Abstract
In mammals, growth hormone-releasing hormone (GHRH) is the most important neuroendocrine factor that stimulates the release of growth hormone (GH) from the anterior pituitary. In nonmammalian vertebrates, however, the previously named GHRH-like peptides were unable to demonstrate robust GH-releasing activities. In this article, we provide evidence that these GHRH-like peptides are homologues of mammalian PACAP-related peptides (PRP). Instead, GHRH peptides encoded in cDNAs isolated from goldfish, zebrafish, and African clawed frog were identified. Moreover, receptors specific for these GHRHs were characterized from goldfish and zebrafish. These GHRHs and GHRH receptors (GHRH-Rs) are phylogenetically and structurally more similar to their mammalian counterparts than the previously named GHRH-like peptides and GHRH-like receptors. Information regarding their chromosomal locations and organization of neighboring genes confirmed that they share the same origins as the mammalian genes. Functionally, the goldfish GHRH dose-dependently activates cAMP production in receptor-transfected CHO cells as well as GH release from goldfish pituitary cells. Tissue distribution studies showed that the goldfish GHRH is expressed almost exclusively in the brain, whereas the goldfish GHRH-R is actively expressed in brain and pituitary. Taken together, these results provide evidence for a previously uncharacterized GHRH-GHRH-R axis in nonmammalian vertebrates. Based on these data, a comprehensive evolutionary scheme for GHRH, PRP-PACAP, and PHI-VIP genes in relation to three rounds of genome duplication early on in vertebrate evolution is proposed. These GHRHs, also found in flounder, Fugu, medaka, stickleback, Tetraodon, and rainbow trout, provide research directions regarding the neuroendocrine control of growth in vertebrates.
Keywords: molecular evolution, genome duplication, pituitary adenylate cyclase-activating polypeptide
Growth hormone-releasing hormone (GHRH), also known as growth hormone-releasing factor, was initially isolated from pancreatic tumors causing acromegaly (1, 2), and hypothalamic human GHRH was shown to be identical to the one isolated from the pancreas tumor (3). Thereafter, the sequences of GHRHs were determined in various vertebrate species and in protochordates (4). In mammals, GHRH is mainly expressed and released from the arcuate nucleus of the hypothalamus (5). The primary function of GHRH is to stimulate GH synthesis and secretion from anterior pituitary somatotrophs via specific interaction with its receptor, GHRH-R (5). In addition, GHRH activates cell proliferation, cell differentiation, and growth of somatotrophs (6–8). There are many other reported activities of GHRH such as modulation of appetite and feeding behavior, regulation of sleeping (9, 10), control of jejunal motility (11), and increase of leptin levels in modest obesity (12).
In mammals, GHRH and pituitary adenylate cyclase-activating polypeptide (PACAP) are encoded by separate genes: GHRH is encoded with a C-peptide with no known function, whereas PACAP and PACAP-related peptide (PRP) are present in the same transcript (13, 14). In nonmammalian vertebrates and protochordates, GHRH and PACAP were believed to be encoded by the same gene and hence processed from the same transcript and prepropolypeptide (15). It was suggested that mammalian GHRH was evolved as a consequence of a gene duplication event of the PACAP gene which occurred just before the divergence that gave rise to the mammalian lineage (4, 16). Before this gene duplication event, PACAP was the true physiological regulator for GH release while its function was progressively taken over by the GHRH-like peptides encoded with PACAP after the emergence of tetrapods (16). The GHRH-like peptide was later evolved to PRP in mammals, and the second copy of the ancestral GHRH-like/PACAP gene, after gene duplication, became the physiological GHRH in mammals (16). This hypothetical evolutionary scheme of GHRH and PACAP genes previously proposed could successfully accommodate and explain most of the information available. However, by data mining of the genomic sequences from several vertebrates, genomic sequences encoding for putative GHRHs and GHRH-Rs from amphibian (Xenopus laevis) and teleost (zebrafish) were found. The corresponding proteins share a higher degree of sequence identity with mammalian GHRH and GHRH-R, when compared with the previously identified GHRH-like peptides and receptors. In this report, by analyzing the phylogeny, chromosomal location, and function of these ligands and receptors, a previously uncharacterized evolutionary scheme for GHRH, PRP-PACAP, and PHI-vasoactive intestinal polypeptide (VIP) genes in vertebrates is proposed. More importantly, the discovery of mammalian GHRH homologues by in silico analysis in other fish provides new research directions regarding the neuroendocrine control of growth in vertebrates.
Results
Predicted Amino Acid Sequences of Fish and Amphibian GHRHs.
