Abstract
In Escherichia coli, the switch between aerobic and anaerobic metabolism is controlled primarily by FNR (regulator of fumarate and nitrate reduction), the protein that regulates the transcription of >100 genes in response to oxygen. Under oxygen-limiting conditions, FNR binds a [4Fe-4S]2+ cluster, generating a transcriptionally active dimeric form. Upon exposure to oxygen the cluster converts to a [2Fe-2S]2+ form, leading to dissociation of the protein into monomers, which are incapable of binding DNA with high affinity. The mechanism of cluster conversion together with the nature of the products of conversion is of considerable current interest. Here, we demonstrate that [4Fe-4S]2+ to [2Fe-2S]2+ cluster conversion, in both native and reconstituted [4Fe-4S] FNR, proceeds via a one electron oxidation of the cluster, to give a [3Fe-4S]1+ cluster intermediate, with the release of one Fe2+ ion and a superoxide ion. The cluster intermediate subsequently rearranges spontaneously to form the [2Fe-2S]2+ cluster, with the release of a Fe3+ ion and, as previously shown, two sulfide ions. Superoxide ion undergoes dismutation to hydrogen peroxide and oxygen. This mechanism, a one electron activation of the cluster, coupled to catalytic recycling of the resulting superoxide ion back to oxygen, provides a means of amplifying the sensitivity of [4Fe-4S] FNR to its signal molecule.
Keywords: DNA regulation, iron-sulfur
In Escherichia coli, the switch between aerobic and anaerobic respiration is primarily controlled by the transcriptional regulator of fumarate and nitrate reduction (FNR) (1–3). The protein belongs to a large family of regulators that modulate physiological changes in response to various environmental and metabolic challenges (4–6). Together with the E. coli cAMP receptor protein (CRP), FNR is a pivotal member of an expanding superfamily of structurally related transcriptional factors (5). The archetypal CRP structural fold provides a versatile system for transducing either environmental or metabolic signals into a physiological response (5–7). Based on sequence homology, FNR, like CRP, consists of two distinct domains that provide DNA-binding and sensory functions (see Fig. 1) (7–9). The C-terminal DNA-binding domain recognizes specific FNR-binding sequences within FNR-controlled promoters (10). The N-terminal sensory domain contains five cysteine residues, four of which (Cys-20, -23, -29, and -122) are essential and capable of binding either a [4Fe-4S]2+ or a [2Fe-2S]2+ cluster (11–13).
Fig. 1.
Predicted structure of a FNR monomer. The proposed structure of a FNR monomer, based on homology with CRP. The locations of important features are shown. The model was generated by using Swiss-Model and the Swiss-PDB viewer (9) with CRP as a template (7).
FNR is activated under anaerobic conditions by the acquisition of one [4Fe-4S]2+ cluster per protein (12–15), which promotes dimerization and enhances site-specific DNA-binding to target promoters (16, 17). Molecular oxygen triggers the conversion of the [4Fe-4S]2+ cluster into a [2Fe-2S]2+ cluster, both in vivo and in vitro, causing a conformational change within the protein that induces monomerization, preventing sequence-specific DNA binding and favorable interactions with the transcription machinery (12, 14–16, 18–20).
The mechanism of the oxygen-mediated cluster conversion is of considerable current interest. Various mechanisms have been proposed for this process, including oxygen reduction to hydrogen peroxide through metal-centered oxidation (15) and oxygen reduction to water through sulfide-based oxidation (21). Recently, we reported the detection of approximately two sulfide ions released per FNR monomer during cluster conversion (22), demonstrating that cluster oxidation is metal based.
Here, we demonstrate that the reaction of oxygen with [4Fe-4S] FNR (either native or reconstituted) in vitro occurs in two steps. The first is a second-order, one-electron oxidation of the [4Fe-4S]2+ cluster, leading to the generation of superoxide ion and a [3Fe-4S]1+ cluster intermediate, with the ejection of one Fe2+. The second step is the spontaneous (first-order) conversion of the [3Fe-4S]1+ cluster to the [2Fe-2S]2+ form, with the release of two sulfides and a Fe3+ ion. Superoxide generated during the first step undergoes, at least in part, dismutation to hydrogen peroxide and oxygen. We propose that catalytic recycling of superoxide/hydrogen peroxide back to oxygen provides a means to amplify the sensitivity of [4Fe-4S] FNR to its signal.
Results
Characterization of Intermediates During the Oxidation of FNR.
