Abstract
Respiratory metabolism plays an important role in energy production in the form of ATP in all aerobically growing cells. However, a limitation in respiratory capacity results in overflow metabolism, leading to the formation of byproducts, a phenomenon known as “overflow metabolism” or “the Crabtree effect.” The yeast Saccharomyces cerevisiae has served as an important model organism for studying the Crabtree effect. When subjected to increasing glycolytic fluxes under aerobic conditions, there is a threshold value of the glucose uptake rate at which the metabolism shifts from purely respiratory to mixed respiratory and fermentative. It is well known that glucose repression of respiratory pathways occurs at high glycolytic fluxes, resulting in a decrease in respiratory capacity. Despite many years of detailed studies on this subject, it is not known whether the onset of the Crabtree effect is due to limited respiratory capacity or is caused by glucose-mediated repression of respiration. When respiration in S. cerevisiae was increased by introducing a heterologous alternative oxidase, we observed reduced aerobic ethanol formation. In contrast, increasing nonrespiratory NADH oxidation by overexpression of a water-forming NADH oxidase reduced aerobic glycerol formation. The metabolic response to elevated alternative oxidase occurred predominantly in the mitochondria, whereas NADH oxidase affected genes that catalyze cytosolic reactions. Moreover, NADH oxidase restored the deficiency of cytosolic NADH dehydrogenases in S. cerevisiae. These results indicate that NADH oxidase localizes in the cytosol, whereas alternative oxidase is directed to the mitochondria.
Keywords: alternative oxidase, Crabtree effect, NADH oxidase, redox metabolism
Redox homeostasis is a fundamental requirement for sustained metabolism and growth in all biological systems. The intracellular redox potential is primarily determined by the NADH/NAD ratio and to a lesser extent by the NADPH/NADP ratio. In Saccharomyces cerevisiae, >200 reactions involve these cofactors spread over a large spectrum of cellular functions (1). Because NADH is a highly connected metabolite in the metabolic network (1), any change in the NADH/NAD ratio leads to widespread changes in metabolism (2). NADH is generated primarily in the cytosol by glycolysis and in the mitochondria by the tricarboxylic acid (TCA) cycle. Because the NADH/NAD redox couple cannot traverse the mitochondrial membrane in S. cerevisiae and other eukaryotic cells (3), distinct mechanisms oxidize NADH to NAD in the cytosol and mitochondria. Cytosolic NADH is oxidized by two external (cytosolic) mitochondrial membrane-bound NADH dehydrogenases encoded by NDE1 and NDE2 genes with catalytic sites facing the cytosol (4). Additionally, glycerol-3-phosphate dehydrogenases (encoded by GPD1 and GPD2) oxidize cytosolic NADH with concomitant glycerol formation when the NADH formation rate surpasses its oxidation rate (5). Mitochondrial NADH is oxidized by one internal mitochondrial membrane-bound NADH dehydrogenase encoded by NDI1 (6).
In many eukaryotic cells, including S. cerevisiae, there is complete respiratory metabolism at low glycolytic fluxes, whereas higher glycolytic fluxes result in overflow metabolism leading to the formation of byproducts. In S. cerevisiae, overflow metabolism begins when the specific glucose uptake rate (or the glycolytic flux) exceeds a threshold rate, and the result is the formation of ethanol and glycerol (7–10). One hypothesis is that this overflow is due to a limitation in capacity of the respiratory pathways (8, 11). The generation of glycolytic NADH beyond the cellular capacity for its oxidation leads to reduced conditions and ultimately reduced coproducts like ethanol and glycerol. Because the fermentative pathways leading to ethanol generate less ATP than the respiratory pathway, cells respond by increasing the glycolytic flux to meet the ATP demand (12), and this may further induce overflow metabolism. Despite many years of study, it is not known whether the Crabtree effect is triggered by a limitation in respiratory capacity, by the onset of glucose repression of the respiratory metabolism, or simply by an overflow metabolism at the pyruvate branchpoint.
Aerobic ethanol and glycerol generation is a ramification of the different capacities of the fermentative and respiratory pathways (7, 13, 14). Glycerol is generated to reoxidize surplus cytosolic NADH that is formed in glycolysis (15, 16). Because rapid consumption of glucose could lead to the accumulation of NADH, decreasing NADH accumulation by elevating either the rate of respiration or the direct oxidation of NADH is a logical approach to reduce overflow metabolism in S. cerevisiae. A previous effort to reduce overflow metabolism in S. cerevisiae by manipulating redox balance included deleting GDH1 (encoding cytosolic NADPH-dependent glutamate dehydrogenase), which slightly reduced glycerol formation (17). The combined overexpression of malic enzyme and pyruvate carboxylase resulted in increased NADPH formation at the expense of NADH and ATP formation, but there was no effect on overflow metabolism (18). These results suggest that NADP(H) has a minor role in controlling overflow metabolism in S. cerevisiae. We therefore increased the direct oxidation of NADH by using two approaches: (i) overexpressing a water-forming NADH oxidase encoded by the Streptococcus pneumoniae nox gene (19) and (ii) increasing respiration by overexpressing an alternative oxidase encoded by the Histoplasma capsulatum AOX1 gene (20). The alternative NADH oxidase decouples the use of NADH for respiratory energy generation by using molecular oxygen to convert NADH to NAD (19). The alternative oxidase mediates the cyanide-resistant, NADH-dependent transport of electrons from the ubiquinone pool to oxygen in many yeasts (21) and is uncoupled with proton translocation (22, 23). By studying the impact of these two oxidases on overflow metabolism in S. cerevisiae and by mapping the physiological and transcriptional responses to these perturbations in the redox metabolism, we demonstrate that the Crabtree effect in S. cerevisiae is a consequence of a limited respiratory capacity and suggest means to overcome it.
