Abstract
The mechanisms of target cell recognition and producer cell self-protection (immunity) are both important yet poorly understood issues in the biology of peptide bacteriocins. In this report, we provide genetic and biochemical evidence that lactococcin A, a permeabilizing peptide–bacteriocin from Lactococcus lactis, uses components of the mannose phosphotransferase system (man-PTS) of susceptible cells as target/receptor. We present experimental evidence that the immunity protein LciA forms a strong complex with the receptor proteins and the bacteriocin, thereby preventing cells from being killed. Importantly, the complex between LciA and the man-PTS components (IIAB, IIC, and IID) appears to involve an on–off type mechanism that allows complex formation only in the presence of bacteriocin; otherwise no complexes were observed between LciA and the receptor proteins. Deletion of the man-PTS operon combined with biochemical studies revealed that the presence of the membrane-located components IIC and IID was sufficient for sensitivity to lactococcin A as well as complex formation with LciA. The cytoplasmic component of the man-PTS, IIAB, was not required for the biological sensitivity or for complex formation. Furthermore, heterologous expression of the lactococcal man-PTS operon rendered the insensitive Lactobacillus sakei susceptible to lactococcin A. We also provide evidence that, not only lactococcin A, but other class II peptide-bacteriocins including lactococcin B and some Listeria-active pediocin-like bacteriocins also target the man-PTS components IIC and IID on susceptible cells and that their immunity proteins involve a mechanism in producer cell self-protection similar to that observed for LciA.
Keywords: antimicrobial peptides, Mannose-PTS, receptor, protein complex, coprecipitation
As the emergence of bacterial antibiotic resistance has become an increasing problem in medical treatments world-wide for decades, there is a pressing need for novel or alternative sources of antimicrobial agents (1, 2). Ribosomally synthesized antimicrobial peptides of bacterial origin, known as bacteriocins, have been regarded as a very promising source of antimicrobials because they are highly potent (being active at nanomolar concentrations) (3, 4, 5). Unlike traditional antibiotics, which mostly act as enzyme inhibitors (2), the majority of peptide–bacteriocins permeabilize the membrane of sensitive cells, leading to leakage of cellular solutes and, eventually, cell death (6, 7). Hence, complementary use of antibiotics together with bacteriocins can potentially be a better approach in preventing emergence of resistant pathogens because it will be much more difficult for a bacterium to acquire resistance to two antimicrobials with different modes of action concurrently.
There exists a wealth of information on different aspects of peptide–bacteriocins, from genetic determinants responsible for production, regulation, and immunity (self-protection) to detailed studies on their three-dimensional structures and modes of action (8–11, 5, 6). With the exception of nisin and a few closely related lantibiotics (6, 12, 13), the mechanisms by which the peptide-bacteriocins specifically recognize their target cells before permeabilization are not well understood. Such knowledge is pivotal for the rational design of novel and more potent antimicrobials and, at least, is required to understand the mechanism behind resistance development. Nisin employs lipid II, a precursor in cell-wall synthesis, as a docking molecule. The subsequent killing of sensitive cells is achieved by a combination of two mechanisms: (i) inhibition of the peptidoglycan biosynthesis through interaction with lipid II and (ii) formation of lethal pores in the cytoplasmic membrane (14). It was proposed that components of the mannose phosphotransferase system (man-PTS) might play a role in target recognition for some class II pediocin-like bacteriocins, because mutants resistant to the pediocin-like bacteriocins displayed reduced expression of the man-PTS (15, 16). The introduction of the Listeria man-PTS genes rendered otherwise insensitive lactococcal cells sensitive to leucocin A, pediocin PA-1, and enterocin A (17). However, whether or not these bacteriocins use the man-PTS components as a receptor per se is not known. In bacteria, the man-PTS is the key pathway for mannose uptake, but it can also accept glucose as a substrate (18). The uptake is coupled with phosphorylation of the incoming carbohydrates through a series of phoshorylation reactions mediated by EI, Hpr, IIAB, IIC, and IID (18). The first two enzymes (EI and Hpr) function in a nonspecific manner as they participate in the initial steps for several different PTSs, whereas the last three proteins (IIAB, IIC, and IID) are dedicated to the man-PTS. IIC and IID together form a membrane-located complex, whereas IIAB resides in the cytoplasm but can also form reversible contacts with a membrane-located part (19).