We previously reported the detection of an EPR-active species, with a signal centered on g ≈ 2.01, after the addition of oxygen to EPR-silent [4Fe-4S] FNR (15). To characterize this species further, [4Fe-4S] FNR was exposed to a near stoichiometric amount of molecular oxygen and rapidly frozen. An S = ½ signal, similar to that previously reported (15), was observed (data not shown). The signal reached a maximum intensity at a temperature of 13.9 ± 0.1 K, after which it rapidly decreased and was broadened beyond detection above ≈30 K (data not shown). The signal saturated at 13.9 K with a P½ value of 1.6 ± 0.3 mW. These properties, together with the similarity of the signal line shape to that of the [3Fe-4S]1+ form of c-aconitase (23), indicate that the signal arises from a [3Fe-4S]1+ cluster. Spin integration revealed a level of ≈15% of the original [4Fe-4S]2+ cluster concentration, consistent with previous observations (15). Identical results were obtained with both native and reconstituted [4Fe-4S] FNR samples (a full comparison of these will be reported elsewhere).
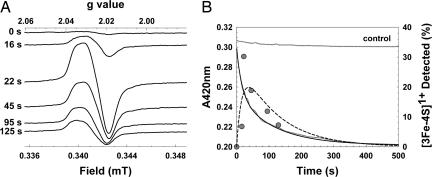
To determine whether the [3Fe-4S]1+ species is an intermediate or a stable dead-end product of an incompletely converted cluster, [4Fe-4S] FNR was mixed with a 10-fold excess of oxygen and aliquots frozen at increasing time periods between 0 and 125 s for EPR analysis (Fig. 2A). The intensity of the [3Fe-4S]1+ S = ½ EPR signal at 15 K was plotted as a function of time (Fig. 2B). The form of the plot demonstrates behavior consistent with that of an intermediate species. A signal at g = 4.3, characteristic of rhombic high-spin Fe3+ ion, was not detected during this time course.
Fig. 2.
Detection of an intermediate cluster during the oxidation of [4Fe-4S] FNR. (A) EPR spectra of reconstituted [4Fe-4S] FNR (20.4 μM), in buffer A, as a function of time after exposure to oxygen (219.5 μM, 21°C). EPR parameters: temperature, 15 K; microwave power, 2.0 mW; frequency, 9.67 GHz; modulation amplitude, 0.5 mT. Spectra are normalized to the same gain. (B) Correlation of EPR observations with optical (420 nm) observations. Optical data are indicated in gray, and EPR data are indicated as filled circles. Double-exponential function fits of the optical and EPR data are shown as a solid line and broken line, respectively (see Data Analysis). Fitting parameters, kobs1 = 0.0611 s−1, kobs2 = 0.0087 s−1.
EPR intensity changes were compared with optical measurements of cluster conversion at 420 nm derived from an identical sample of [4Fe-4S] FNR after the addition of a 10-fold excess of oxygen (Fig. 2B). The time course of the data could not be fitted to a single exponential but fitted well to a double-exponential function (Fig. 2B). The EPR kinetic data subsequently were also fitted to a double-exponential function with rate constants identical to those determined from the 420-nm decay (Fig. 2B). Thus, the intermediate is formed and decays at rates consistent with the overall optical decay process, indicating that the same processes are being observed.
Addition of oxygen results in the initial conversion of [4Fe-4S]2+ to [3Fe-4S]1+, which here is a pseudo-first-order reaction with kobs ≈ 0.06 s−1. Division of the observed rate constant by the oxygen concentration gives an estimate of the apparent second-order rate constant, k1, to be ≈278 M−1·s−1 at 21°C. This reaction is followed by the conversion of the [3Fe-4S]1+ cluster to the [2Fe-2S]2+ form, a first-order process with rate constant k2 ≈ 0.0087 s−1 at 21°C.
Detection and Quantitation of Superoxide.
The reduction of oxidized cytochrome c by superoxide is a single-electron process that can be monitored with great sensitivity because of the large change in the optical properties of the heme α/β absorption bands upon reduction (Δε550nm of 21,000 M−1·s−1) (24).
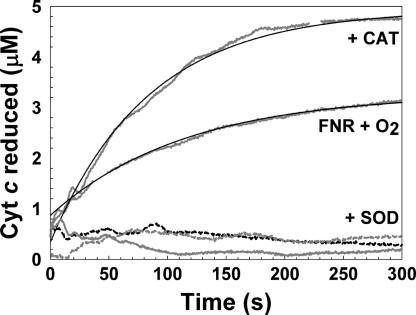
To investigate whether the conversion of [4Fe-4S] to [2Fe-2S] FNR by oxygen generates superoxide, we modified the reported cytochrome c reduction assay (25) (see Materials and Methods). Under pseudo-first-order conditions (in which cytochrome c was in excess), addition of oxygen to 9 μM [4Fe-4S] FNR resulted in 3.3 ± 0.9 μM reduced cytochrome c, indicating that superoxide was produced (0.37 O2−• per [4Fe-4S]) (Fig. 3A). To confirm the specificity of the assay, the experiment was repeated in the presence of superoxide dismutase (SOD) (Fig. 3A). Here, only ≈0.5 μM reduced cytochrome was detected, consistent with measurements under anaerobic conditions [with and without catalase/SOD (Fig. 3B)]. Hydrogen peroxide, which may result from the spontaneous dismutation of superoxide, reoxidizes reduced cytochrome c (24). To assess whether hydrogen peroxide is generated, the assay was repeated in the presence of catalase. Under these conditions 5.2 ± 0.2 μM reduced cytochrome was detected (0.58 superoxide ions per [4Fe-4S]) (Fig. 3A), indicating that some hydrogen peroxide is indeed produced during the oxygen reaction of [4Fe-4S] FNR (15).