Recently, several nonmammalian vertebrate genome databases of avian [Gallus gallus (chicken)], amphibian [Xenopus tropicalis (African clawed frog)], and fish [Danio rerio (zebrafish), Takifugu rubripes (Fugu), and Tetraodon nigroviridis (pufferfish)] species were completely or partially released. Using these valuable resources, we performed bioinformatics analyses to look for previously uncharacterized GHRHs and GHRH-Rs in amphibian and fish. Putative genes for GHRH were predicted from X. tropicalis, goldfish (gfGHRH), and zebrafish (zfGHRH), and with the help of these sequences, we successfully cloned their full-length cDNAs. [supporting information (SI) Figs. 6–8]. By aligning all known vertebrate GHRH precursor sequences (SI Fig. 9), it was found that only the N-terminal region (1–27) of GHRHs is conserved. Human shares 81.5% and 74.1% sequence identity with goldfish/zebrafish and X. laevis GHRHs, respectively (SI Fig. 10). Within the first 7 aa, there is only one amino acid substitution in goat (position 1), human (position 1), mouse (position 1), and Xenopus (position 2). These observations clearly show that a strong selective pressure has acted to preserve the GHRH sequence in evolution, indicating that this peptide plays important functions from fish to mammals. It should be noted that within the first 27 aa, the previously named GHRH-like peptides share much lower level of sequence identity with these fish and mammalian GHRHs (51.9–59.3% for gfPRPsalmon, and 37–44.4% for gfPRPcatfish; SI Fig. 10). Instead, these GHRH-like peptides are structurally homologous to mammalian PRPs (Fig. 1A).
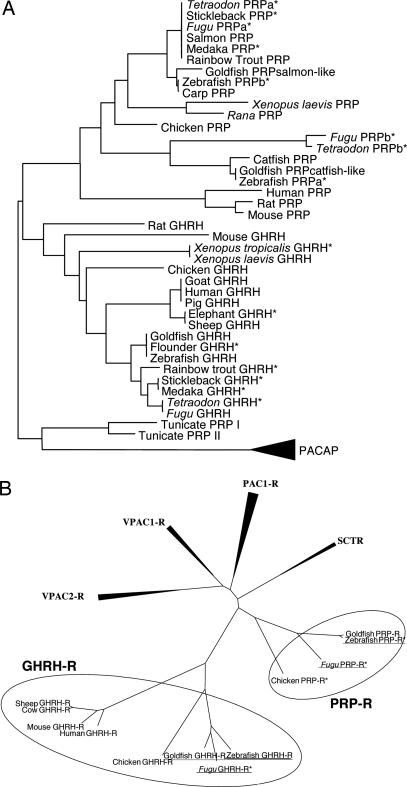
Fig. 1.
Phylogenetic analysis of GHRH, PRP, and GHRH-like peptide (1–27), GHRH-Rs, and PRP-Rs. Predicted peptide and receptor sequences are marked with asterisks. (A) PACAP sequences were used as an out-group. (B) Receptors identified in this report are underlined.
Cloning of GHRH-Rs from Zebrafish and Goldfish.
Putative GHRH-R cDNAs were cloned from zebrafish and goldfish (zfGHRH-R and gfGHRH-R) (see SI Figs. 11 and 12). The amino acid sequences of these GHRH-R receptors share 78.4% identity among themselves, and 51.1% and 49.7%, respectively, with the human GHRH-R (SI Fig. 13). Kyte-Doolittle hydrophobicity plots of these receptors indicated the presence of seven hydrophobic transmembrane regions that are conserved in all class IIB receptors (SI Fig. 14). In addition, the eight cysteine residues in the N termini of fish and human GHRH-Rs, which presumably structurally maintain the ligand binding pocket (17), are identical. These receptors also contain the basic motifs in the third endoloop, indicating that they can also couple to the cAMP pathway, similar to other members in the same PACAP/glucagon receptor family (18).
A comparison of the fish GHRH-R with other class IIB receptors (SI Fig. 13) clearly shows that fish GHRH-Rs, sharing 64.8–78.4% sequence identity, are distinct from the previously identified GHRH-like receptors (PRP-Rs). The sequence identity between goldfish GHRH-R and PRP-R is 39.7%, which is not different (36.4–38.3%) from other goldfish receptors in the same gene family, including PAC1-R, VPAC1-R, and PHI-R. The sequence identity shared by goldfish, avian, and mammalian GHRH-Rs is 48.9–51.1%, which is significantly higher than any other receptors in the same class IIB family, whether in goldfish or in human.
Phylogenetic Analysis of the Novel GHRH and GHRH-Rs.
Based on the previously uncharacterized GHRH sequences, a revised phylogenetic tree for GHRH, GHRH-like, and PRP peptides was constructed by using PACAP as the out-group (Fig. 1A). The identified GHRH peptides from fish and amphibian were grouped together with all other previously cloned or predicted GHRH sequences from mammals to fish. Interestingly, the previously named GHRH-like peptides were phylogenetically closer to mammalian PRPs and resembled to form a distinct branch together. Within the GHRH grouping, GHRHs clustered together according to their class with the exception of rodents. Within all PRPs, avian, amphibian, and fish PRPsalmon-like (formerly GHRHsalmon-like) clustered to form a sub-branch, whereas, the fish PRPcatfish-like peptides formed a different group. Apparently, there are two forms of PRPs in fish, but only the salmon-like PRPs are observed in avian and amphibian species. Catfish-like PRP, therefore, is uniquely present in fish and hence must have evolved by gene duplication after the split that gave rise to tetrapod and ray-finned fish.