Results
Batch Responses to Engineering Redox Balance.
Batch culture growth was compared for the control strain (CON), for the strain overexpressing NADH oxidase (NOX), and for the strain overexpressing alternative oxidase (AOX). The maximum specific growth rate (μmax) for NOX was 10% lower than for CON, but AOX grew at a rate indistinguishable from CON (Table 1). However, byproduct formation differed between the strains. The ethanol yield and specific productivity were similar for CON and NOX but were ≈70% lower for AOX. In contrast, glycerol yield and specific productivity were similar for CON and AOX but were 6-fold lower for NOX (Table 1). Clearly, in batch cultures the NADH oxidase influences glycerol generation, whereas the alternative oxidase affects ethanol generation. Aerobic glycerol generation in S. cerevisiae results from excess NADH in the cytosol (15, 16).
Table 1.
Physiological characterization of S. cerevisiae having perturbations in redox metabolism
| Strain | μmax, h−1 | Yield from glucose, g/g |
||
|---|---|---|---|---|
| Biomass | Ethanol | Glycerol | ||
| CON | 0.33 | 0.11 | 0.31 | 0.05 |
| NOX | 0.29 | 0.10 | 0.26 | 0.01 |
| AOX | 0.34 | 0.09 | 0.08 | 0.05 |
| ΔΔΔ | 0.22 | 0.06 | 0.16 | 0.24 |
| ΔΔΔ-NOX | 0.29 | 0.08 | 0.27 | 0.02 |
All values were calculated in batch culture during exponential growth phase on glucose, identified by the linear relationship between the natural logarithm of biomass and culture time.
Normally, cytosolic NADH in S. cerevisiae is oxidized by two external (cytosolic) NADH dehydrogenases, encoded by NDE1 and NDE2 (4), or electrons are shuttled to the mitochondria by the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) shuttle involving GUT2 (24). To confirm that reduced glycerol formation in NOX is due to elevated oxidation of cytosolic NADH by NADH oxidase, glycerol generation in a triple deletion mutant ΔΔΔ (nde1Δ nde2Δ gut2Δ) with and without the nox gene was compared. The glycerol yield and the specific glycerol production for ΔΔΔ were >4-fold greater than with CON, whereas for ΔΔΔ-NOX, glycerol production was close to the levels observed for NOX (Table 1). Interestingly, ΔΔΔ also exhibited low ethanol generation, but ΔΔΔ-NOX generated ethanol at a level similar to CON (Table 1).
Steady-State Responses to Engineering Redox Balance.
Steady-state cultivations permitted comparative analysis of metabolic characteristics between strains at identical specific growth rates. The metabolism of CON, NOX, and AOX at a specific growth rate (dilution rate) of 0.1 h−1, using glucose (carbon) or ammonium (nitrogen) as limiting nutrients, was studied. Under carbon limitation, the biomass yield was 10% lower for NOX and 5% lower for AOX compared with CON (Table 2). The presence of NADH oxidase or alternative oxidase increased the specific uptake rate of glucose (rS) and oxygen (rO2), reflecting faster glucose oxidation and subsequent metabolism. The specific CO2 evolution rate (rCO2) also was higher for NOX and AOX than in CON (Table 2).
Table 2.
Steady-state comparison of S. cerevisiae grown in chemostats under carbon or nitrogen limitation
| Strain | Dilution rate, h−1 | Limitation | rS* | Yield† | rO2* | rCO2* | reth* | rgly* | Carbon recovery |
|---|---|---|---|---|---|---|---|---|---|
| CON | 0.10 | C | 1.12 (0.02) | 0.49 (0.00) | 2.75 (0.05) | 2.70 (0.00) | ND | ND | 99.2 (0.8) |
| 0.27 | C | 3.27 (0.07) | 0.46 (0.01) | 7.46 (0.32) | 8.32 (0.22) | ND | ND | 97.5 (3.2) | |
| 0.10 | N | 4.83 (0.32) | 0.11 (0.00) | 4.42 (0.35) | 12.11 (0.75) | 6.16 (0.70) | 0.04 (0.01) | 98.4 (0.5) | |
| NOX | 0.10 | C | 1.25 (0.00) | 0.45 (0.00) | 3.01 (0.04) | 3.14 (0.10) | ND | ND | 95.5 (1.8) |
| 0.26 | C | 4.04 (0.13) | 0.39 (0.01) | 10.77 (0.86) | 11.70 (0.58) | ND | ND | 94.3 (0.1) | |
| 0.10 | N | 5.73 (0.39) | 0.09 (0.00) | 5.28 (0.08) | 17.70 (0.99) | 5.26 (0.35) | 0.00 (0.00) | 93.8 (1.5) | |
| AOX | 0.10 | C | 1.18 (0.06) | 0.47 (0.00) | 2.99 (0.04) | 3.00 (0.02) | ND | ND | 98.1 (2.8) |
| 0.32 | C | 3.87 (0.14) | 0.43 (0.01) | 9.82 (0.36) | 10.92 (0.60) | ND | ND | 98.6 (0.4) | |
| 0.10 | N | 5.27 (0.05) | 0.11 (0.01) | 5.54 (0.39) | 17.20 (0.93) | 4.21 (0.04) | 0.05 (0.00) | 94.1 (1.2) |
The values represent the averages from two independent chemostats; the standard deviation is given in parentheses. ND, not detected.