Fig. 3.
Detection of superoxide during [4Fe-4S] FNR oxidation. Cytochrome c reduction measured at 21°C in the presence of reconstituted [4Fe-4S] FNR (≈9 μM), oxidized cytochrome c (74 μM) and oxygen (219.5 μM), oxygen and catalase (CAT), or oxygen and SOD (dashed black line), as indicated. Anaerobic (dashed gray line) and anaerobic with CAT and SOD (solid gray) served as controls. Reactions were carried out in buffer A. Single exponential fits are drawn in (solid black lines).
The data at 550 nm fitted well to a single exponential function, giving a pseudo-first-order rate constant, kobs, of 0.0107 ± 0.0012 s−1. Division of the observed rate constant by the cytochrome c concentration estimated the apparent second-order rate constant, kcyt, to be ≈145 M−1·s−1 at 21°C.
Oxygen Dependence of [4Fe-4S]2+ to [3Fe-4S]1+ Cluster Conversion Measured by Absorbance at A420nm.
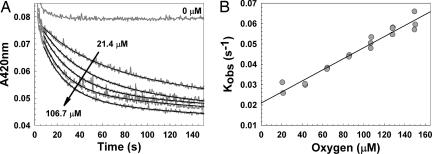
If the initial reaction of [4Fe-4S]2+ FNR with oxygen is a second-order reaction, then the initial rate constant measured under pseudo-first-order conditions (where oxygen is in excess) should exhibit a linear dependence on oxygen concentration. To test this hypothesis, 420-nm decays were measured at increasing concentrations of oxygen (Fig. 4A). A single-exponential function did not provide good fits, but the data fit well to a double-exponential function. Observed pseudo-first-order rate constants for the first exponential phase, kobs, corresponding to [4Fe-4S]2+ to [3Fe-4S]1+ cluster conversion, were plotted against the oxygen concentration (Fig. 4B). A linear dependence of kobs on oxygen concentration is observed, giving an apparent second-order rate constant, k1, of ≈300 M−1·s−1 at 25°C {for both native (data not shown) and reconstituted [4Fe-4S] FNR}.
Fig. 4.
Oxygen dependence of cluster conversion measured by optical absorbance. (A) Reconstituted [4Fe-4S] FNR (5 μM) was mixed with aliquots of buffer B containing varying concentrations of dissolved oxygen at 25°C. Loss of the [4Fe-4S] cluster was monitored at 420 nm as a function of time (in gray). Double- exponential function fits of the data are indicated by solid black lines. Arrows indicate the direction of response with increasing initial oxygen concentrations. (B) Plot of the first observed (first order) rate constants obtained from the data in A and similar experiments as a function of oxygen concentration (filled circles). A least-squares linear fit (black line) of the data is drawn in.
Rate constants for the second exponential phase, corresponding to [3Fe-4S]1+ to [2Fe-2S]2+ cluster conversion, showed some variability between data sets but were essentially oxygen independent.
Determination of the Oxidation State of Iron Released During Cluster Conversion.
The Fe2+-specific chelator 5,5′(3-(2-pyridyl)-1,2,4-triazine-5,6-diyl)-bis-2-furansulfonate (Ferene), which forms a complex, [Fe(II)(Ferene)3]4− (hereafter denoted as Fe2+-Ferene), with intense absorbance at 593 nm, was used to monitor the kinetics of cluster conversion, specifically detecting Fe2+ upon release of iron from the cluster (21). Under anaerobic conditions, both native and reconstituted [4Fe-4S]2+ clusters in FNR are stable to excess Ferene (see 0 μM oxygen, Fig. 5A). A slow reaction was observed, but ≥16 h was required to observe full breakdown of the cluster. Thus, rapid formation of the colored Ferene complex after the addition of oxygen to [4Fe-4S] FNR results from the oxygen-mediated release of iron from the cluster. Control experiments established that ≥98% of Fe2+ ions [as (NH4)2Fe(II)(SO4)2] introduced under anaerobic or aerobic conditions could be recovered as Fe2+-Ferene essentially instantaneously (data not shown), provided that the concentration of Ferene is maintained in an ≈10-fold excess over Fe2+. Control reactions between Ferene (100 μM) and Fe3+ (6.4 μM as FeCl3) resulted in the detection of only 0.14 μM iron as the Fe2+-Ferene complex. Thus, we conclude that Ferene can be used to report accurately on Fe2+ ion release from the [4Fe-4S]2+ cluster.