Lactococcin A is a member of class II bacteriocins, which are nonlantibiotic peptides (8), and its mode of action has been studied in great detail (20). The bacteriocin acts exclusively on lactococci by permeabilizing the cytoplasmic membrane in a proton-motive-force (pmf)-independent manner, causing leakage (both efflux and influx) of solutes across the membrane and, thereby, cell death. Based on studies with liposomes and vesicles from sensitive cells, it has been suggested that a specific membrane receptor is required for sensitivity (21, 22). However, the nature of such a receptor, if any, has remained elusive. In this study, we demonstrate, genetically and by protein–protein interaction analyses, that lactococcin A indeed employs the proteins IIC and IID of the man-PTS as a receptor on target cells and that the cognate immunity protein (LciA) is tightly associated with the bacteriocin–receptor complex to render producer cells immune. We also show that several other class II bacteriocins, including pediocin-like bacteriocins, employ a similar mechanism to target their susceptible cells. A model of the mechanisms underlying receptor recognition and producer-cell immunity for these bacterciocins is presented.
Results
Formation of Protein Complex with the Immunity Protein.
Lactococcin A production depends on the expression of four genes: (i) lcnA, encoding a prelactococcin A, (ii) lciA, encoding a hydrophobic protein that confers immunity to the cognate bacteriocin, (iii and iv) lcnC and lcnD, encoding an ABC-transporter and an accessory protein that, together, form the complete machinery dedicated to maturation (proteolytic removal of the leader peptide) and export of the bacteriocin (20, 23, 24). It has previously been shown that the lactococcal strain IL1403, which is sensitive to lactococcin A, acquires immunity when expressing the cloned gene lciA (20). We reasoned that an immunity protein has to interact with its cognate bacteriocin or/and a receptor on target cells to confer immunity. If true, purification of the immunity protein might allow identification of the proteins/factors that complex with the immunity protein. To facilitate the detection and purification of LciA, an amino acid sequence (MDYKDDDDKL; so-called flag-tag) containing an epitope recognized by the antibody M2, was genetically fused to the N terminus of LciA. The resulting gene, flciA, was expressed alone or in combination with the bacteriocin gene lcnA in Lactococcus lactis Il1403, which expresses an ABC-transport system capable of processing and exporting lactococcin A (25). The clone B268, which expresses only flciA, was found to be immune, whereas B244, which expresses both lcnA and flciA, was both bacteriocin-producing and immune (data not shown). As expected, both clones produced an immunodetectable fLciA that conferred immunity to lactococcin A (Fig. 1A), demonstrating that the N-terminal flag-tag did not impair the immunity function of the fusion protein.
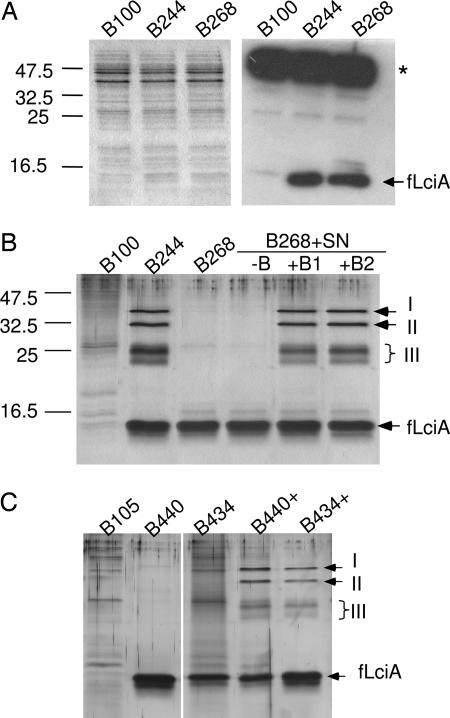
Fig. 1.
Expression of the fusion immunity gene flciA and purification of proteins that form complex with fLciA. (A) Coomassie-stained SDS/PAGE gel (Left) and immunoblot (Right) showing the presence of the fusion protein fLciA in the cell extracts (each 100 μg) of L. lactis Il1403-derived clones: B244 expressing lcnA and flciA and B268 expressing only flciA; B100, containing the empty vector, was used as negative control. (B) Silver-stained SDS/PAGE gel showing fLciA and its copurified proteins from Il1403-derived clones. B100, B244, and B268 were grown in the absence (first four lanes) or presence (last two lanes) of an exogenously added bacteriocin concentrate (final 50 BU/ml in +B1 and 200 BU/ml in +B2). In lane (−B), a nonbacteriocin concentrate from B100 was added as a control. (C) Silver-stained SDS/PAGE gel showing fLciA and its copurified proteins from MG1363-derived clones: B105 containing empty vector (control), B440 expressing lcnA and flciA, and B434 expressing only flciA. Samples were purified from cultures grown in the absence (B105, B440, and B434) or presence (B440+ and B434+) of added lactococcin A (100 BU/ml). Bars signify protein molecule weight markers, arrows signify fLciA and the copurified proteins (I, II, and III). For B and C, equal volumes of the eluted fractions (each 5 μl of total 100 μl) were applied to wells for direct comparison. ∗, a protein(s) that cross-reacted with the antibody M2.