*The uptake and generation rates are given in mmol/g dry cell weight per hour.
†Calculated as the biomass formed (in grams) for 1 g of glucose consumed.
Nitrogen-limited chemostats allowed steady-state study of the strains under glucose-repressing conditions. These typically respirofermentative conditions were accompanied by the production of ethanol and glycerol, neither of which was produced in the carbon-limited cultures at this low dilution rate. In the presence of NADH oxidase or alternative oxidase, the values of rS and rO2 were both greater, reflecting higher rates of glucose oxidation. Similar to results for the batch conditions, the specific ethanol production rate was elevated for AOX but only slightly for NOX, whereas NOX produced 80% less glycerol (Table 2). The rO2 was 20% greater and the rCO2 was 40% greater for both NOX and AOX compared with CON under nitrogen-limiting conditions.
Effect of Engineering the Redox Balance on the Critical Dilution Rate.
Steady-state experiments at a low dilution rate of 0.1 h−1 under nitrogen limitation demonstrated that NADH oxidase or alternative oxidase affects the formation of ethanol and glycerol. The significant differences prompted us to determine the critical dilution rate (Dcrit) for each strain under carbon limitation (the specific glucose uptake rate at the critical dilution rate represents the glycolytic flux at the onset of overflow metabolism). There was no significant difference in Dcrit for CON (mean ± standard deviation; 0.29 ± 0.01 h−1) and NOX (0.27 ± 0.02 h−1). However, the Dcrit for AOX was 0.32 ± 0.007 h−1, a 10% increase compared with CON. The increase in the Dcrit indicates higher respiratory capacity of S. cerevisiae in the presence of the alternative oxidase.
The three strains were each grown at a specific growth rate slightly lower than the respective Dcrit to ensure completely respiratory conditions (Table 2). At this dilution rate, physiological differences between the three strains were more evident than at the dilution rate of 0.1 h−1. Compared with the lower dilution rate, NOX exhibited a 14% lower biomass, whereas biomass for CON and AOX was only 7% lower. Moreover, the values of rS, rO2, and rCO2 were 3-fold greater for CON and almost 3.5-fold greater for NOX and AOX compared with a dilution rate of 0.1 h−1. The presence of either of the oxidases increased the capacity of NADH oxidation by 25%, as reflected in the increase in the value of rS and rO2 (Table 2).
Enzymatic Analysis of the Response to Engineering Redox Balance.
Because physiological changes in response to the introduction of NADH oxidase or alternative oxidase occurred in glycerol and ethanol production, the activities of key redox-dependent enzymes (glycerol-3-phosphate dehydrogenase, G3PDH; alcohol dehydrogenase, ADH; and isocitrate dehydrogenase, ICDH) were measured under carbon limitation at a dilution rate of 0.1 h−1 and at the Dcrit for CON, NOX, and AOX. An important difference observed was that generally the activities of cytosolic G3PDH and ADH were greater at Dcrit, whereas the activity of mitochondrial ICDH was greater at a dilution rate of 0.1 h−1 (Fig. 1). Moreover, the enzyme activities followed the product formation profile in the three strains at Dcrit. G3PDH activity was 60% lower in NOX and 16% lower in AOX than in CON and correlated with the observation that glycerol production was lowest in NOX. ADH activity was lower for both NOX and AOX compared with CON, whereas ICDH activity was 60% greater for NOX and doubled for AOX (Fig. 1).
Fig. 1.
Specific activities (in units/mg protein) of NADH-dependent glycerol-3-phosphate dehydrogenase (G3P DH), NADH-dependent alcohol dehydrogenase (ADH), and NAD-dependent isocitrate dehydrogenase (IDH) in CON, NOX, and AOX during carbon-limited chemostats grown at a dilution rate of 0.1 h−1 (yellow bars) and at respective Dcrit (brown bars).
The NADH oxidation capacity and the intracellular NADH and NAD concentrations at steady state were also measured. The assay for determining NADH oxidation is not specific for NADH oxidase or alternative oxidase and includes native activity that S. cerevisiae possesses (e.g., NADH dehydrogenases). Under carbon limitation, this basal level of total NADH oxidation in CON was almost twice as high at Dcrit compared with a dilution rate of 0.1 h−1. However, under nitrogen limitation this basal level was 50% lower (Fig. 2A). NOX and AOX consistently exhibited greater NADH oxidation activity than CON under all conditions. Interestingly, unlike in CON, the activity was not lower under nitrogen limitation in NOX and AOX (Fig. 2A). Generally, the intracellular concentrations of NADH and NAD correlated with the total NADH oxidation activity. Specifically, for either carbon-limited or nitrogen-limited conditions, the NADH/NAD ratio was 20–50% lower for NOX and AOX than for CON (Fig. 2B).
Fig. 2.
Measurement of indicators of the redox metabolism during chemostat cultivations under carbon or nitrogen limitation in CON, NOX, and AOX. (A) Total specific NADH oxidation activity. (B) Ratio of intracellular NADH/NAD. Carbon-limited chemostats were operated at a dilution of 0.1 h−1 (white bars) or at the respective Dcrit (gray bars), and the nitrogen-limited chemostat (black bars) was operated at a dilution rate of 0.1 h−1.