Fig. 5.
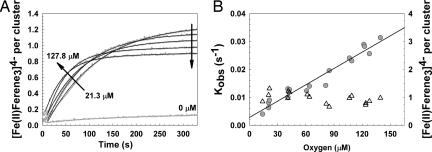
Oxygen dependence of the rate of Fe2+ release from [4Fe-4S] FNR upon oxidation. (A) Reconstituted [4Fe-4S] FNR (2 μM) was mixed with aliquots of buffer B containing varying concentrations of dissolved oxygen at 25°C. Release of Fe2+ was monitored with Ferene (100 μM) at 593 nm (in gray). Single- exponential function fits of the data are indicated by solid black lines. Arrows indicate the direction of response with increasing initial oxygen concentrations. (B) Plot of the first observed (first order) rate constants obtained from the data in A and similar experiments as a function of oxygen concentration (filled circles, left ordinate). A least-squares linear fit (black line) of the data is drawn in. The dependence of Fe2+ recovered per [4Fe-4S] by Ferene is plotted as a function of the oxygen concentration (open triangles, right ordinate).
Kinetic experiments were repeated in the presence of excess Ferene, with Fe2+-Ferene monitored at 593 nm. Data from both native and reconstituted [4Fe-4S] FNR (Fig. 5A) fit well to a single-exponential function. A plot of the observed rate constant as a function of the oxygen concentration (Fig. 5B, left ordinate) is linear, giving an apparent second-order rate constant, k1, of ≈200 M−1·s−1. Hence, monitoring cluster conversion at 420 nm and detection of Fe2+ release by Ferene complex formation are consistent, giving an apparent second-order rate constant of k1 = 250 ± 50 M−1·s−1 at 25°C. These observations are also consistent with the kinetic modeling of the intermediate [3Fe-4S]1+ cluster.
From the Ferene assay data, the total concentration of Fe2+ detected during cluster conversion was determined. A plot of Fe2+ recovered as a function of oxygen concentration is shown in Fig. 5B (right ordinate). This plot demonstrates clearly that ≈1 Fe2+ ion per cluster is recovered. The slight downward trend indicates that somewhat less Fe2+ is detected at higher oxygen concentration, presumably because of an increasing propensity of Fe2+ toward oxidation. In equivalent experiments, the inclusion of the reductant sodium ascorbate (4 μM) resulted in the detection of 2 Fe2+ ions per cluster (data not shown). Thus, we conclude that Fe2+ is released during step 1, and Fe3+ is released during step 2.
Discussion
The mechanism by which the global transcriptional regulator FNR converts from an active DNA-binding [4Fe-4S]2+ form to an inactive [2Fe-2S]2+ form is currently the subject of debate (21, 26). Here, we have studied the kinetics of the conversion process and sought to identify the intermediates generated during it. A nearly complete description of the process emerges from this study.
The data demonstrate that the oxygen-induced conversion of the [4Fe-4S]2+ cluster of FNR to the [2Fe-2S]2+ form proceeds in at least two steps, with the first leading to an EPR-active [3Fe-4S]1+ intermediate species. The rate of this step increased linearly with oxygen, indicating an overall second-order reaction with an apparent rate constant, k1 = 250 ± 50 M−1·s−1 at 25°C.
The conversion of [4Fe-4S] to [3Fe-4S] clusters is well known. Some ferredoxins (Fds), such as FdI and FdIII from Azotobacter vinelandii and Desulfovibrio africanus, respectively, have labile clusters that interconvert by a reversible, Fe2+ concentration-dependent equilibrium (27, 28). This process is not the reaction in FNR because the [4Fe-4S]2+ cluster is stable toward the addition of the Fe2+ chelator Ferene. Instead, the reaction of the [4Fe-4S]2+ cluster is an oxidative one. Oxidative conversion of [4Fe-4S] to [3Fe-4S] clusters was first observed in simple bacterial Fds (29) and quantified by using electrochemical methods (30). In studies of Clostridium pasteurianum 8Fe Fd, a one-electron oxidation pulse was observed to take the two clusters transiently to the superoxidized state, [4Fe-4S]3+, which labilized an Fe2+ ion to generate the oxidized form, [3Fe-4S]1+ (31); see Eq. 1:
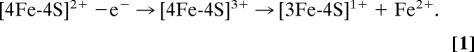 |
This reaction may provide a good model for the [4Fe-4S] FNR reaction observed here. A one-electron oxidation reaction of this type would result in an Fe2+ ion and the one-electron reduction product of oxygen in superoxide. Alternatively, the Fe2+ may also be oxidized, resulting in the two-electron reduction product, hydrogen peroxide. We have previously detected hydrogen peroxide after the reaction of [4Fe-4S] FNR with oxygen (15).
