Abstract
Here, we report a catalytic beacon sensor for uranyl (UO22+) based on an in vitro-selected UO22+-specific DNAzyme. The sensor consists of a DNA enzyme strand with a 3′ quencher and a DNA substrate with a ribonucleotide adenosine (rA) in the middle and a fluorophore and a quencher at the 5′ and 3′ ends, respectively. The presence of UO22+ causes catalytic cleavage of the DNA substrate strand at the rA position and release of the fluorophore and thus dramatic increase of fluorescence intensity. The sensor has a detection limit of 11 parts per trillion (45 pM), a dynamic range up to 400 nM, and selectivity of >1-million-fold over other metal ions. The most interfering metal ion, Th(IV), interacts with the fluorescein fluorophore, causing slightly enhanced fluorescence intensity, with an apparent dissociation constant of ≈230 μM. This sensor rivals the most sensitive analytical instruments for uranium detection, and its application in detecting uranium in contaminated soil samples is also demonstrated. This work shows that simple, cost-effective, and portable metal sensors can be obtained with similar sensitivity and selectivity as much more expensive and sophisticated analytical instruments. Such a sensor will play an important role in environmental remediation of radionuclides such as uranium.
Keywords: DNA, DNAzyme, fluorescence, deoxyribozyme, catalytic DNA
Designing sensors that rival the most sensitive and selective analytical instrument has been a grand challenge for a long time. This challenge is especially imposing for metal sensing (1–5), because many metal ions are very similar and sometimes even identical in charge, ionic radius, and other properties, making them difficult to detect at ultra-low concentrations with no interference by other metal ions. Meeting such a challenge is critical in advancing a number of fields including chemistry, biology, environmental engineering, and medicine. A primary example is sensing of uranium, which is a naturally occurring radionuclide that exists ubiquitously in the environment (6). In the past half-century, uranium has been significantly enriched and widely used in nuclear power plants, missiles, and nuclear weapons, and its usage is likely to grow as an important energy source. A method for simple, fast, on-site, and real-time detection and quantification of uranium will be helpful in environmental remediation and minimization of uranium exposure to humans and related adverse health effects, such as radiation and kidney damage, because enriched uranium has been or has the potential to be released into the environment in different parts of the world. Assessment of uranium contamination problems and monitoring the effectiveness of uranium remediation (7–12) require a simple and portable sensing method with high spatial and temporal resolution.
The uranyl ion (UO22+) is the most stable chemical form of uranium in water and is therefore highly bioavailable to pose the greatest risk to human health (13, 14). Current uranium detection relies mainly on instrumental analysis methods, which are based on intrinsic physical properties of the element, such as atomic absorption (15), emission (16), phosphorescence (17, 18), mass (19), or redox potential (20). Most instrumental analysis methods detect only total uranium with poor portability. Being very sensitive, the costs and sophisticated operation procedures of instruments keep uranium detection mostly in well equipped analytical laboratories. Therefore, although on-site and real-time detection of uranium is difficult (21), it is also the most desirable for assessing uranium contamination problems and the effectiveness of remediation strategies. Chemical and biological sensors for UO22+, although available, cannot match the above instrumental analysis methods in sensitivity or selectivity (22–26). Perhaps the most challenging aspect of this sensor design is selectivity over Th(IV), because Th(IV) almost always coexists with uranium. Because of the similar chemical and physical properties of the two actinides, most sensors, and even some analytical instruments [e.g., inductively coupled plasma atomic emission spectroscopy (ICP-AES)] can hardly distinguish between the two metals.