Transcription-Based Identification of Metabolic Modules.
The genome-wide transcription response of CON, NOX, and AOX correlated with the physiological changes observed at Dcrit. When the three strains were grown in duplicate cultures at dilution rates just below their respective Dcrit (Table 3), 229 genes (for NOX) and 389 genes (for AOX) exhibited differential expression compared with CON (P < 0.01). The products of the genes exhibiting differential expression in NOX and AOX relative to CON had functions that included carbohydrate metabolism, amino acid biosynthesis, and stress response. The transcriptional changes were superimposed on the metabolic network to identify metabolic units that changed in response to the overexpression of the two oxidases. By using an algorithm that detects metabolic modules based on biologically significant changes in gene expression (25), “reporter metabolites” were identified around which significant coordinated gene expression changes occur. The top reporter metabolites were cytosolic NAD(H) in NOX and mitochondrial NAD(H) in AOX (Table 3). Additionally, several key metabolites in glycerol synthesis, fatty acid metabolism, and amino acid transport were also significantly affected in NOX. For alternative oxidase overexpression, several metabolites belonging to mitochondrial processes such as the TCA cycle, respiration, and acetaldehyde metabolism were affected (Table 3). Differences in the localization of the reporter metabolites clearly indicate compartment-specific functionality of the two oxidases: NADH oxidase predominantly affects the cytosol, and alternative oxidase is active in the mitochondria.
Table 3.
Reporter metabolites around which most significant gene expression changes occurred in response to overexpression of NADH oxidase or alternative oxidase in S. cerevisiae
| Reporter metabolites | P value* |
|---|---|
| In response to NADH oxidase | |
| NADH | 5.38E-09 |
| NAD | 2.18E-08 |
| sn-glycerol 3-phosphate | 3.16E-03 |
| 3-phosphonooxypyruvate | 3.98E-03 |
| NADH–mitochondrial | 5.17E-03 |
| Acetaldehyde–mitochondrial | 6.06E-03 |
| 3-phospho-d-glyceroyl phosphate | 9.18E-03 |
| Acetaldehyde | 9.50E-03 |
| In response to alternative oxidase | |
| NADH–mitochondrial | 7.48E-08 |
| NAD–mitochondrial | 6.20E-05 |
| Orotate | 1.37E-03 |
| CoA–mitochondrial | 1.53E-03 |
| Oxaloacetate–mitochondrial | 2.23E-03 |
| Acetaldehyde–mitochondrial | 2.50E-03 |
| Oxygen–mitochondrial | 2.85E-03 |
| 2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinone–mitochondrial | 2.85E-03 |
* Probability that normalized transcription activity around a metabolite is the same as that in the background.
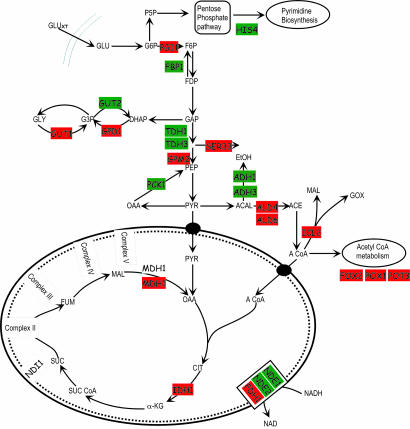
To further analyze the transcription results, the genome-scale model of S. cerevisiae (1) was expressed as an enzyme-interaction graph on which enzymes were represented as nodes, with the common metabolites connecting the nodes. The metabolic modules that had the highest score of interaction (after correcting for background distribution) indicate the metabolic subnetwork in which maximal transcriptional differences occurred. In general, the overexpression of NADH oxidase caused up-regulation of genes whose products catalyze the synthesis of NADH in the cytosol. For example, the induction of genes involved in the synthesis of NADH via the formate dehydrogenase pathway was observed, but significant reduction was noted in the glycerol and ethanol synthesis pathways (Fig. 3). The glycerol assimilation pathway, involving the GUT1 and GUT2 genes, was up-regulated, whereas the synthesis pathway was down-regulated upon overexpression of NADH oxidase. Interestingly, the conversion of acetaldehyde to ethanol was down-regulated, whereas the conversion of acetaldehyde to acetate was stimulated (Fig. 3). Genes in the native pathway for NADH oxidation were severely down-regulated, suggesting higher affinity of bacterial NADH oxidase toward NADH than toward the two native NADH dehydrogenases. In contrast to NADH oxidase, the overexpression of the alternative oxidase had more pronounced effects in the mitochondria (Fig. 4). Several genes in the TCA cycle and in mitochondrial processes were significantly up-regulated. Furthermore, genes involved in ethanol generation from acetaldehyde (ADH1, ADH2, and ADH3) were down-regulated. Alternative oxidase overexpression down-regulated genes in the glyoxylate shunt and fatty acid oxidation pathway. The native NADH oxidation pathway in S. cerevisiae, mediated by the NDI1 gene, was down-regulated in the presence of alternative oxidase. Gluconeogenesis genes were generally down-regulated when either of the oxidases was overexpressed.
Fig. 3.
Analysis of transcription responses of NOX vs. CON superimposed on the metabolic network, as described previously (25). Shown are the key enzymes that were identified in the central metabolism as part of the subnetwork. Genes that were up-regulated are indicated in red; those that were down-regulated are indicated in green.
Fig. 4.