To resolve this crucial question, we sought to determine the nature of the reduced oxygen species and the oxidation state of the iron released during step 1 of the reaction. A modified version of the cytochrome c reduction method of McCord and Fridovich (25) demonstrated that a significant quantity of superoxide (0.37 ions per cluster) is generated during the oxygen reaction of [4Fe-4S]. In the presence of added SOD, the reduction of cytochrome c is essentially prevented, confirming the specificity of the reaction.
Superoxide ions are generated in cells as a by-product of aerobic respiration, in which oxygen is reduced to water (32). The process is not 100% efficient, and partially reduced oxygen species are generated that can be extremely toxic (33). The cell is equipped with SODs and catalases/peroxidases to deal with them (32, 33). Our observations here demonstrate that, in its reaction with oxygen, FNR generates superoxide as an essential step in its mechanism of action, rather than as a by-product. We are not aware of any other well characterized examples of the deliberate generation of superoxide.
The O2/O2−• couple is not powerfully oxidizing (Eo < 0 V) (34), even when the concentration of oxygen far exceeds that of superoxide. The fact that the one-electron oxidation reaction proceeds therefore indicates that the potential of the [4Fe-4S]3+/2+ couple in FNR must be low, and the observed reaction rate suggests that the protein environment is exquisitely tuned for reaction of the cluster with oxygen.
The concentration of superoxide ion detected was increased further in the presence of catalase (0.58 superoxide ions per cluster), demonstrating that released superoxide disproportionates to hydrogen peroxide and oxygen. The presence of catalase inhibits the reoxidation of reduced cytochrome c by hydrogen peroxide (24), leading to an increase in the total concentration of reduced cytochrome detected. Even with catalase, substoichiometric amounts of superoxide were detected, indicating that an intrinsic dismutase activity competes with the reaction of superoxide with cytochrome c. This observation is consistent with our previous study (15) in which ≈0.5 mol of H2O2 per cluster were detected. We note that superoxide will readily disproportionate to H2O2 and O2 at a substantial rate, especially in the presence of Fe2+ ions (k ≥ 106 M−1·s−1 at pH 7.0) (35).
The apparent second-order rate constant for the reaction of superoxide with oxidized cytochrome c determined here (kcyt ≈ 145 M−1·s−1) is significantly lower than that reported in ref. 36. This finding indicates that this reaction is not the rate-determining step. It is similar to that measured here for step 1 (≈250 M−1·s−1), indicating that the rate-determining step is either the generation of superoxide or its release from the protein. Further experiments are required to establish which of these is the case. The latter possibility is of interest in relation to the intrinsic dismutation activity exhibited by the protein.
To address the question of the oxidation state of the iron released during the first step, we used the strong Fe2+-chelator Ferene, which has been used previously to monitor iron release from FNR (21). A comparison of data obtained from the Ferene assay with that obtained in the absence of Ferene (at 420 nm) shows that there is close agreement. Thus, Ferene does not perturb the rate of Fe2+ released by FNR and therefore acts as reporter of the rate of the first step of the reaction. In addition, the Ferene data fit well to a single exponential, indicating that under the conditions used, the assay reports only on the first step of cluster conversion. Importantly, the total amount of Fe2+ ions detected by the Ferene assay was ≈1 Fe2+ ion per cluster, demonstrating that iron is released as Fe2+ in the first step.
Thus, we have identified and quantified each of the products involved in the first step of the reaction between [4Fe-4S] FNR and oxygen, which can be written as in Eq. 2:
The second step of the reaction involves the spontaneous conversion of [3Fe-4S] to [2Fe-2S] FNR. We recently reported the release of two sulfides (and two irons) per cluster during cluster conversion, ruling out sulfide oxidation (22). The [3Fe-4S]1+ and [2Fe-2S]2+ clusters contain three and two Fe3+ ions, respectively; thus, this step involves the release of one Fe3+.
The lack of an EPR signal in the g ≈ 4.3 region, characteristic of rhombic high-spin Fe3+, would be consistent with the conclusion that only Fe2+ ions are released from the cluster after exposure to oxygen. However, we note that Fe3+ and S2− ions can form an EPR invisible species, e.g., Fe2S3 (37, 38). Furthermore, the total amount of Fe2+ released during cluster conversion, i.e., approximately one per cluster, and the detection of 2 Fe2+ ions per cluster in the presence of reductant implies that the second iron is indeed released as Fe3+.
