RankL gene as well as the VDRE sequence itself is highly evolutionarily conserved within vertebrate genomes including that of the human (Fig. 5a). To explore whether the corresponding RANKL D5 region (hRLD5) in the human RANKL gene might similarly mediate the actions of 1,25(OH)2D3 and DEX on RANKL gene expression, we cloned approximately 0.8 kb of the human hRLD5 region into the TK promoter, transfected this plasmid or the mouse mRLD5 TK plasmid into MG63 human osteoblastic cells and evaluated transcriptional response to both 1,25(OH)2D3, DEX or both ligands. The results in Fig. 5b reveal that the hRLD5 region is comparable to the mouse version in its capacity to mediate response to both 1,25(OH)2D3 and DEX. DEX, however, appears able to exert a small but measurable effect directly on the hRLD5 region even in the absence of 1,25(OH)2D3. Importantly, mutagenesis of two of the putative human VDRE half-sites within the context of hRLD5 abrogated response to 1,25(OH)2D3, thus confirming the role of these elements in 1,25(OH)2D3 dependent induction (data not shown). In fact, we saw VDR binding on the human D5 region by ChIP as well as EMSA, indicating that this region indeed functions for the gene activation mediated by 1,25(OH)2D3 in human (data not shown). These results clearly establish the ability of the hRLD5 region to mediate the actions of 1,25(OH)2D3 and DEX on the human RANKL gene, and provide additional supportive data for the hypothesis that the conserved enhancer activity of RLD5 is essential to the regulation of both mouse and human RANKL genes.
Figure 5.
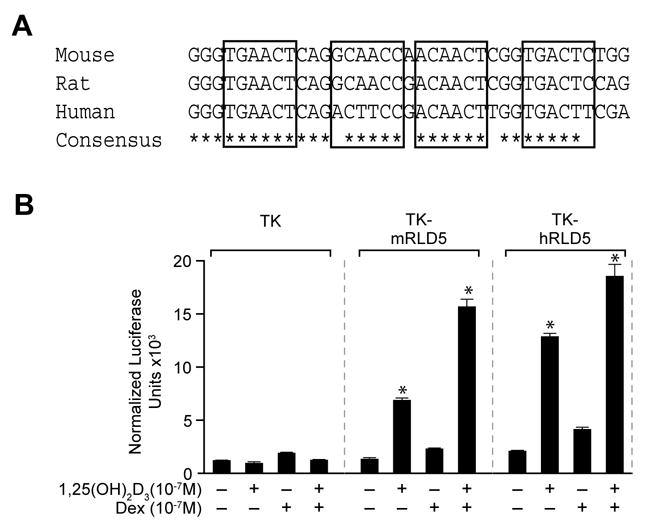
The highly evolutionarily conserved hRLD5 region mediates the transcriptional activity of 1,25(OH)2D3 in the human RANKL gene. A, Sequence conservation of the mRL-VDRE site within mRLD5 in the RankL gene. Nucleotide sequence and conservation surrounding the mRL-VDRE within mRLD5 for multiple species is depicted (Feb 2006 Assembly). B, The hRLD5 region of the hRANKL gene mediates transcriptional induction by 1,25(OH)2D3 and DEX. MG63 cells were transfected with pCH110-βgal (50 ng), pcDNA-hVDR (50 ng), and either the pTK control vector (250 ng), pTK-mRLD5 or pTK-hRLD5. Cells were treated with either vehicle, 1,25(OH)2D3 (10−7 M), DEX (10−7 M) or DEX (10−7 M) plus 1,25(OH)2D3 (10−7 M) and evaluated after 24 hr for both luciferase and β-gal activity. Each point represents the normalized RLU average ± SEM for a triplicate set of transfections.
Acknowledgments
We thank the members of the Pike Laboratory for their helpful discussion. This work was supported by National Institutes of Health Grant DK-74993 (to J.W.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lacey DL, Timm E, Tan HL, Kelly MJ, Dunstan CR, Burgess T, Elliott R, Clombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 2.Jimi E, Akiyama S, Tsurukai T, Okahashi N, Kobayashi K, Udagawa N, Nishihara T, Takahashi N, Suda T. Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J Immunol. 1999;163:434–442. [PubMed] [Google Scholar]
- 3.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-SantosA J, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 5.Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitazawa S, Kitazawa R, Maeda S. Promoter structure of mouse RANKL/TRANCE/ODF gene. Biochem Biophys Acta. 1999;1445:134–141. doi: 10.1016/s0167-4781(99)00032-9. [DOI] [PubMed] [Google Scholar]
- 7.Fan X, Roy EM, Murphy TC, Nanes MS, Kim S, Pike JW, Rubin J. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J Cell Biochem. 2004;93:807–818. doi: 10.1002/jcb.20217. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien CA, Kern B, Gubrij I, Karsenty G, Manolagas SC. Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone. 2002;30:453–462. doi: 10.1016/s8756-3282(01)00692-5. [DOI] [PubMed] [Google Scholar]
- 9.Shevde NK, Plum LA, Clagett-Dame M, Yamamoto H, Pike JW, DeLuca HF. A potent analog of 1alpha,25-dihydroxyvitamin D3 selectively induces bone formation. Proc Natl Acad Sci U S A. 2002;99:13487–13491. doi: 10.1073/pnas.202471299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 11.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim TH, Barrera LO, Qu C, Van Calcar S, Trinklein ND, Cooper SJ, Luna RM, Glass CK, Rosenfeld MG, Myers RM, Ren B. Direct isolation and identification of promoters in the human genome. Genome Res. 2005;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberley MJ, Tsao J, Yau P, Farnham PJ. High throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Method Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 15.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, Derisi JL, Gerton JL. Gemnome-wide mapping of the cohesion complex in the yeast Saccharaomyces cerrevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto H, Shevde NK, Warrier A, Plum LA, DeLuca HF, Pike JW. 2-Methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J Biol Chem. 2003;278:31756–31765. doi: 10.1074/jbc.M304737200. [DOI] [PubMed] [Google Scholar]