Similarly, the discovered gfGHRH-R and zfGHRH-R, and the predicted GHRH-R from chicken and Fugu, branched together with mammalian GHRH-Rs, whereas the fish PRP-Rs (formerly GHRH-like receptor) formed a separate branch (Fig. 1B). These data confirmed that, together with GHRHs, receptors that are structurally closely related to mammalian GHRH-Rs also exist in other classes. It is interestingly to note that PRP-R might have been lost in mammals (see below). As the function of PRP in nonmammalian vertebrates is not known, the implications of losing PRP-R in mammals is unclear.
Chromosomal Synteny of GHRH and GHRH-R Genes.
According to the latest genome assembly versions, the locations, gene structures, and organizations of neighboring genes of GHRHs and GHRH-Rs in zebrafish (zebrafish assembly Version 4, Zv4) and Xenopus (X. tropicalis 4.1) were determined (SI Fig. 15). All exon-intron splice junctions agree with the canonical GT/AG rule. Both fish and amphibian GHRH genes have four exons and the mature peptides are located in exon 3. In mammals, GHRH genes also have four exons, whereas the mature peptides are found in exon 2 instead of exon 3. Also, exon 2 and its encoded sequences in frog and goldfish are different and are unique in their own species (SI Fig. 9). This information indicated that, although there were frequent exon rearrangements, the mature peptide-encoding exon, in terms of sequence and structure, have been relatively well preserved during evolution. Finally, the GHRH genes are also structurally distinct from PRP-PACAP genes (SI Fig. 15), in which the three exons encoding cryptic peptide, PRP and PACAP are arranged in tandem. Thus, the organizations of GHRH and PRP-PACAP genes also indicate that they have different origins in evolution.
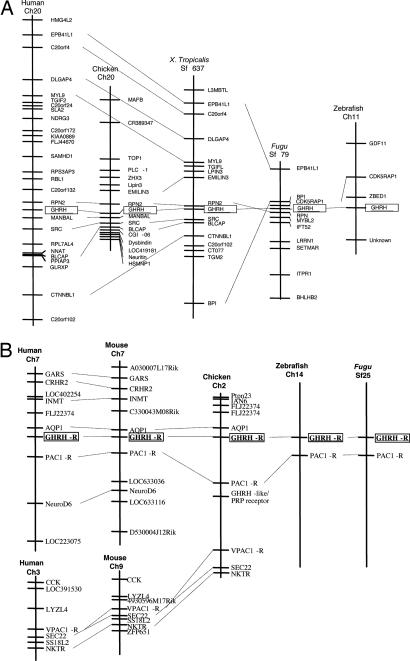
Fig. 2 summarizes the genomic locations of GHRH and GHRH-R in various vertebrates. For the GHRH genes, the nearest neighboring genes of GHRH are RPN2 and MANBAL in human and chicken. In fact, the RPN2 genes are found also in similar locations in all species analyzed except zebrafish (Fig. 2A). Other than RPN2, EPB41L1, C20orf4, DLGAP4, MYL9, and CTNNBL1 are also closely linked to the GHRH gene loci in human and frog. In Fugu and Xenopus, BPI and EPB41L1 genes are both linked to GHRH gene, whereas in Fugu and zebrafish, the CDK5RAP1 gene is found close to the GHRH gene locus. These kinds of similarities in chromosomal syntenic linkage were not observed when comparing the GHRH and PRP-PACAP genes in vertebrate genomes, again confirming the idea that two different genes encoding for GHRH and PACAP exist from fish to mammal.
Fig. 2.
Chromosomal locations of GHRH and GHRH-R in various vertebrate species. Genes adjacent to GHRH and GHRH-R in different genomes are shown. The genes are named according to their annotation in the human genome. GHRH and GHRH-R genes are boxed.
Regarding the receptor, the GHRH-R and PAC1-R gene loci are in close proximity in human, chicken, zebrafish, and Fugu (Fig. 2B), indicating that they are homologues in different species. Interestingly, we found that the PRP-R locus is also linked to PAC1-R gene in chicken. Despite our efforts, no PRP-R gene could be found in mammalian genomes (human, chimpanzee, mouse, rat, and rabbit), and hence it is likely that the PRP-R gene was lost after the divergence that gave rise to the mammalian lineage.
Comparison of Fish GHRH and PRP in Activating GHRH-R and PRP-R.