Analysis of transcription responses of AOX vs. CON superimposed on the metabolic network, as described previously (25). Shown are the key enzymes that were identified in the central metabolism as part of the subnetwork. Genes that were up-regulated are indicated in red; those that were down-regulated are indicated in green.
Discussion
To gain insight into the molecular processes underlying the Crabtree effect in S. cerevisiae, we studied the metabolic and transcriptional responses to the overexpression of either NADH oxidase or alternative oxidase. NADH oxidase mediates the nonrespiratory dissipation of NADH, whereas alternative oxidase recruits parts of the respiratory system in the transfer of electrons from NADH to oxygen. In general, physiological differences due to overexpression of either of the oxidases were more prominent at Dcrit, because at lower dilution rates (0.1 h−1) there is no overflow and presumably no “excess” NADH. The reduction in biomass yield as a result of overexpression of either of the oxidases is likely due to increased energy dissipation. Neither of the enzymes is coupled with proton translocation, and each therefore dissipates free energy captured in NADH which might otherwise have been available for oxidative phosphorylation. Neither NADH oxidase nor alternative oxidase was repressed by glucose, as indicated by the high activity for NOX and AOX in the presence of excess glucose (Fig. 2). Given that native S. cerevisiae genes involved in respiration are repressed by glucose (26), these two heterologous oxidases offer a potential solution to maintaining high respiratory capacity even in the presence of high glucose concentrations.
NADH oxidase primarily impacted glycerol production, whereas alternative oxidase affected ethanol production. Normally, cytosolic NADH is reoxidized by two external (cytosolic) NADH dehydrogenases, (15, 16), and when glycolytic NADH generation surpasses the rate at which these dehydrogenases can act, S. cerevisiae activates the glycerol synthesis pathway as another outlet for NADH consumption (24, 27). The reduced glycerol generation in NOX in batch (Table 1) and continuous (Table 2) cultures, as well as the reduced G3PDH activity (Fig. 1), demonstrates that NADH oxidase relieves the need to activate the glycerol pathway. Moreover, the substantially reduced glycerol generation in ΔΔΔ-NOX illustrates that bacterial NADH oxidase could functionally replace the native NADH dehydrogenases and is predominantly localized in the cytosol in S. cerevisiae. The cytosolic localization of NADH oxidase was further confirmed by identifying cytosolic NAD(H) as the top reporter metabolite around which a majority of the transcription changes occurred (Table 4). Furthermore, transcription analysis revealed that GUT1 and GPD1 were up-regulated and GUT2 was down-regulated in NOX relative to CON, suggesting a shift toward glycerol catabolism, compared with glycerol production in CON (Fig. 3).
Table 4.
List of plasmids and strains used in this study
| Plasmid/strain | Genotype | Source |
|---|---|---|
| Plasmid | ||
| pYX212 | 2μ, TPI promoter, AMPR | R & D Systems |
| pYX212-NOX | pYX212 with nox from Str. pneumoniae | This study |
| pYX212-AOX | pYX212 with AOX1 from H. capsulatum | This study |
| Strain | ||
| CEN.PK113–7D | MATa URA3 HIS3 LEU2 TRP1 MAL2-8 SUC2 | P. Kötter* |
| CEN.PK113–5D | CEN.PK113-7D ura3-52 | P. Kötter* |
| CEN.PK398–12B | CEN.PK113-5D nde1(41–1659)::loxP-kanMX4-loxP nde2(51–100)::loxP-kanMX4-loxP gut2(41–2010)::loxP-kanMX4-loxP | P. Kötter* |
| CON | CEN.PK113-5D/pYX212 | This study |
| NOX | CEN.PK113-5D/pYX212-NOX | This study |
| AOX | CEN.PK113-5D/pYX212-AOX | This study |
| ΔΔΔ | CEN.PK398-12B/pYX212 | This study |
| ΔΔΔ-NOX | CEN.PK398-12B/pYX212-NOX | This study |
*Institut für Mikrobiologie der Johann Wolfgang Goethe Universität, Frankfurt, Germany.
Overexpression of alternative oxidase reduced ethanol generation. Transcription analysis revealed that the overexpression of alternative oxidase up-regulated almost every step of the TCA cycle (Fig. 4). An increase in TCA cycle activity and amino acid biosynthesis was recently reported from proteome data when the alternative oxidase gene from Hansenula anomala (HaAOX1) was overexpressed in S. cerevisiae (28). Moreover, the heterologous alternative oxidases from plants expressed in various yeasts were directed to the mitochondria (21, 29). That the alternative oxidase is present in the mitochondria is illustrated by the identification of mitochondrial NAD(H) and quinones as the top reporter metabolites, as well as several other metabolites of mitochondrial origin around which coordinated transcriptional changes occurred (Table 3). The coordinated transcriptional activity is reflected in the significant transcriptional changes in metabolic subnetworks in the mitochondria in AOX (Fig. 4).