We are interested in finding a new alternative method to design highly sensitive and selective sensors for UO22+, and we chose to use DNAzymes, because they are a promising metal sensing platform. DNA is known to most people as a genetic material to carry and pass heritable information from one generation to the next. In 1994, DNA was first shown to have catalytic activities (27), and such catalytically active DNA are called “DNAzymes” in this work (also known as catalytic DNA, DNA enzymes, or deoxyribozymes elsewhere). To date, no naturally occurring DNAzymes have been identified, and all known DNAzymes have been isolated with a combinatorial biology method called in vitro selection in the laboratory (28–32). Most DNAzymes require metal ion cofactors for structure and function, and many DNAzymes show high metal-binding affinity and specificity (33–38). For example, the 8-17 DNAzyme can be activated by a trace amount of Pb2+ (39), and it has been transformed into sensors for Pb2+ (39–42). Despite the initial success, no metal sensor has reached the detection limit and selectivity of instrumental analysis. Here, we report a DNAzyme-based catalytic beacon that can detect UO22+ in <2 min at ambient temperatures with 11 parts-per-trillion (45 pM) sensitivity and >1-million-fold selectivity over any other metal ions. Practical application of the sensor in detecting uranium in contaminated soil samples is also demonstrated.
Results and Discussion
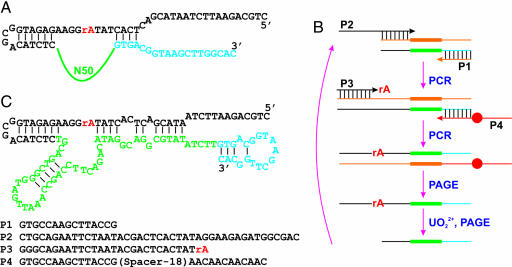
To search for DNAzymes specific for UO22+, in vitro selections with UO22+ as a metal cofactor were carried out. The DNA pool contained a 50-nucleotide random region (N50 in Fig. 1A) flanked by two constant regions (in blue and black). In the middle of this DNA strand, a ribo-adenosine (rA) was introduced to serve as the putative cleavage site for in vitro selection, because a ribonucleotide is ≈100,000 fold more susceptible to hydrolytic cleavage than a deoxyribonucleotide. The selection scheme is presented in Fig. 1B, and a total of four primers were used in two polymerase chain reactions (PCR). In the first PCR, P2 and P1 were used to generate a full-length pool; in the second PCR, rA was introduced through P3. P4 contained a PEG spacer (Spacer-18; denoted as a red dot) that was incorporated into the negative strand without rA. The purpose of the spacer was to stop the PCR extension (43). As a result, two strands of unequal lengths were produced, and the positive strand was purified by denaturing PAGE. UO22+ was added to search for DNAzymes that can perform the self-cleavage reaction, and the cleaved products were separated by PAGE to seed the next round of selection. Initially, 5 mM UO22+ with a 5 h reaction time was used, which gradually decreased to 0.1 mM and 15 min in round 11. The round 10 pool was cloned, and 86 sequences were obtained (see supporting information (SI) Fig. 6). After performing activity assays on individual clones, clone 39 was chosen for uranium sensing. The secondary structure of clone 39 predicted by Mfold (44) is re-drawn in Fig. 1C. The N50 region is shown in green. After truncation and rational design of substrate binding sequences, a transcleavage DNAzyme was constructed (Fig. 2A). The strand that contains rA is the substrate (39S), and the other strand is the enzyme (39E). In the presence of UO22+, 39S is cleaved by 39E and breaks into two fragments.
Fig. 1.
In vitro selection of a UO22+-specific DNAzyme. (A) The sequence of the DNA pool used for the selection, which contained a 50-nucleotide random region (N50) and a cleavage site (rA). (B) Scheme of the selection procedure. (C) The sequence of clone 39 before truncation. P1–P4 are listed from 5′ to 3′.
Fig. 2.