From 420-nm absorption measurements, the observed rate constant for the second-step reaction was found to be oxygen independent, consistent with it being the rate-limiting step. Hence, step 2 is a first-order reaction with an apparent rate constant, k2, of ≈0.008 s−1 (≈1 order of magnitude lower than step 1) and can be written as in Eq. 3:
The overall reaction is given as Eq. 4:
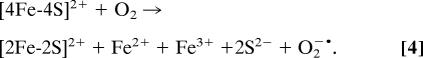 |
The model presented here is not consistent with that proposed by other workers, in which a concerted single-step conversion mechanism was favored (21). The detection and characterization of a [3Fe-4S]1+ intermediate, along with the production of superoxide ion, shows clearly that the conversion reaction does not occur in a single step. However, ejection of one Fe2+ and one Fe3+ during the conversion (Eq. 4) is consistent with previous observations by Mössbauer spectroscopy both in vivo and in vitro (12, 18).
Our model indicates that the first step, the bimolecular reaction of the [4Fe-4S]2+ cluster with oxygen, is the sole oxidative step. This reaction results in a change in the cluster, from one that can bind four cysteine thiols, [Fe4S4(Cys)4]2−, to one capable of binding only three, [Fe3S4(Cys)3]2−. This process is likely to initiate a conformational change and is reminiscent of the ligand exchange at the heme group of CooA (a CRP/FNR family member) that triggers the conformational changes needed to induce DNA binding in the presence of carbon monoxide (39).
In FNR the required reducing equivalent is released during the [4Fe-4S]2+ to [3Fe-4S]1+ conversion. However, unlike bacterial Fds, the [3Fe-4S]1+ cluster itself is thermally unstable, expelling a further iron as Fe3+ in a nonoxidative reaction. This instability enables the protein to complete the conformational change, initiated in step 1, which rearranges the cysteine ligands to accommodate the [2Fe-2S]2+ cluster. We note that a mechanism similar to that described here for FNR may occur in the oxygen-induced degradation of the [4Fe-4S]2+ cluster of biotin synthase and other radical SAM enzymes, which degrade to a semistable [2Fe-2S]2+ cluster (40, 41).
The generation of superoxide ion in step 1 appears to be a hazardous strategy for a transcriptional regulator, given the possibility of DNA damage (42). However, when considered as a regulator with a requirement for a high sensitivity to oxygen levels, we note that, because the process is triggered by a one-electron reaction (step 1), one oxygen molecule could oxidize four [4Fe-4S] clusters provided that the product, superoxide, is rapidly dismutated back to oxygen. In the reaction of four [4Fe-4S] FNR molecules with oxygen, the resulting four equivalents of superoxide undergo dismutation, generating two equivalents each of oxygen and hydrogen peroxide (Eq. 5):
Hydrogen peroxide itself then undergoes dismutation, generating one equivalent of oxygen and two equivalents of water (Eq. 6):
The sum of reactions is then given by Eq. 7:
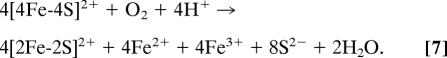 |
Therefore, one oxygen molecule accounts for the conversion of four [4Fe-4S]2+ clusters. We have demonstrated (here and in ref. 15) that FNR, during cluster conversion, has a significant intrinsic dismutase activity. We also note that, even under anaerobic conditions, E. coli contains SOD and catalase enzymes (SodB and KatE, respectively), which rapidly deal with reactive oxygen species present in the cytosol (43, 44). Thus, the rapid recycling of superoxide/hydrogen peroxide back to the primary signal molecule, O2, provides a feedback mechanism to amplify the sensitivity of [4Fe-4S] FNR to oxygen.
Materials and Methods
Purification of Native and Reconstituted [4Fe-4S] FNR.
Native FNR protein was overproduced in aerobic cultures of JRG5369 (E. coli BL21λ DE3 pGS1859) as described (22). GST-FNR fusion protein was overproduced in aerobically grown E. coli BL21λDE3 pGS572. FNR was purified and reconstituted in vitro as described (15) by using 25 mM Hepes/2.5 mM CaCl2/100 mM NaCl/100 mM NaNO3, pH 7.5 (buffer A).
Quantitative Methods.
FNR concentrations were determined by using the method of Bradford (Bio-Rad, Hercules, CA) (45) with BSA as the standard (14). The iron and acid labile sulfide content were determined as described (22, 46). Based on the analyses, both native and reconstituted [4Fe-4S] FNR samples exhibited ε405 nm values of 16,220 ± 135 M−1·cm−1, in close agreement with reported values (21). Concentrations of dissolved oxygen in buffer solutions were determined as described (15).