Aerobic ethanol formation in S. cerevisiae is the result of a limitation in electron transport from NADH to oxygen (9) and is accompanied by reduced activity of glycolytic enzymes phosphoglycerate kinase, enolase, and triosephosphate isomerase (7) and ethanol-assimilating enzymes such as NAD-dependent alcohol dehydrogenase NAD-acetaldehyde dehydrogenase (8), as well as increased activity of pyruvate decarboxylase (7). The presence of alternative oxidase increased respiratory capacity by elevating the rate of NADH oxidation and rCO2 and thereby facilitating greater coupling between glucose oxidation and respiratory pathways. The 2-fold increase at Dcrit in the activity of NADH-generating ICDH (Fig. 1), and the up-regulation of several TCA cycle genes (Fig. 4), demonstrate a greater capacity of the TCA cycle in AOX than with CON. In the absence of alternative oxidase (CON), the activity of ICDH decreased as the specific growth rate approached Dcrit, an effect likely due to inhibition caused by an increasing NADH/NAD ratio, as observed previously for Yarrowia lipolitica (30). Allosteric inhibition of pyruvate dehydrogenase (31, 32) and of other key TCA cycle enzymes (ICDH, α-ketoglutarate dehydrogenase, and malate dehydrogenase) by NADH (or by the NADH/NAD ratio) restricts the entry of glycolytic carbon from pyruvate into the mitochondria. Under these circumstances, carbon is shunted to acetaldehyde, and subsequently to ethanol, by coordinated action of pyruvate decarboxylase and NAD-dependent aldehyde dehydrogenases, routes that avoid additional NADH accumulation. AOX provided an additional NADH-ubiquinol sink, relieving this restriction in the TCA cycle. The resulting increased capacity of the TCA cycle permitted more glycolytic carbon to enter the TCA cycle and reduced the diversion of carbon to ethanol. Thus, our results clearly show that the onset of overflow metabolism toward ethanol is due to a limited capacity of the respiratory system involved in oxidation of mitochondrial NADH.
Previously it was shown that fusing Hxt7 (a high-affinity hexose transporter) with Hxt1 (a low-affinity hexose transporter) reduced aerobic ethanol generation by modulating rS (33). This fusion not only resulted in decreased rS, but also decreased the specific growth rate. In the present study, we demonstrated that by engineering the redox balance of the cytosol and the mitochondria independently through NADH oxidase and alternative oxidase, it is possible to the control byproduct formation without sacrificing the rate of glucose consumption or specific growth rate. Increased nonrespiratory assimilation of NADH in the cytosol by NADH oxidase leads to reduction in glycerol production. Increased alternative oxidase in the mitochondria reduced ethanol production. The introduction of these two pathways in S. cerevisiae therefore suggests viable metabolic engineering strategies to modulate aerobic byproduct formation.
Materials and Methods
Yeast Strains and Plasmid Construction.
The strains and plasmids used in this study are listed in Table 4. The NADH oxidase gene (nox) from Str. pneumoniae was PCR-amplified with pPANOX7 as template (M.-C. Trombe, Université Paul Sabatier, Toulouse, France) by using the Expand High Fidelity PCR system (Roche Applied Science, Indianapolis, IN). The primers were designed based on the published Str. pneumoniae gene sequence (GenBank Accession no. AF014458) (19) and contained a BamHI site (underlined) in the forward primer, 5′-TACGGATCCAGGAGGAACAGCTATGAGTAAAATCGTTGTAGTCGGTGC-3′ (the ATG start site is in bold) and a SalI site (underlined) in the reverse primer, 5′-ACGGGTCGACTTATTTTTCAGCCGTAAGGGCAGCCA-3′. Similarly, the primers for the H. capsulatum alternative oxidase gene (AOX1) and its flanking regions were designed based on the published gene sequence (GenBank Accession no. AF133236) (20) and contained a NcoI site (underlined) in the forward primer, 5′-ATCGCCCCATGGTCAGCACTGCCATTACTAATACACCTCACTTCC-3′ and a SacI site (underlined) in the reverse primer, 5′-TACTCGGAGCTCGTTTTGTTTAAGCTGATGCAATTTTTT-GCCG-3′. The plasmid pAOX_3_1_1 was used as the template (20). The 1.4-kb nox fragment and the 1.7-kb AOX1 fragment with its flanking regions were gel-isolated, digested with the appropriate restriction enzymes, and ligated into the pYX212 plasmid to construct pYX212-NOX and pYX212-AOX, respectively. These three plasmids were transformed into the host strain, CEN.PK113-5D, and the yeast strains containing these plasmids are designated CON, NOX, and AOX, respectively (Table 4). The pYX212 and pYX212-NOX plasmids were also transformed into CEN.PK398-12B, and the resulting strains are designated ΔΔΔ and ΔΔΔ-NOX (Table 4).
Media and Growth Conditions.
The three strains (CON, NOX, and AOX) were maintained on agar plates made from synthetic complex medium (standard dropout medium) lacking uracil (SC-Ura). The mineral salts medium for batch and chemostat cultivations was prepared as described in ref. 34. For glucose-limited chemostat cultivations, 10 g/liter was fed. The medium for nitrogen-limited chemostats had 1.5 g/liter (NH4)2SO4 supplemented with 5.3 g/liter K2SO4. Glucose concentration was adjusted so that its concentration in effluent was ≈15 g/liter. Aerobic batch cultivations of 4 liter were carried out as described earlier (17, 18) with 40 g/liter initial glucose. At least two independent chemostat cultivations of 1 liter at the desired dilution (specific growth) rate were operated as described in refs. 17 and 18, and steady state was achieved when seven volume changes occurred since the last perturbation in conditions, and the CO2 evolution rate, O2 consumption rate, and biomass concentration remained constant during at least two volume changes (±3%). The critical dilution rate (Dcrit, the specific growth rate at which ethanol formation commences) was determined by using a glucose-limited chemostat with an online ethanol sensor and was operated as a productostat (18). This sensor (Figaro TGS 822; Hammer Electronic, Elsinore, Denmark) detected ethanol in the off-gas and signaled the controller to adjust the nutrient feed rate so that the effective dilution rate was maintained close to Dcrit (35). Concentrations of residual glucose and any products were measured with HPLC (18).