Design of a catalytic beacon to detect UO22+. (A) The secondary structure of an in vitro-selected DNAzyme specific to UO22+, which contains a substrate (39S) and an enzyme (39E). (B) Secondary structure of a Pb2+-specific DNAzyme composed of a substrate (17S) and an enzyme (17E). (C) Design of a catalytic beacon with a fluorophore and two quenchers. Cleavage of the substrate in the presence of UO22+ enhances the fluorescence. (D) Fluorescence signal in the absence and presence of 400 nM UO22+ after 10 min. (E) Biochemical assay of the 39E DNAzyme. Lane 1, substrate alone with 1 μM UO22+; lanes 2–7: time 0 (no UO22+), 1, 2, 5, 10, and 30 min after addition of 200 nM UO22+ to the DNAzyme. The upper and lower bands are uncleaved and cleaved substrates, respectively. (F) A sensor array containing both the UO22+ and Pb2+ sensors. The analytes (0.4 μM UO22+ and/or 2 μM Pb2+, or none) added to each well are indicated at the top of the figure. The Pb2+ sensor was in Tris acetate buffer (pH 8.2) with 100 mM NaCl, whereas the UO22+ sensor was in Mes buffer (pH 5.5) with 300 mM NaCl. The image was scanned 10 min after addition of metal ions.
To signal the reaction, a catalytic beacon was constructed by labeling a fluorophore (6-carboxylfluorescein or FAM) and a quencher (Black Hole Quencher-1) at the 5′ and 3′ ends of 39S, respectively. A second quencher was linked to the 3′ end of 39E (Fig. 2C). The dual quencher method was used to minimize background fluorescence resulting from incomplete hybridization (45). When the fluorescently modified 39S and 39E were hybridized, the fluorescence signal was low because of the proximity between the fluorophore and quencher (Fig. 2D, dashed curve). Upon addition of UO22+, the emission increased >15 times (solid curve), which was attributable to the release of the FAM-labeled fragment after cleavage. The signal generation mechanism is similar to that of molecular beacons (46), except that a catalytic reaction is involved, benefiting from enzymatic turnovers for signal amplification, and is thus termed a catalytic beacon (42).
To confirm that the observed fluorescence increase was due to DNAzyme-catalyzed cleavage, several gel-based assays were performed (Fig. 2E). For this study, 39S was labeled only with a FAM on the 5′ end (no quencher) to eliminate artifacts associated from quenching. The first lane contained only the substrate 39S and 1 μM UO22+, and no cleavage was observed. Similarly, no cleavage was observed for the 39S/39E complex in the absence of UO22+ (lane 2). Upon addition of UO22+ (200 nM) to the 39S/39E DNAzyme (2 μM), the cleavage band intensity started to increase with time (lanes 3–7). This experiment shows that the increased fluorescence was indeed due to DNAzyme-catalyzed cleavage with UO22+ as a cofactor. Because the DNAzyme was in excess in the experiment, the experiment showed the multiple turnover property of UO22+. From the gel, it was calculated that each uranyl ion turned over two DNAzyme cleavage reactions in 1 min. With very high fluorescence increase in the presence of UO22+, the system can be used as a fluorescent sensor for UO22+.
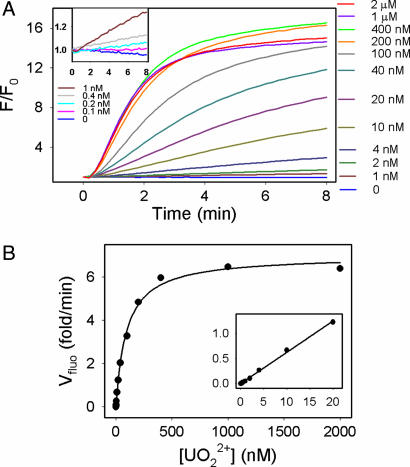
To test the performance of the sensor, the kinetics of fluorescence increase in the presence of varying UO22+ concentrations was monitored (Fig. 3A). The y axis is fluorescence enhancement over background fluorescence before addition of UO22+. A higher rate of fluorescence increase was observed with a higher UO22+ level. The responses of the sensor to low UO22+ concentrations are shown in Fig. 3A Inset. To quantify UO22+, the initial rate of fluorescence increase was calculated and plotted in Fig. 3B. The rate increased with UO22+ concentration until saturation at 400 nM. An apparent dissociation constant of 97 nM was obtained by fitting the curve to binding of one UO22+. There is a linear relationship between fluorescence enhancement and UO22+ concentration until 20 nM (Fig. 3B Inset), and the detection limit was determined to be 11 parts per trillion or 45 pM (3σ/slope). The quenching of FAM by UO22+ was insignificant when the UO22+ concentration was <2 μm (see also SI Fig. 7).