The detection and quantitation of superoxide was carried out by using a modified version of the cytochrome c reduction procedure reported by McCord and Fridovich (25). Briefly, cytochrome c (74 μM; Sigma, St. Louis, MO) in aerobic buffer A (219.5 μM O2, 21°C) was injected with [4Fe-4S] FNR (9 μM), mixed by inversion, and incubated at room temperature (21°C). Reduction of cytochrome c was monitored at 550 nm by using Δε550 nm = 21,000 M−1·cm−1 (24). To confirm the specificity of the assay, experiments were repeated in the presence of catalase (234 units) and/or SOD (16 units).
Spectroscopy.
Absorbance was measured by using a V550 UV-visible spectrophotometer (Jasco, Easton, MD). EPR measurements were made as described (15). To study the initial products of FNR reaction with oxygen, [4Fe-4S] FNR (20.4 μM) was mixed with aerobic buffer A (21°C) via injection, mixed by inversion, loaded into an EPR tube, and frozen rapidly to 77 K. Spin concentrations were estimated as described (15).
Kinetic Measurements.
Kinetic measurements were performed under pseudo-first-order conditions (with oxygen in excess) at 25°C by combining varying ratios of aerobic and anaerobic buffer (2 ml total volume) essentially as described by Sutton et al. (21), except that absorbance changes were monitored within an anaerobic cabinet via a fiber-optic link (Hellma, Forest Hills, NY), and all aerobic buffers contained dissolved atmospheric oxygen. The reaction was initiated by the injection of native or reconstituted [4Fe-4S] FNR, and the mixture was stirred throughout. The dead time of mixing was ≈5 s. Changes in absorbance at 420 or 593 nm were used to track cluster conversion or formation of the [Fe(II)(Ferene)3]4− complex, respectively. Diffusion of dissolved atmospheric oxygen from the buffer into the head space of the sealed cuvette was assumed to be proportional to each initial starting oxygen concentration (21). [Fe(II)(Ferene)3]4− exhibited a ε593nm value of 39,600 M−1·cm−1 in 50 mM potassium phosphate/400 mM KCl/10% glycerol, pH 6.8 (buffer B) (21, 22).
Data Analysis.
Kinetic absorbance data (at 420 or 593 nm) or EPR data (following signals due to the [3Fe-4S] FNR intermediate) were fitted either to a single- or double-exponential function. For absorbance data, observed rate constants (kobs) obtained from the fits (in the case of fitting to a double-exponential function, the rate constant of the first reaction phase was used) were plotted against the corresponding initial concentration of oxygen to obtain the apparent second-order rate constant. Fitting of kinetic data was performed by using Origin (Microcal, Amherst, MA), and kinetic modeling of the [3Fe-4S]1+ intermediate species was performed by using Dynafit (47).
Acknowledgments
We thank Prof. Dennis Dean (Virginia Tech, Blacksburg, VA) for plasmid pDB551 and Belle Technology (Dorset, U.K.) for their assistance with the anaerobic glove box fridge-freezer. This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/C500360/1 and BB/C500836/1 and a Wellcome Trust award from the Joint Infra-structure Fund for equipment.