Enzyme Activity.
Enzyme activity was measured by extracting 10 ml of culture into 35 ml of ice, immediately centrifuging (4,000 × g at 1°C for 1 min), and storing the pellet at −80°C. For analysis, the pellet was thawed on ice, and cell-free extracts were prepared by lysing cells (Fastprep FP120; Savant Instruments, Farmingdale, NY) (36). After centrifugation (20,000 × g at 0°C for 20 min), the supernatant was used to determine activity. Total NADH oxidase activity was assayed spectrophometrically at 25°C in 50 mM potassium phosphate buffer (pH 7.0), 0.29 mM NADH, and 0.3 mM EDTA (37). A unit of activity was the quantity that catalyzed the oxidation of 1 μmol of NADH per min. Activities were also determined for NADH-dependent G3PDH (38), NADH-dependent ADH (8), and NAD-dependent ICDH (39). Protein was quantified by the Bradford method using BSA as a standard.
Quantification of Intracellular NADH/NAD.
Metabolism was rapidly quenched by extracting two 10-ml aliquots into 35 ml of methanol, prechilled in a dry ice/ethanol bath. Cells were centrifuged (4,000 × g at −20°C for 1 min) and immediately resuspended in 0.25 ml of 0.2 M HCl (for NAD) or 0.2 M NaOH (for NADH). These suspensions were boiled for 1 min, and cell debris was removed by centrifugation (5,000 × g for 5 min). The cycling assay (40) containing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), phenazine sulfate (PES), yeast ADHII, and ethanol was used to determine the concentrations.
Transcription Analysis.
Global gene expression at Dcrit was measured by growing each strain in duplicate to a steady state slightly below its respective Dcrit and extracting three 10-ml aliquots into ice. After immediate centrifugation (4,000 × g at 0°C for 1 min), the pellet was frozen in liquid nitrogen and stored at −80°C. The extraction of mRNA, cDNA synthesis, cRNA synthesis, and labeling, as well as array hybridization to Affymetrix (Santa Clara, CA) Yeast Genome 2.0 arrays were performed as described in ref. 41. Washing and staining of arrays were performed by using the GeneChip Fluidics Station 400 (Affymetrix) and arrays were scanned with the Affymetrix GeneArray scanner. Data acquisition and quantification of array images, and preliminary data analysis, were performed using Microarray Suite version 4.0.1 (Affymetrix).
Identification of Redox-Sensitive Metabolic Modules.
Arrays were globally scaled to a target value of 500 by using the average signal from all gene features. The microarrays contain probe sets representing 9,335 distinct transcription features. After excluding all probe sets not assigned yORF abbreviations, as identified in the Saccharomyces Genome Database (www.yeastgenome.org), and all probe sets representing groups of genes, already represented as singletons, 5,650 probe sets remained. Student's t test was used to identify genes that changed significantly between CON, NOX, and AOX. Gene expression changes were mapped on the genome-scale metabolic model of S. cerevisiae (1) to identify metabolic hubs, based on transcriptional regulation (25).
Acknowledgments
We thank Jérôme Maury, Jette Mortensen, Tina Johanssen, and Martin Nielsen for technical assistance with the research; Peter Kötter and Clayton Johnson for kindly providing the yeast deletion mutants and the AOX1 cDNA, respectively; and Jochen Förster and Thomas Grotkjær for helpful comments on the manuscript. G.N.V. acknowledges a fellowship (DE-FG36-01ID14007) from the U.S. Department of Energy.
Abbreviations
- ADH
alcohol dehydrogenase
- G3PDH
glycerol-3-phosphate dehydrogenase
- ICDH
isocitrate dehydrogenase
- TCA
tricarboxylic acid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE6267).