Fig. 3.
Sensitivity of the catalytic beacon-based UO22+ sensor. (A) Kinetics of fluorescence increase over background fluorescence at varying UO22+ concentrations. The DNAzyme sensor concentration was 60 nM, and the buffer contained 50 mM Mes (pH 5.5) and 300 mM NaNO3. (Inset) Sensor responses to low concentrations of UO22+. (B) Plot of the initial rate of fluorescence enhancement vs. UO22+ concentration. (Inset) Low UO22+ concentration range with linear responses.
The sensitivity of this sensor rivals those achieved by the most sensitive analytical instruments (see SI Table 1). The U.S. Environmental Protection Agency (EPA) defined the toxic UO22+ level in drinking water to be 130 nM, which is well within the sensor dynamic range. The ultrahigh sensitivity was attributable to several factors associated with the catalytic beacon-based sensing strategy. First, the DNAzyme itself binds UO22+ very strongly. To the best of our knowledge, a dissociation constant of 97 nM represents the highest metal affinity for nucleic acids. Second, the sensor design allows very low background fluorescence or background variation. Low background offers high signal increases (high slope), whereas low background variation gives low noise (small standard variation). Third, and probably most unique, the signal can be amplified through catalytic turnovers. Each UO22+ can react with multiple catalytic beacons and thus drives the sensitivity up. Because the DNAzyme concentration was lower in the sensor (60 nM) compared to that used in the gel-based assay (2 μM; see Fig. 2E), the turnover number for each UO22+ is smaller. For example, with 10 nM UO22+, each UO22+ turned over approximately two catalytic beacons in the 8-min time window monitored, whereas in the gel-based assay, each UO22+ turned over twice every minute. Therefore, to further increase sensitivity, higher concentrations of the catalytic beacon can be used. The catalytic rate of the DNAzyme is fast (≈1 min−1), which gives quick sensor response. As can be observed in Fig. 3A, detection can be accomplished in <2 min.
To test the selectivity of the sensor for UO22+ vs. other metal ions, the sensor response to 19 competing metal salts at concentrations of 10 μM, 200 μM, and 1 mM was tested. Most metals induced little fluorescence change (see SI Fig. 8 for original kinetic traces), whereas some metals induced strong quenching to FAM, such as Cu2+, Fe2+, Fe3+, Hg2+, and Tb3+. Similarly, the rate of fluorescence change was calculated. To eliminate artifacts associated with initial quenching, the first three data points were omitted. As shown in Fig. 4, at all three concentrations, none of the metals [except Th(IV)] showed a response higher than that of 1 nM UO22+. Some Th(IV)-dependent fluorescence increase was observed. However, gel-based assays showed no Th(IV)-dependent cleavage even with 1 mM Th(IV) (Fig. 4 Inset). In a control experiment where a noncleavable FAM-labeled DNA was used, similar fluorescence increase was observed in the presence of Th(IV) (Fig. 4 Inset).
Fig. 4.
Selectivity of the catalytic beacon-based UO22+ sensor. Sensor responses to all competing metal ions at three concentrations (10 μM, 200 μM, and 1 mM) were tested. (Left Inset) The kinetics of fluorescence enhancement in the presence of 1 mM Th(IV) by the sensor (blue curve), a noncleavable control (red curve), and the sensor in the presence of 10 nM UO22+ (green curve). (Right Inset) Gel-based cleavage assay of the DNAzyme by 10 μM (lane 1), 200 μM (lane 2), 1 mM Th(IV) (lane 3), and 1.7 μM UO22+ (lane 4). The reaction time was 10 min.