To confirm the functional identity of fish GHRH and its receptor, zfGHRH-R-transfected CHO cells were exposed to 100 nM of the various related peptides (Fig. 3A). Both fish and frog (1 –27) GHRHs were able to stimulate cAMP accumulation above background levels (no peptide or pcDNA3.1-transfected cells). Graded concentrations of fishGHRH and xGHRH induced dose-dependent stimulation of zfGHRH-R with EC50 values of 3.7 × 10−7 and 8.4 × 10−7 M, respectively, the fish peptide being 2.3 folds more efficacious than xGHRH (Fig. 3B). In contrast, gfPRPsalmon-like and gfPRPcatfish-like did not induce cAMP production in zfGHRH-R-transfected cells. Interestingly, in gfPRP-R-transfected cells, gfPRPsalmon-like, fishGHRH, and xGHRH could all stimulate intracellular cAMP production dose-dependently with EC50 values of gfPRPsalmon-like 5.8 × 10−9M > fishGHRH 1.8 × 10−7M > xGHRH 4.8 × 10−7 M (Fig. 3C).
Fig. 3.
Functional assays. (A) Intracellular cAMP accumulation in CHO-zfGHRH-R cells stimulated with 100 nM peptides: fish GHRH, Xenopus GHRH, goldfish PRPsalmon-like, goldfish PRPcatfish-like, goldfish PHI, goldfish glucagon, zebrafish GIP, human PACAP-27, and human PACAP-38. (B and C) Effect of graded concentrations (from 10−11 to 10−5 M) of various GHRHs and gfPRPs on cAMP accumulation in CHO-zfGHRH-R cells (B) and CHO-gfPRP-R cells (C). Values represent means ± SE from three independent experiments each in duplicate. (D and E) Effect of graded concentrations (from 10−11 to 10−6 M) of fishGHRH (D) and gfPRPsalmon-like (E) on GH release from cultured goldfish pituitary cells. Cells were incubated for 4 h with fishGHRH (D; n = 12) or gfPRPsalmon-like (E; n = 4). Values represent means ± SEM from at least two experiments performed in duplicate.
To test whether fish GHRH and gfPRPsalmon-like can exert a regulatory role at the pituitary level to modulate GH secretion, goldfish pituitary cells were challenged with graded concentrations of goldfish GHRH and PRP. A 4-h incubation of cells with goldfish GHRH induced a dose-related increase in GH release, the minimum effective dose being 10 nM (Fig. 3D) whereas gfPRPsalmon-like, at concentrations up to 1 μM was totally inactive (Fig. 3E).
Tissue Distribution of GHRH and GHRH-R in Goldfish and X. laevis.
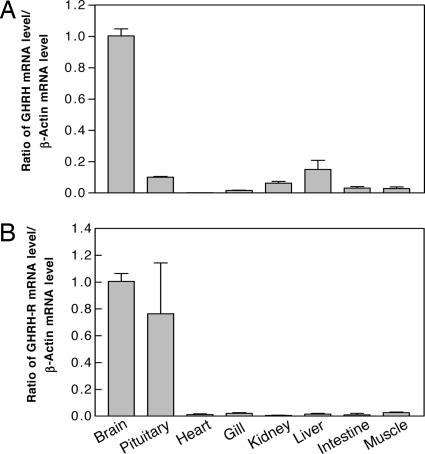
The physiological relevance of GHRH and its cognate receptor in goldfish was tested by measuring their transcript levels using real-time PCR (Fig. 4). GHRH mRNA was detected mostly in the brain, whereas GHRH-R mRNA was found in both brain and pituitary. The expression patterns of these genes in the brain-pituitary axis further support the functional role of the previously uncharacterized GHRH acting to stimulate GH release from the anterior pituitary.
Fig. 4.
Relative abundance of GHRH (A) and GHRH-R (B) transcripts in different tissues in goldfish. A relative abundance of 1 was set arbitrarily for both genes in the brain. Data are from at least three experiments performed in duplicate. Values are expressed as means ± SEM.
Discussion
The Newly Identified GHRHs, but Not GHRH-Like Peptides, Are Mammalian GHRH Homologues.
The primordial gene for the PACAP/VIP/glucagon gene family arose >650 million years ago, and through exon duplication and insertion, gene duplication, point mutation, and exon loss, the family of peptides has developed into the forms that are now recognized. Based on sequence comparison, phylogenetic study and chromosomal locations of GHRH and GHRH-R genes in vertebrates, we here show that the previously named GHRH-like peptides are homologues of mammalian PRPs, and hence should be renamed as PRP from now on. Moreover, the tissue distribution of these PRPs in the fish brain is different when compared with mammalian GHRHs (19). Functionally, PRPs were unable to stimulate zfGHRH-R, as well as incapable of activating GH release from fish pituitary as shown in this and previous studies (20–22).
More importantly, the present report demonstrates the presence of authentic GHRH peptides in nonmammalian vertebrates. In addition to goldfish and zebrafish, several ray-finned fish GHRHs were identified in medaka, strickland, Fugu, Tetraodon, flounder, and rainbow trout (see SI Fig. 16) by in silico analysis. The mature GHRH sequences are almost identical in these eight fish species, with only one substitution in rainbow trout and medaka GHRHs at position 25 (S) and 26 (I), respectively. Moreover, the peptide cleavage sites (R and GKR) are also conserved. This information clearly suggests an important physiological function of GHRH in such diversified fish species and shows that the discovered GHRHs are homologues of mammalian GHRHs. In parallel with the evolution of GHRHs, previously uncharacterized GHRH-Rs that are homologous to their mammalian counterparts in structure, function, chromosomal localization, and tissue distribution are also found in fish. We have therefore shown that, unlike what was hypothesized in previous reports (15, 16) both GHRH and its receptor genes actually existed before the split for tetrapods and ray-finned fish.