We purified fLciA from cell extracts by immunoprecipitation, and the samples were analyzed by SDS/PAGE. As shown in Fig. 1B, in addition to fLciA, which was found in both flciA-expressing clones, a few additional proteins were copurified with fLciA in the sample derived from B244 (the clone expressing both lcnA and flciA). These copurified proteins were not found in the sample derived from B268, which expressed flciA in the absence of lcnA (Fig. 1B). Two of the copurified proteins gave distinct bands (I and II) in the size range of 32–35 kDa, whereas a group of proteins gave two to three bands (III) in the area close to 25 kDa.
Given that the protein complex formation with fLciA takes place only in cultures producing bacteriocin, we examined whether this phenomenon could be established with the immune and nonproducing clone B268 if exogenous bacteriocin was added to its cultures. The result from this experiment showed the presence of an apparently identical protein complex (Fig. 1B). Thus, purified samples from cultures of B268 that had been grown in the presence of added lactococcin A (as a bacteriocin concentrate), gave rise to the same protein pattern as B244 when analyzed by SDS/PAGE. This pattern could not be established with a concentrate from a nonbacteriocin producer (Fig. 1B). To assess this property further, we used a second L. lactis strain, MG1363, as an expression host. This strain is different from Il1403 in that it lacks an active ABC-transport system specific for export of double-glycine leader-containing bacteriocins (such as lactococcin A). Two derivatives of MG1363, B440 containing the cloned genes lcnA and flciA and B434 containing only flciA, were both found to be immune and bacteriocin-negative (data not shown), and, as expected, the purified fLciA samples from these two clones did not contain the copurified proteins as found with B244 (Fig. 1C). Only when exogenous bacteriocin was added to their cultures, did the corresponding copurified proteins appear in the purified samples of fLciA (Fig. 1C). This is an important piece of evidence indicating that the protein complex formation with fLciA can be achieved only in the presence of mature bacteriocin in the extracellular environments but not with an intracellular and unprocessed prebacteriocin (LcnA).
Components of the Man-PTS Copurify with fLciA.
The copurified protein bands were subsequently isolated from SDS/PAGE gels, trypsinized, and subjected to MS for peptide identification. By using the peptide mass fingerprinting (PMF) data alone, the spectrum representing band I resulted in a single significant (P < 0.05) hit, namely the mannose-specific PTS component IIAB from L. lactis. A total of 11 peptides in the query matched the tryptic map of this protein, representing 43% of the total sequence. In a second round of database searches, in which the PMF data were combined with MS/MS data from selected peptides, single and significant hits were obtained for all three bands. The identified proteins were all components of the L. lactis man-PTS, namely IIAB in band I, IID in band II, and IIC in band III [see supporting information (SI) Fig. 6]. The calculated sizes for IIAB, IIC, and IID from Il1403 are 35,064.5, 27,562.6, and 33,646.7 Da, respectively, which correspond well with their electrophoretic size as shown by SDS/PAGE (Fig. 1) (see SI Methods for discussion on multiple proteins in band III).
Deletion of the Man-PTS Operon (ptn) Confirms Its Involvement as Receptor for Lactococcin A.
The man-PTS operon (ptn) in IL1403 is composed of three genes, which have been annotated as ptnAB, -C and -D (GenBank accession no. AE005176). To examine whether the encoded proteins, IIAB, IIC, and IID, could function as a receptor for lactococcin A, we deleted the ptn operon of the sensitive strain Il1403 by homologous recombination using the targeting plasmid pD1 (Fig. 2A). The removal of the ptn operon in the resulting deletion mutants was confirmed by PCR (Fig. 2A), and it was also reflected in the reduced ability of the mutant (B464) to grow on glucose or mannose, sugars that largely depend on the man-PTS for uptake (see SI Fig. 7). As shown in Fig. 2B and SI Methods, the resulting ptn mutant (B464) was no longer sensitive to the bacteriocin. When the complete ptn operon was reintroduced as cloned genes in the deletion mutant (resulting in B515), sensitivity to the bacteriocin was reestablished, an observation further supporting the notion that the man-PTS components act as a receptor/target for lactococcin A.