In a further control experiment, fluorescein alone without any DNA attached to it was titrated with Th(IV), and increased emission was also observed (see SI Fig. 9). An apparent dissociation constant of ≈230 μM was obtained by fitting the data. To understand the nature of Th(IV) and fluorescein interactions, the emission spectra of fluorescein in the presence or absence of 1 mM Th(IV) were collected, and a shift in the spectra was observed, suggesting specific metal/fluorophore interactions that changed the emission property of fluorescein. Because Th(IV) is the only stable form of thorium in solution, it is unlikely for redox reactions to occur. At pH 5.5, fluorescein is present as a mixture of monoanionic and neutral forms, and the anion form could interact with the highly charged Th(IV) to form a complex. Therefore, the change in fluorescence by Th(IV) was attributed to Th(IV) interacting with the fluorophore instead of cleavage. Unlike UO22+-dependent fluorescence enhancement, the fluorescence increase was <1-fold in the presence of Th(IV), which can be used to distinguish between the two metals. The highest concentration of competing metal tested was 1 mM. Therefore, the selectivity of the sensor for UO22+ is at least 1-million-fold higher compared to Th(IV) or any other metal ions. The DNAzyme sensor described here relies on both binding and cleavage activity of metal ions for sensing. Results obtained so far cannot rule out the possibility that other cations can bind to the DNAzyme sensors. In all gel-based biochemical assays performed up to now, however, cleavage activity was observed only in the presence of UO22+ but not other metal ions, which further confirmed that the high selectivity of the sensor was indeed due to the high selectivity of the DNAzyme instead of artifacts.
With the ultrahigh sensitivity and selectivity of the sensor, we further tested the application of the sensor in contaminated soil samples from the Field Research Center of Environmental Remediation Science Program (Office of Science) at the Department of Energy Y-12 National Security Complex in Oak Ridge, TN. Samples from three different locations were tested. UO22+ was extracted from ≈1.9 g of soil by a 20-ml mixture of 0.25 M NaHCO3 and 0.25 M Na2CO3 at room temperature in air for 20 h according to an established protocol (13). The extracted solution was filtered through a 0.2-μm filter. The DNAzyme sensor responses to these extracted solutions (after diluting 300 times) are shown in Fig. 5. Soil no. 1 contained no uranium, whereas soil no. 3 contained the highest amount of uranium. The UO22+ concentrations calculated from the sensor response are comparable with those obtained from ICP within 20% difference (Fig. 5 Inset).
Fig. 5.
Detection and quantification of uranium in contaminated soil samples. Three soil samples (1, 2, and 3) were extracted and tested. The three kinetics curves are the responses of the sensor to extracted solutions diluted 300-fold. (Inset) The results from the sensor were compared with those from ICP-MS. Error bars indicate variations from three sensor measurements.
We previously reported fluorescent Pb2+ sensors based on an in vitro selected Pb2+-specific DNAzyme (Fig. 2B). The secondary structures of the two DNAzymes show great similarities: both are transcleaving enzymes with the catalytic core flanked by two substrate recognition arms. Such similarities allow a general sensor design to be applied to both DNAzymes. For example, we also attached fluorophore and quenchers in the same way as shown in Fig. 2C to the Pb2+-specific DNAzyme, and a small sensor array was prepared with the UO22+ sensor in the upper row and the Pb2+ sensor in the lower row (Fig. 2F). After addition of metal ions, the sensors lit up only in the presence of the cognate target, whereas no cross-reaction or inhibition was observed. Because DNAzymes specific for many other metal ions can be obtained by in vitro selection, the same signal transduction methodology can be applied to conveniently construct sensor arrays to detect and quantify many metal ions simultaneously.
In summary, we have obtained a UO22+-specific DNAzyme and demonstrated a highly sensitive and selective UO22+ sensor based on the DNAzyme. With a detection limit (45 pM) approaching those of the most sensitive instruments, and with >1-million-fold selectivity over other metal ions (better than instruments such as ICP-AES), the sensor can be a simple portable alternative to instrument analysis. It shows that the DNAzyme sensing platform has an enormous potential for detection and quantification of many metal ions. With wide availability of portable fluorometers, such a highly sensitive and selective UO22+ sensor will find wide applications in on-site and real-time environmental monitoring.