Another interesting observation is the existence of PRP-specific receptors from fish (23–25) to chicken, suggesting that PRP potentially is a physiological regulator in these species. However, despite our efforts, we were unable to find the PRP-R gene in any of the mammalian genome databases (in total, 12 databases were analyzed), including the almost completed human and mouse genomes, indicating that PRP-R (but not PRP) was lost in the mammalian lineage. A possible explanation is that PRP-R was lost in a chromosomal translocation event. As shown in Fig. 3B, in chicken, GHRH-R, PAC1-R, PRP-R, and VPAC1-R genes are clustered in the same chromosomal region, whereas, in mammals, VPAC1-R is translocated to a different chromosome and the DNA sequences between VPAC1-R and PAC1-R containing the PRP-R, somehow disappeared.
Implications of the Newly Discovered GHRHs on our Understanding of the Evolution of GHRH and PRP-PACAP Genes.
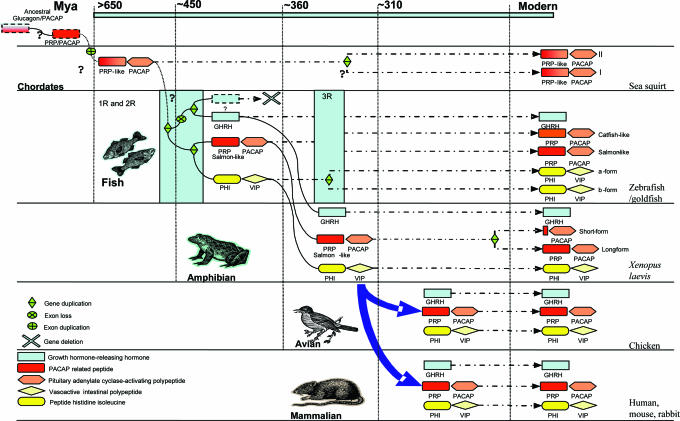
The “one-to-four” rule is the most widely accepted and prevalent model to explain the evolution of many vertebrate genes. This model is based on the “two round genome duplication” hypothesis (26). According to this 2R hypothesis, a first genome duplication occurred as early as in deuterostomes and this was followed by a second round of genome duplication, that took place before the origin of gnathostomes, resulting in having four copies of the genome. Based on this hypothesis and our findings, we propose a scheme for the evolution of GHRH and PRP-PACAP genes in vertebrates (Fig. 5). In the beginning, the ancestral chordates possessed one copy of the PACAP-like gene which encoded a single peptide, and by exon duplication, the PRP-PACAP ancestral gene was formed. As a result of two rounds of genome duplication (1R and 2R), the ancestral gene produced four paralogous genes, but only three of them, PRP-PACAP, PHI-VIP, and GHRH genes, persisted in later lineages. One of the duplicated copies could have been lost via a global process called diploidization (27–29). Although there are two highly conserved PRP-PACAP genes in fish and tunicates, they are produced neither by 1R nor 2R, and this will be discussed later.
Fig. 5.
A proposed evolutionary scheme of GHRH, PRP-PACAP, and PHI-VIP genes with respect to several rounds of genome duplication. Labeling of peptides and gene organizations are shown in the key. The genome duplication events are highlighted by light-blue boxes. Unknown or unclear paths are marked with question marks. Time for divergence in millions of years ago was taken from ref. 41.
The earliest PRP-PACAP genes were identified in protochordates (Chelyosoma productum) (15), in which two PRP-PACAP genes with little differences in gene structure were found. Protochordates are invertebrates that split from vertebrates before the two rounds of genome duplication. Therefore, if these two PRP-PACAP genes existed before 1R and 2R, there should be eight PRP-PACAP paralogues in vertebrates. Also, the tunicate PRP-PACAP genes share very high levels of similarity among themselves in terms of gene structure and peptide sequence. Hence, these duplicated copies of PRP-PACAP genes are the result of a more recent gene duplication event that occurred after the initial divergence. Interestingly, in the phylogenetic tree, the tunicate PRP-like peptide belongs neither to GHRH nor to PRP groupings. The N termini of these peptides are similar to PACAP, whereas the C termini share higher similarity with PRP. Therefore, it is likely that these tunicate PRP-like peptides are evolved from the same ancestral sequences of PRP and/or PACAP.