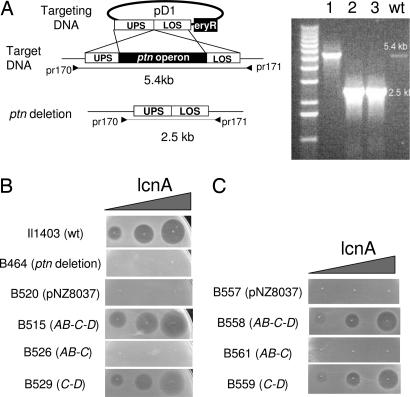
Fig. 2.
Deletion and complementation studies of the man-PTS operon. (A) Deletion of the ptnAB-C-D operon was obtained through a double cross-over event between the chromosome (target DNA) and the targeting plasmid (pD1). pD1 is composed of four modules: upper flanking sequence (UPS; 1kb), lower flanking sequence (LOS; 1.2 kb), the eryR unit for selection, and pCR-TOPO as vector. The double cross-over event was verified by PCR, using the primer pair pr170 and pr171. As shown (Right), the mutant clones (lanes 2, 3) gave rise to a 2.5-kb PCR product, whereas the WT and a revertant (lane 1) yielded the full-length (5.4 kb) product. (B) Sensitivity of the various L. lactis clones to lactococcin A: Il1403 (WT strain), B464 (ptn deletion mutant), B520 (ptn deletion mutant expressing the empty vector), B515, B526, B529, ptn deletion mutants expressing cloned ptnAB-C-D, ptnAB-C, and ptnC-D, respectively. (C) Sensitivity of L. sakei Lb790 derivatives to lactococcin A: B557 expressing the empty vector, B558, B561, and B559, expressing the lactococcal ptnAB-C-D, ptnAB-C, and ptnC-D, respectively. Nisin (5 ng/ml) was used to express cloned genes in lactococcal clones, whereas in Lactobacillus clones, where leakage in gene expression was sufficient for our purpose, nisin was omitted. In B and C, increasing amounts of lactococcin A (0.5, 5, and 50 BU) were directly spotted onto lawns of indicator cells. Growth inhibition by bacteriocin is seen as clear zones.
To identify which component(s) of the man-PTS serve(s) as receptor for lactococcin A, we cloned and expressed the ptn genes in the deletion mutant, either as single genes or as pairs of genes. For this purpose, we used the regulated NICE (Nisin-controlled gene expression) system to fine-tune expression of the ptn genes, because high expression of PTS genes often is toxic to cells. Expression of the single genes did not cause sensitivity to the bacteriocin (data not shown) nor did the expression of the gene pair ptnAB and -C (B526 in Fig. 2B). However, the expression of the gene pair ptnC and -D did establish bacteriocin sensitivity (B529 in Fig. 2B). Taken together, the results strongly suggest that the encoded proteins of ptnC and -D form the receptor for lactococcin A and that ptnAB is not required for this function.
Expression of the Lactococcal Man-PTS Genes in a Lactococcin A-Resistant Lactobacillus sakei Strain Renders It Sensitive.
Like most Lactobacillus strains, Lb790 is not sensitive to lactococcin A, although its genome also contains a man-PTS highly homologous to the lactococcal counterpart. To investigate whether the lactococcal man-PTS could render the Lb. sakei strain sensitive to lactococcin A, the complete ptn operon as well as the gene pairs ptnC and -D, ptnAB and -C, and the individual genes were separately expressed in Lb. sakei Lb790. It was revealed that only the clone expressing the complete ptn operon (B558) or the gene pair ptnC and -D (B559) became sensitive to lactococcin A, whereas any of the other clones did not (Fig. 2C; data for the individual genes are not shown). These results not only demonstrate that the lactococcal man-PTS receptor/target is lactococcin A-specific but also confirm that the expression of the gene pair ptnC and -D is sufficient to form a functional receptor for lactococcin A.
fLciA Can Form Complex with IIC and IID in the Absence of IIAB, and the Bacteriocin Itself Is Part of the Complex.