Materials and Methods
Materials.
all DNA samples were purchased from Integrated DNA Technologies (Coralville, IA) and were purified by HPLC. For the uranium sensor, Black Hole Quencher-1 quenchers (structure not available because it is proprietary information from the commercial vendor) were used; for the lead sensor, the quencher on the enzyme strand is a Dabcyl, and the quencher on the substrate is a Black Hole Quencher-1. Uranium acetate dihydrate was purchased from Fisher Scientific (Hampton, NH) and was dissolved to make a 50 mM stock solution in water or in 100 mM sodium citrate. Subsequent dilutions were all made in water.
In Vitro Selection.
The sequences of the template and primers are shown in Fig. 1A. The initial selection pool was generated through template-directed extension followed by PCR amplification. The extension was carried out with 200 pmol of DNA template and 400 pmol of primer P3 in 20 × 100-μl reaction mixtures for seven thermal cycles (1 min each at 92°C, 52°C, and 72°C). The reaction buffer also included 0.05 units/μl TaqDNA polymerase (from Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris·HCl (pH 8.3 at 25°C), 0.01% gelatin, and 0.2 mM each dNTP. After seven cycles of extension, 1 nmol each of P3 and P4 were added to the reaction mixtures for seven additional amplification cycles. The PCR products were gel purified, precipitated with ethanol, and dissolved in 100 μl of buffer (50 mM Mes, pH 5.5, and 250 mM NaNO3). The uranyl acetate stock solution was made in a 2× sodium citrate solution. Eleven rounds of selection were carried out, and the metal ion concentration was gradually decreased from 5 mM to 100 μM UO22+. The reaction time was decreased from 5 h to 15 min. An internal 32P label was incorporated into the PCR product after each round, and the size of the PCR product was checked by running the PCR product mixture next to standard DNA markers of the same size as the PCR product and the cleavage product on a polyacrylamide gel. The bands were then visualized by autoradiography and cut out to purify the DNA. The round 10 DNA pool was then cloned by using the TA-TOPO Cloning Kit (Invitrogen). The vector was transformed into Escherichia coli competent cells through heat-shock. DNA was extracted from cells containing the vector by using miniprep kits (Promega, Madison, WI) and eluted into nanopure water. The concentrations of 86 randomly selected clones were determined by measuring A260 (1 A260 = 50 ng/μl double-stranded DNA). All of the clones were then diluted to a concentration of ≈140 ng/μl and were submitted to the W. M. Keck Center at the University of Illinois for sequencing. Sequences were analyzed by using the Chromas software package (Technelysium Pty, Ltd., Helensville, Queensland, Australia), and sequence alignments were performed by using the program MultAlin (47).
Sensor Preparation and Uranyl Detection.
A total of 60 nM FQ-39S and 60 nM Q-39E (see Fig. 2C) were annealed in a buffer containing 300 mM NaNO3 and 50 mM Mes (pH 5.5) by warming the solution to 70°C for 2 min and subsequently cooling to 20°C in 1 h. The volume for the annealing was 3 ml, and 500 μl of the above prepared sensor was transferred to a quartz cuvette with a 0.5 cm path length on each side. The cuvette was placed in a fluorometer (FluoroMax-P; Horiba Jobin Yvon, Edison, NJ) thermostated at 20°C. The excitation was set at 490 nm and emission at 520 nm was monitored at 12-s intervals. After the initial reading, the cuvette was taken out, and a small volume of concentrated metal solution was added to initiate the cleavage reaction. After vortexing, the cuvette was put back into the fluorometer to continue the kinetics measurement. Other metal salts used included the following: AgNO3, MgCl2, CaCl2, SrCl2, BaCl2, VOSO4, Mn(OAc)2, Fe(NH4)2(SO4)2, FeCl3, CoCl2, NiCl2, CuCl2, ZnCl2, Cd(ClO4)2, Hg(ClO4)2, Pb(NO3)2, TbCl3, EuCl3, and Th(NO3)4.
Gel-Based Activity Assay.