Based on our model, the evolution of GHRH, PRP-PACAP, and PHI-VIP genes could be explained by using the concept of duplication–degeneration–complementation (30, 31). After the first round of genome duplication (1R), the ancestral PRP-PACAP gene duplicated into two and each of them independently mutated. One of the genes had mutations mainly in the PRP-like region, thus giving rise to the PRP-PACAP gene with the function of the ancestral gene mostly preserved in the PACAP domain. Together with PRP's changes, a duplicated PACAP receptor evolved to interact specifically with PRP. Although we have no information regarding the function of PRP from fish to birds, the presence of a specific receptor that is expressed in a tissue-specific manner strongly suggests that this peptide exerts some physiological role. As the primary function was secured by the conservation of PACAP, in the other copy, PRP is free to mutate to GHRH while PACAP sequences were lost by exon deletion, producing the GHRH gene in vertebrates. After 2R, the duplicated gene copy for PRP-PACAP was free to mutate and hence to acquire new functions to become PHI-VIP, and consistently, PHI and VIP are similar to PRP and PACAP, respectively. Until now, we have not been able to find a second GHRH homologue in vertebrates, and we thus propose that the duplicated GHRH gene may have been deleted or lost in the process of genome rediploidization.
The final question is the origin of the two forms of PRP, salmon-like and catfish-like, and correspondingly two PRP-PACAP genes, in fish. We named these PRPcatfish-like and salmon-like as there are clearly two branches in the phylogenetic tree in which the avian and amphibian PRPs are more similar to fish PRPsalmon-like. Based on previous reports and sequence analysis, we found that, in fact, all bony fish possess a PRPsalmon-like gene, supporting the idea that PRPsalmon-like is the ancestral form, whereas PRPcatfish-like existed before the tetrapod/fish split, a duplicated copy arising from a more recent duplication event that occurred after the appearance of the teleost lineage (32). This is confirmed by the presence of duplicated PHI-VIP and glucagon genes in the Fugu and zebrafish genomes, as well as two genes for PAC1-R, VPAC1-R, and VPAC2-R in fish (33). There is further evidence supporting the hypothesis of a third round of genome duplication; for example, zebrafish possesses seven HOX clusters on seven different chromosomes, instead of four clusters found in mammals (34). Also, there are many duplicated segments in the zebrafish genome where similar duplication are not found in the mammalian genome. This information suggests that some of the genome was duplicated in an ancient teleost. Based on this, we hypothesize that catfish-like and salmon-like PRP encoding genes were produced from the actinopterygian-specific genome duplication at ≈300–450 million years ago.
Materials and Methods
Animals and Peptides.
Zebrafish and goldfish were purchased from local fish markets, and X. laevis was bought from Xenopus I. Peptides including X. laevis GHRH (xGHRH), zebrafish or goldfish GHRH (fish GHRH), goldfish PRPsalmon-like (previously gfGHRHsalmon-like) (24), and goldfish PRPcatfish-like (previously gfGHRHcatfish-like) (24) were synthesized by the Rockefeller University.
Data Mining and Phylogenetic Analysis.
By searching genome databases (Ensembl genome browse and NCBI database) of chicken (Gallus gallus), Xenopus (X. tropicalis), zebrafish (Danio rerio), and Fugu (Fugu rubripes), putative GHRH and GHRH-R sequences were found. These sequences were used for phylogenetic analyses by MEGA3.0 to produce neighbor-joining trees with Dayhoff matrix (35). Predicted protein sequences were then aligned by using ClustalW. Sequence-specific or degenerate primers were designed accordingly for cDNA cloning by PCR amplifications.
Molecular Cloning of GHRHs (X. laevis, Zebrafish, and Goldfish) and GHRH-Rs (Zebrafish and Goldfish).
Total RNAs from brain (zebrafish and goldfish) and pituitary (X. laevis) were extracted according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). RACE (5′ and 3′) was performed by using the 5′ and 3′ RACE amplification kits (Invitrogen). Full-length cDNA clones encompassing the 5′ to 3′ untranslated regions were produced by PCR with specific primers and confirmed by DNA sequencing. Full-length GHRH-R cDNAs were subcloned into pcDNA3.1+ (Invitrogen) for functional expression. Primers used in this study are listed in SI Fig. 17.
Tissue Distribution of GHRH and GHRH-R in Goldfish.
Goldfish GHRH and GHRH-R transcript levels in various tissues were measured by real-time RT-PCR. First-strand cDNAs from various tissues of sexually matured goldfish were prepared by using the oligo(dT) primer (Invitrogen). GHRH and GHRH-R levels were determined by using the SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA) together with specific primers (sequences listed in SI Fig. 18). Fluorescence signals were monitored in real-time by the iCycler iQ system (Bio-Rad, Hercules, CA). The threshold cycle (Ct) is defined as the fractional cycle number at which the fluorescence reaches 10-fold standard deviation of the baseline (from cycle 2 to 10). The ratio change in target gene relative to the β-actin control gene was determined by the 2−ΔΔCt method (36).
Functional Studies of Fish GHRH-R and PRP-R.