Abbreviations
- FNR
regulator of fumarate and nitrate reduction
- Fd
ferredoxin
- Ferene
5,5′(3-(2-pyridyl)-1,2,4-triazine-5,6-diyl)-bis-2-furansulfonate
- SOD
superoxide dismutase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Green J, Paget MS. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 2.Guest JR. Philos Trans R Soc London B. 1995;350:189–202. doi: 10.1098/rstb.1995.0152. [DOI] [PubMed] [Google Scholar]
- 3.Guest JR. J Gen Microbiol. 1992;138:2253–2263. doi: 10.1099/00221287-138-11-2253. [DOI] [PubMed] [Google Scholar]
- 4.Green J, Scott C, Guest JR. Adv Microb Physiol. 2001;44:1–34. doi: 10.1016/s0065-2911(01)44010-0. [DOI] [PubMed] [Google Scholar]
- 5.Korner H, Sofia HJ, Zumft WG. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 6.Spiro S. Antonie Van Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 7.Schultz SC, Shields GC, Steitz TA. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 8.Shaw DJ, Rice DW, Guest JR. J Mol Biol. 1983;166:241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- 9.Guex NP, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 10.Spiro S, Gaston KL, Bell AI, Roberts RE, Busby SJ, Guest JR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 11.Green J, Sharrocks AD, Green B, Geisow M, Guest JR. Mol Microbiol. 1993;8:61–68. doi: 10.1111/j.1365-2958.1993.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 12.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiley PJ, Beinert H. FEMS Microbiol Rev. 1999;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 14.Green J, Bennett B, Jordan P, Ralph ET, Thomson AJ, Guest JR. Biochem J. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crack J, Green J, Thomson AJ. J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 16.Khoroshilova N, Beinert H, Kiley PJ. Proc Natl Acad Sci USA. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazazzera BA, Bates DM, Kiley PJ. Genes Dev. 1993;7:1993–2005. doi: 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- 18.Popescu CV, Bates DM, Beinert H, Münck E, Kiley PJ. Proc Natl Acad Sci USA. 1998;95:13431–13435. doi: 10.1073/pnas.95.23.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan PA, Thomson AJ, Ralph ET, Guest JR, Green J. FEBS Lett. 1997;416:349–352. doi: 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 20.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 21.Sutton VR, Mettert EL, Beinert H, Kiley PJ. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crack JC, Green J, Le Brun NE, Thomson AJ. J Biol Chem. 2006;281:18909–18913. doi: 10.1074/jbc.C600042200. [DOI] [PubMed] [Google Scholar]
- 23.Beinert H, Kennedy MC, Stout CD. Chem Rev. 1996;96:2335–2374. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 24.Vandewalle PL, Petersen NO. FEBS Lett. 1987;210:195–198. doi: 10.1016/0014-5793(87)81336-4. [DOI] [PubMed] [Google Scholar]
- 25.McCord JM, Fridovich I. J Biol Chem. 1968;243:5753–5760. [PubMed] [Google Scholar]
- 26.Unden G, Schirawski J. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 27.Gao-Sheridan HS, Kemper MA, Khayat R, Tilley GJ, Armstrong FA, Sridhar V, Prasad GS, Stout CD, Burgess BK. J Biol Chem. 1998;273:33692–33701. doi: 10.1074/jbc.273.50.33692. [DOI] [PubMed] [Google Scholar]
- 28.Busch JL, Breton JL, Bartlett BM, Armstrong FA, James R, Thomson AJ. Biochem J. 1997;323:95–102. doi: 10.1042/bj3230095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beinert H, Thomson AJ. Arch Biochem Biophys. 1983;222:333–361. doi: 10.1016/0003-9861(83)90531-3. [DOI] [PubMed] [Google Scholar]
- 30.Camba R, Jung YS, Hunsicker-Wang LM, Burgess BK, Stout CD, Hirst J, Armstrong FA. Biochemistry. 2003;42:10589–10599. doi: 10.1021/bi035021v. [DOI] [PubMed] [Google Scholar]
- 31.Tilley GJ, Camba R, Burgess BK, Armstrong FA. Biochem J. 2001;360:717–726. doi: 10.1042/0264-6021:3600717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imlay JA, Fridovich I. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 33.Park SYX, Imlay JA. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood PM. Biochem J. 1988;253:287–289. doi: 10.1042/bj2530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolann BJ, Ulvik RJ. FEBS Lett. 1993;318:149–152. doi: 10.1016/0014-5793(93)80010-r. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi M, Asada K. J Biochem (Tokyo) 1982;91:889–896. doi: 10.1093/oxfordjournals.jbchem.a133777. [DOI] [PubMed] [Google Scholar]
- 37.Douglas T, Dickson D, Betteridge S, Charnock J, Garner C, Mann S. Science. 1995;269:54–57. doi: 10.1126/science.269.5220.54. [DOI] [PubMed] [Google Scholar]
- 38.Griffith R, Morcom A. J Chem Soc. 1945;1945:786–790. doi: 10.1039/jr9450000786. [DOI] [PubMed] [Google Scholar]
- 39.Lanzilotta W, Schuller D, Thorsteinsson MV, Kerby R, Roberts G, Poulos T. Nat Struct Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 40.Cosper MM, Jameson GNL, Hernandez HL, Krebs C, Huynh BH, Johnson MK. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- 41.Broderick JB, Duderstadt RE, Fernandez DC, Wojtuszewski K, Henshaw TF, Johnson MK. J Am Chem Soc. 1997;119:7396–7397. [Google Scholar]
- 42.Imlay JA. Adv Microb Physiol. 2002;46:111–153. doi: 10.1016/s0065-2911(02)46003-1. [DOI] [PubMed] [Google Scholar]
- 43.Kargalioglu Y, Imlay JA. J Bacteriol. 1994;176:7653–7658. doi: 10.1128/jb.176.24.7653-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schellhorn HE, Hassan HM. J Bacteriol. 1988;170:4286–4292. doi: 10.1128/jb.170.9.4286-4292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 46.Beinert H. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 47.Kuzmic P. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]