Two micromolar 5′-FAM, singly labeled 39S, and nonmodified 39E were annealed in 300 mM NaNO3 and 50 mM Mes (pH 5.5). After taking a 5-μl aliquot out as a zero time point, UO22+ was added to a final concentration of 200 nM to the remaining solution, and aliquots were taken out at designated time points and quenched with a solution containing 90% formamide, 1 mM EDTA, and 50 mM Tris acetate (pH 8.5). The zero time point sample was mixed with the quenching solution at the end of the experiment to see the effect in the absence of UO22+. A sample containing only the substrate and 1 μM UO22+ was also prepared. The cleaved and uncleaved substrates were separated by 20% PAGE, and the gel was analyzed by a fluorescence imager (FLA-3000G; Fuji, Tokyo, Japan) by exciting at 473 nm.
Detection and Quantification of UO22+ in Field Samples.
Approximately 1.9 g of soil samples from three different locations were weighed. The samples were placed in a beaker, and 10 ml of 0.5 M Na2CO3 and 10 ml of 0.5 M NaHCO3 were added to extract UO22+ for 20 h in air. The extraction solution was collected by filtering through a 0.2-μm filter. The solutions were sent to ICP-MS (microanalysis laboratory, University of Illinois) for uranium content analysis and were also tested by the catalytic beacon-based sensor. A total of 100 μl of the extracted solution was transferred to a tube, and 200 μl of 2% HNO3 was added to react with the carbonate and bicarbonate species in solution. The solution was then mixed with an equal volume of 500 mM Mes to adjust the pH to 5.5. Ten microliters of this solution from samples 1 and 2, or 2 μl of this solution from sample 3 was added to 500 μl of sensor. To quantify the UO22+ concentration, a single point standard addition method was used. Two and 5 μM UO22+ were spiked into the initial 100 μl of extracted solutions for soil samples 2 and 3, respectively. Other procedures for sensing were the same. The UO22+ content from soil sample 1 was estimated to be zero from the standard curve, because the soil was not contaminated.
Supplementary Material
Acknowledgments
This work was supported by the Office of Science (BER), U. S. Department of Energy Grant DEFG02-01-ER63179, National Science Foundation Water CAMPWS Science and Technology Center Grant CTS-0120978, and by National Institutes of Health Small Business Innovation Research Phase I Grant ES014125.
Abbreviations
- ICP-AES
inductively coupled plasma atomic emission spectroscopy
- rA
ribo-adenosine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607875104/DC1.
References
- 1.Tsien RY. In: Fluorescent Chemosensors for Ion and Molecule Recognization. Czarnik AW, editor. Vol 538. Washington, DC: Am Chem Soc; 1993. pp. 130–146. (ACS Symposium Series). [Google Scholar]
- 2.Hitomi Y, Outten CE, O'Halloran TV. J Am Chem Soc. 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- 3.Nolan EM, Lippard SJ. J Am Chem Soc. 2003;125:14270–14271. doi: 10.1021/ja037995g. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z, Fahrni CJ. J Am Chem Soc. 2004;126:8862–8863. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Greenberg B, Taghavi S, Romano C, van der Lelie D, He C. Angew Chem Int Ed. 2005;44:2715–2719. doi: 10.1002/anie.200462443. [DOI] [PubMed] [Google Scholar]
- 6.Gongalsky Konstantin B. Environ Monit Assess. 2003;89:197–219. doi: 10.1023/a:1026031224658. [DOI] [PubMed] [Google Scholar]
- 7.Lovley DR. Science. 2001;293:1444–1446. doi: 10.1126/science.1063294. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Kelly SD, Kemner KM, Banfield JF. Nature. 2002;419:134. doi: 10.1038/419134a. [DOI] [PubMed] [Google Scholar]
- 9.Yan T, Fields MW, Wu L, Zu Y, Tiedje JM, Zhou J. Environ Microbiol. 2003;5:13–24. doi: 10.1046/j.1462-2920.2003.00393.x. [DOI] [PubMed] [Google Scholar]