For functional studies, zfGHRH-R cDNA in pcDNA3.1 (Invitrogen) was permanently transfected into CHO cells (American Type Culture Collection, Manassas, VA) by using the GeneJuice reagent (Novagen, Darmstadt, Germany) and followed by G418 selection (500 μg/ml) for 3 weeks. Several clones of receptor-transfected CHO cells were produced by dilution into 96-well plates. After RT-PCR to monitor the expression levels of individual clones, the colony with the highest expression was expanded and used for cAMP assays. A gfPRP-R-transfected (previously gfGHRH-R) (25) permanent CHO cell line was also used for comparison in functional expression studies.
To measure cAMP production upon ligand activation, 2 days before stimulation, 2.5 × 105 CHO-zfGHRH-R or CHO-gfPRP-R cells were seeded onto six-well plates (Costar). The assay was performed essentially as described (37) by using the Correlate-EIA immunoassay kits (Assay Design, Ann Arbor, MI).
Measurement of GH Release from Goldfish Pituitary Cells.
Goldfish in late stages of sexual regression were used for the preparation of pituitary cell cultures. Fish were killed by spinosectomy after anesthesia in 0.05% tricane methanesulphonate (Syndel, Vancouver, BC, Canada). Pituitaries were excised, diced into 0.6-mm fragments, and dispersed by using the trypsin/DNA II digestion method (38) with minor modifications (39). Pituitary cells were cultured in 24-well cluster plates at a density of 0.25 × 106 cells per well. After overnight incubation, cells were stimulated for 4 h, and culture medium was harvested for GH measurement by using a RIA previously validated for goldfish GH (40).
Data Analysis.
Data from real-time PCR are shown as the means ± SEM of duplicated assays in at least three independent experiments. All data were analyzed by one-way ANOVA followed by a Dunnett's test in PRISM (Version 3.0; GraphPad, San Diego, CA).
Supplementary Material
Acknowledgments
This work was supported by Hong Kong Government RGC Grants HKU7639/07M, HKU7566/06M, and CRCG10203410 (to B.K.C.C.), and by the Institut National de la Santé et de la Recherche Médicale (H.V.).
Abbreviations
- GH
growth hormone
- GHRH
growth hormone-releasing hormone
- GHRH-R
GHRH receptor
- PACAP
pituitary adenylate cyclase-activating polypeptide
- PAC1-R
PACAP receptor
- PHI
peptide histidine isoleucine
- PRP
PACAP-related peptide
- PRP-R
PACAP-related peptide receptor
- VIP
vasoactive intestinal polypeptide.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ991243–DQ991247 and ABJ55977–ABJ55981).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611008104/DC1.
References
- 1.Guillemin R, Brazeau P, Böhlen P, Esch F, Ling N, Wehrenberg WB. Science. 1982;218:585–587. doi: 10.1126/science.6812220. [DOI] [PubMed] [Google Scholar]
- 2.Thorner MO, Perryman RL, Cronin MJ, Rogol AD, Draznin M, Johanson A, Vale W, Horvath E, Kovacs K. J Clin Invest. 1982;70:965–977. doi: 10.1172/JCI110708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling N, Esch F, Böhlen P, Brazeau P, Wehrenberg WB, Guillemin R. Proc Natl Acad Sci USA. 1984;81:4302–4306. doi: 10.1073/pnas.81.14.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherwood NM, Krueckl SL, McRory JE. Endocr Rev. 2000;21:619–670. doi: 10.1210/edrv.21.6.0414. [DOI] [PubMed] [Google Scholar]
- 5.Bloch B, Brazeau P, Ling N, Bohlen P, Esch F, Wehrenberg WB, Benoit R, Bloom F, Guillemin R. Nature. 1983;301:607–608. doi: 10.1038/301607a0. [DOI] [PubMed] [Google Scholar]
- 6.Billestrup N, Swanson LW, Vale W. Proc Natl Acad Sci USA. 1986;83:6854–6857. doi: 10.1073/pnas.83.18.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayo KE, Hammer RE, Swanson LW, Brinster RL, Rosenfeld MG, Evans RM. Mol Endocrinol. 1988;2:606–612. doi: 10.1210/mend-2-7-606. [DOI] [PubMed] [Google Scholar]
- 8.Lin C, Lin SC, Chang CP, Rosenfeld MG. Nature. 1992;360:765–768. doi: 10.1038/360765a0. [DOI] [PubMed] [Google Scholar]
- 9.Ehlers CL, Reed TK, Henriksen SJ. Neuroendocrinology. 1986;42:467–474. doi: 10.1159/000124489. [DOI] [PubMed] [Google Scholar]