References
- 1.Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen J. J Bacteriol. 2003;185:7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Jagow G, Klingenberg M. Eur J Biochem. 1970;12:583–592. doi: 10.1111/j.1432-1033.1970.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 4.Luttik MA, Overkamp KM, Kötter P, de Vries S, van Dijken JP, Pronk JT. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 5.Larsson C, Pahlman IL, Ansell R, Rigoulet M, Adler L, Gustafsson L. Yeast. 1998;14:347–357. doi: 10.1002/(SICI)1097-0061(19980315)14:4<347::AID-YEA226>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Marres CA, de Vries S, Grivell LA. Eur J Biochem. 1991;195:857–862. doi: 10.1111/j.1432-1033.1991.tb15775.x. [DOI] [PubMed] [Google Scholar]
- 7.van Hoek P, van Dijken JP, Pronk JT. Appl Environ Microbiol. 1998;64:4226–4233. doi: 10.1128/aem.64.11.4226-4233.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postma E, Verduyn C, Scheffers WA, van Dijken JP. Appl Environ Microbiol. 1989;55:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Deken RH. J Gen Microbiol. 1966;44:149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- 10.Fiechter A, Fuhrmann GF, Kappeli O. Adv Microb Physiol. 1981;22:123–183. doi: 10.1016/s0065-2911(08)60327-6. [DOI] [PubMed] [Google Scholar]
- 11.Rieger M, Kappeli O, Fiechter A. J Gen Microbiol. 1983;129:653–661. [Google Scholar]
- 12.Gancedo JM. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kappeli O. Adv Microb Physiol. 1986;28:181–209. doi: 10.1016/s0065-2911(08)60239-8. [DOI] [PubMed] [Google Scholar]
- 14.Sonnleitner B, Kappeli O. Biotechnol Bioeng. 1986;28:927–937. doi: 10.1002/bit.260280620. [DOI] [PubMed] [Google Scholar]
- 15.Rigoulet M, Aguilaniu H, Averet N, Bunoust O, Camougrand N, Grandier-Vazeille X, Larsson C, Pahlman IL, Manon S, Gustafsson L. Mol Cell Biochem. 2004;256–257:73–81. doi: 10.1023/b:mcbi.0000009888.79484.fd. [DOI] [PubMed] [Google Scholar]
- 16.Bakker BM, Overkamp KM, van Maris AJ, Kötter P, Luttik MA, van Dijken JP, Pronk JT. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 17.Moreira dos Santos M, Thygesen G, Kötter P, Olsson L, Nielsen J. FEMS Yeast Res. 2003;4:59–68. doi: 10.1016/S1567-1356(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 18.Moreira dos Santos M, Raghevendran V, Kötter P, Olsson L, Nielsen J. Metab Eng. 2004;6:352–363. doi: 10.1016/j.ymben.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi AD, Le Thomas I, Garel JR, Paton JC, Trombe MC. Mol Microbiol. 1999;34:1018–1028. doi: 10.1046/j.1365-2958.1999.01663.x. [DOI] [PubMed] [Google Scholar]
- 20.Johnson CH, Prigge JT, Warren AD, McEwen JE. Yeast. 2003;20:381–388. doi: 10.1002/yea.968. [DOI] [PubMed] [Google Scholar]
- 21.Vanlerberghe GC, McIntosh L. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- 22.Maresca B, Lambowitz AM, Kobayashi GS, Medoff G. J Bacteriol. 1979;138:647–649. doi: 10.1128/jb.138.2.647-649.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berthold DA, Andersson ME, Nordlund P. Biochim Biophys Acta. 2000;1460:241–254. doi: 10.1016/s0005-2728(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 24.Overkamp KM, Bakker BM, Kötter P, van Tuijl A, de Vries S, van Dijken JP, Pronk JT. J Bacteriol. 2000;182:2823–2830. doi: 10.1128/jb.182.10.2823-2830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patil KR, Nielsen J. Proc Natl Acad Sci USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson M. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 27.Pahlman IL, Gustafsson L, Rigoulet M, Larsson C. Yeast. 2001;18:611–620. doi: 10.1002/yea.709. [DOI] [PubMed] [Google Scholar]
- 28.Mathy G, Navet R, Gerkens P, Leprince P, De Pauw E, Sluse-Goffart CM, Sluse FE, Douette P. J Proteome Res. 2006;5:339–348. doi: 10.1021/pr050346e. [DOI] [PubMed] [Google Scholar]
- 29.Affourtit C, Albury MS, Crichton PG, Moore AL. FEBS Lett. 2002;510:121–126. doi: 10.1016/s0014-5793(01)03261-6. [DOI] [PubMed] [Google Scholar]
- 30.Morgunov IG, Solodovnikova NY, Sharyshev AA, Kamzolova SV, Finogenova TV. Biochemistry (Mosc) 2004;69:1391–1398. doi: 10.1007/s10541-005-0086-3. [DOI] [PubMed] [Google Scholar]
- 31.Wais U, Gillmann U, Ullrich J. Hoppe-Seylers Z Physiol Chem. 1973;354:1378–1388. doi: 10.1515/bchm2.1973.354.2.1378. [DOI] [PubMed] [Google Scholar]
- 32.Keha EE, Ronft H, Kresze GB. FEBS Lett. 1982;145:289–292. doi: 10.1016/0014-5793(82)80185-3. [DOI] [PubMed] [Google Scholar]
- 33.Otterstedt K, Larsson C, Bill RM, Stahlberg A, Boles E, Hohmann S, Gustafsson L. EMBO Rep. 2004;5:532–537. doi: 10.1038/sj.embor.7400132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verduyn C, Postma E, Scheffers WA, van Dijken JP. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 35.Lei F, Olsson L, Jorgensen SB. Biotechnol Bioeng. 2003;82:766–777. doi: 10.1002/bit.10624. [DOI] [PubMed] [Google Scholar]
- 36.Moller K, Bro C, Piskur J, Nielsen J, Olsson L. FEMS Yeast Res. 2002;2:233–244. doi: 10.1111/j.1567-1364.2002.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 37.Lopez de Felipe F, Kleerebezem M, de Vos WM, Hugenholtz J. J Bacteriol. 1998;180:3804–3808. doi: 10.1128/jb.180.15.3804-3808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valadi A, Granath K, Gustafsson L, Adler L. J Biol Chem. 2004;279:39677–39685. doi: 10.1074/jbc.M403310200. [DOI] [PubMed] [Google Scholar]
- 39.Lin AP, McAlister-Henn L. J Biol Chem. 2002;277:22475–22483. doi: 10.1074/jbc.M202534200. [DOI] [PubMed] [Google Scholar]
- 40.Bernofsky C, Swan M. Anal Biochem. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- 41.Affymetrix. Affymetrix GeneChip Expression Analysis Technical Manual. Santa Clara, CA: Affymetrix; 2000. [Google Scholar]