- 10.Istok JD, Senko JM, Krumholz LR, Watson D, Bogle MA, Peacock A, Chang YJ, White DC. Environ Sci Technol. 2004;38:468–475. doi: 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- 11.Wan J, Tokunaga TK, Brodie E, Wang Z, Zheng Z, Herman D, Hazen TC, Firestone MK, Sutton SR. Environ Sci Technol. 2005;39:6162–6169. doi: 10.1021/es048236g. [DOI] [PubMed] [Google Scholar]
- 12.Wu W-M, Carley J, Gentry T, Ginder-Vogel MA, Fienen M, Mehlhorn T, Yan H, Caroll S, Pace MN, Nyman J, et al. Environ Sci Technol. 2006;40:3986–3995. doi: 10.1021/es051960u. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P, Gu B. Environ Sci Technol. 2005;39:4435–4440. doi: 10.1021/es0483443. [DOI] [PubMed] [Google Scholar]
- 14.Sheppard SC, Evenden WG. Arch Environ Con Tox. 1992;23:117–124. [Google Scholar]
- 15.Abbasi SA. Int J Environ Anal Chem. 1989;36:163–172. doi: 10.1080/03067318108071554. [DOI] [PubMed] [Google Scholar]
- 16.Huff EA, Bowers DL. Appl Spectrosc. 1990;44:728–729. [Google Scholar]
- 17.Brina R, Miller AG. Anal Chem. 1992;64:1413–1418. [Google Scholar]
- 18.Kaminski R, Purcell FJ, Russavage E. Anal Chem. 1981;53:1093–1096. [Google Scholar]
- 19.Boomer DW, Powell MJ. Anal Chem. 1987;59:2810–2813. doi: 10.1021/ac00150a019. [DOI] [PubMed] [Google Scholar]
- 20.Mlakar M, Branica M. Anal Chim Acta. 1989;221:279–287. [Google Scholar]
- 21.Rao TP, Metilda P, Gladis JM. Talanta. 2006;68:1047–1064. doi: 10.1016/j.talanta.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Rohwer H, Rheeder N, Hosten E. Anal Chim Acta. 1997;341:263–268. [Google Scholar]
- 23.Sessler JL, Melfi PJ, Seidel D, Gorden AEV, Ford DK, Palmer PD, Tait CD. Tetrahedron. 2004;60:11089–11097. [Google Scholar]
- 24.Safavi A, Bagheri M. Anal Chim Acta. 2005;530:55–60. [Google Scholar]
- 25.Blake RC, II, Pavlov AR, Khosraviani M, Ensley HE, Kiefer GE, Yu H, Li X, Blake DA. Bioconjug Chem. 2004;15:1125–1136. doi: 10.1021/bc049889p. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Jones RM, Blake DA. Int J Environ Anal Chem. 2005;85:817–830. [Google Scholar]
- 27.Breaker RR, Joyce GF. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 28.Breaker RR. Nat Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 29.Sen D, Geyer CR. Curr Opin Chem Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y. Chem Eur J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Joyce GF. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 32.Achenbach JC, Chiuman W, Cruz RPG, Li Y. Curr Pharm Biotechnol. 2004;5:312–336. doi: 10.2174/1389201043376751. [DOI] [PubMed] [Google Scholar]
- 33.Cuenoud B, Szostak JW. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 34.Santoro SW, Joyce GF. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown AK, Li J, Pavot CMB, Lu Y. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 38.Nelson KE, Bruesehoff PJ, Lu Y. J Mol Evol. 2005;61:216–225. doi: 10.1007/s00239-004-0374-3. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Lu Y. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]
- 40.Liu J, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Liu J, Li J, Bruesehoff PJ, Pavot CMB, Brown AK. Biosens Bioelectron. 2003;18:529–540. doi: 10.1016/s0956-5663(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Lu Y. Meth Mol Biol. 2006;335:275–288. doi: 10.1385/1-59745-069-3:275. [DOI] [PubMed] [Google Scholar]
- 43.Williams KP, Bartel DP. Nucleic Acids Res. 1995;23:4220–4221. doi: 10.1093/nar/23.20.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Lu Y. Anal Chem. 2003;75:6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
- 46.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 47.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.