- 10.Obal F, Jr, Alfoldi P, Cady AB, Johannsen L, Sary G, Krueger JM. Am J Physiol. 1988;255:R310–R316. doi: 10.1152/ajpregu.1988.255.2.R310. [DOI] [PubMed] [Google Scholar]
- 11.Bueno L, Fioramonti J, Primi MP. Peptides. 1985;6:403–407. doi: 10.1016/0196-9781(85)90104-4. [DOI] [PubMed] [Google Scholar]
- 12.Cai A, Hyde JF. Endocrinology. 1999;140:3609–3614. doi: 10.1210/endo.140.8.6925. [DOI] [PubMed] [Google Scholar]
- 13.Mayo KE, Cerelli GM, Lebo RV, Bruce BD, Rosenfeld MG, Evans RM. Proc Natl Acad Sci USA. 1985;82:63–67. doi: 10.1073/pnas.82.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosoya M, Kimura C, Ogi K, Ohkubo S, Miyamoto Y, Kugoh H, Shimizu M, Onda H, Oshimura M, Arimura A, et al. Biochim Biophys Acta. 1992;1129:199–206. doi: 10.1016/0167-4781(92)90488-l. [DOI] [PubMed] [Google Scholar]
- 15.McRory J, Sherwood NM. Endocrinology. 1997;138:2380–2390. doi: 10.1210/endo.138.6.5167. [DOI] [PubMed] [Google Scholar]
- 16.Montero M, Yon L, Kikuyama S, Dufour S, Vaudry H. J Mol Endocrinol. 2000;25:157–168. doi: 10.1677/jme.0.0250157. [DOI] [PubMed] [Google Scholar]
- 17.DeAlmeida VI, Mayo KE. Mol Endocrinol. 1998;12:750–765. doi: 10.1210/mend.12.5.0102. [DOI] [PubMed] [Google Scholar]
- 18.Laburthe M, Couvineau A, Gaudin P, Maoret JJ, Rouyer-Fessard C, Nicole P. Ann NY Acad Sci. 1996;805:94–109. doi: 10.1111/j.1749-6632.1996.tb17476.x. [DOI] [PubMed] [Google Scholar]
- 19.McRory JE, Parker DB, Ngamvongchon S, Sherwood NM. Mol Cell Endocrinol. 1995;108:169–177. doi: 10.1016/0303-7207(94)03467-8. [DOI] [PubMed] [Google Scholar]
- 20.Montero M, Yon L, Rousseau K, Arimura A, Fournier A, Dufour S, Vaudry H. Endocrinology. 1998;139:4300–4310. doi: 10.1210/endo.139.10.6239. [DOI] [PubMed] [Google Scholar]
- 21.Parker DB, Power ME, Swanson P, Rivier J, Sherwood NM. Endocrinology. 1997;138:414–423. doi: 10.1210/endo.138.1.4830. [DOI] [PubMed] [Google Scholar]
- 22.Melamed P, Eliahu N, Levavi-Sivan B, Ofir M, Farchi-Pisanty O, Rentier-Delrue F, Smal J, Yaron Z, Naor Z. Gen Comp Endocrinol. 1995;97:13–30. doi: 10.1006/gcen.1995.1002. [DOI] [PubMed] [Google Scholar]
- 23.Chan KW, Yu KL, Rivier J, Chow BK. Neuroendocrinology. 1998;68:44–56. doi: 10.1159/000054349. [DOI] [PubMed] [Google Scholar]
- 24.Wong AO, Li WS, Lee EK, Leung MY, Tse LY, Chow BK, Lin HR, Chang JP. Biochem Cell Biol. 2000;78:329–343. [PubMed] [Google Scholar]
- 25.Kee F, Ng SS, Vaudry H, Pang RT, Lau EH, Chan SM, Chow BK. Gen Comp Endocrinol. 2005;140:41–51. doi: 10.1016/j.ygcen.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Hughes AL. J Mol Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe KH. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 28.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 29.Kellis M, Patterson N, Birren B, Berger B, Lander ES. J Comput Biol. 2004;11:319–355. doi: 10.1089/1066527041410319. [DOI] [PubMed] [Google Scholar]
- 30.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch M, Force A. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van de Peer Y, Taylor JS, Meyer A. J Struct Funct Genomics. 2003;3:65–73. [PubMed] [Google Scholar]
- 33.Cardoso JC, Power DM, Elgar G, Clark MS. J Mol Endocrinol. 2004;33:411–428. doi: 10.1677/jme.1.01575. [DOI] [PubMed] [Google Scholar]
- 34.Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Kwok YY, Chu JY, Vaudry H, Yon L, Anouar Y, Chow BK. Gen Comp Endocrinol. 2006;145:188–196. doi: 10.1016/j.ygcen.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Chang JP, Cook H, Freedman GL, Wiggs AJ, Somoza GM, de Leeuw R, Peter RE. Gen Comp Endocrinol. 1990;77:256–273. doi: 10.1016/0016-6480(90)90310-i. [DOI] [PubMed] [Google Scholar]
- 39.Lee EK, Chan VC, Chang JP, Yunker WK, Wong AO. J Neuroendocrinol. 2000;12:311–322. doi: 10.1046/j.1365-2826.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 40.Marchant TA, Fraser RA, Andrews PC, Peter RE. Regul Pept. 1987;17:41–52. doi: 10.1016/0167-0115(87)90031-0. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Hedges SB. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.