Given that ptnAB is not required for the lactococcin A-induced sensitivity, we wanted to examine whether IIAB coprecipitated with fLciA as a mere cargo through binding to a membrane-located part or as a result of direct contact with the immunity protein. flciA was therefore coexpressed with individual or pairs of genes of the ptn operon. When the complete ptn operon was introduced together with flciA into the ptn deletion clone B464 resulting in B607, proteins of size corresponding to IIAB, IIC, and IID were found to copurify with fLciA from the bacteriocin-treated B607 as expected (Fig. 3). Similarly, the clone B612 expressing flciA together with ptnC and -D (but without ptnAB) gave rise to coprecipitation of IIC and IID. By contrast, neither the coexpression of flciA with the individual genes nor the coexpression of flciA with the gene pair ptnAB and -C gave rise to coprecipitated PTS components. These results indicate that fLciA can form a complex with IIC and IID without the involvement of the cytoplasmic component IIAB and that IIAB itself probably does not form any strong contact with fLciA. Note that a protein of size similar to IID was also present in the samples derived from clones lacking ptnD (B520, B609, B613, and B615); however, the intensity of this band is somewhat weaker than in clones expressing ptnD (B268, B607, and B612). Based on these observations, we conclude that this background protein in the ptnD-negative samples had come along during immunoprecipitation as a contaminant.
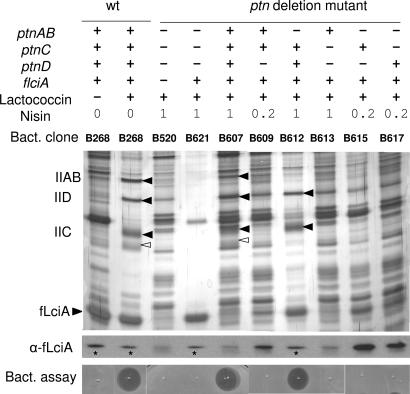
Fig. 3.
Only IIC and IID of the man-PTS are required for complex-formation with fLciA and lactococcin A. The presence (+) or absence (−) of ptnAB, ptnC, ptnD, and flciA in the various clones (Bact. clone) is indicated in the (Top) as is the presence (+) or absence (−) of added lactococcin A (50 BU/ml) and nisin (0.2 or 1 ng/ml for induction of the cloned genes). After overnight growth, fLciA and its copurified proteins were collected by immunoprecipitation and analyzed by SDS/PAGE and silver staining (Middle). For immunodetection of fLciA with the antibody M2 (α-fLciA; only area containing fLciA is shown here), purified samples were diluted 5- or 25-fold (∗; to avoid oversaturated signals) before 10 μl of each diluted sample was analyzed by Western blotting. (Bottom). Coprecipitated bacteriocin activity was analyzed by applying 5 μl of the eluted fractions directly onto lawns of the indicator stain Il1403. Growth inhibition is seen as clear zones. Filled arrowheads denote the man-PTS components, and open arrowheads (in bacteriocin-treated B268 and B607) indicate a protein that was coprecipitated only when the complete man operon was expressed.
More importantly, only the purified samples containing coprecipitated PTS components (clones B268, B607, and B612) displayed bacteriocin activity when tested on lawns of indicator cells, whereas other samples lacking coprecipitated man-PTS components (B520, B621, B609, B613, B615, and B617) did not (Fig. 3). The latter result strongly indicates that the bacteriocin is not only required for the fLciA-man-PTS complex formation but also is part of the complex.
Other Class II Bacteriocins Targeting Man-PTSs.
To evaluate whether the involvement of the man-PTS as a receptor is a unique case for lactococcin A or whether it may serve similar function for other class II bacteriocins as well, we analyzed a number of different bacteriocins using a similar approach as for lactococcin A. Lactococcins A and B are both produced by the multibacteriocin producer L. lactis LMG2130 (26). Like lactococcin A, lactococcin B inhibits mainly the growth of other lactococci by permeabilizing the cytoplasmic membrane; however, lactococcins A and B share no obvious homology to each other at the amino acid level (26). We found that lactococcin B was active against the lactococcal wild-type strain Il1403 and the ptn deletion clone expressing the reintroduced ptn operon (B515) or the gene pair ptnC and ptnD (B529) but failed to kill the isogenic ptn deletion mutant (clone B520) or the derivative clones lacking both ptnC and ptnD (e.g., B526, B619, B538, and B541 expressing ptnAB and -C, ptnAB, ptnC, or ptnD, respectively). (Details of these clones is provided in SI Table 1). This observation suggests that lactococcin B probably also uses the same receptor as lactococcin A, namely the man-PTS components IIC and IID, to target susceptible cells.
The Listeria-active pediocin-like bacteriocins, enterocin P (27), sakacin A (28), pediocin PA1 (29), and the newly discovered penocin A (30), share a consensus sequence (YGNGV) at their N terminus, but none of them share any obvious sequence homology with lactococcin A or B. These pediocin-like bacterciocins are not active against L. lactis IL1403, but they display inhibitory activity against several L. sakei strains including Lb790, 23K, and NCDO2714. To examine whether these pediocin-like bacteriocins could specifically target a Lactobacillus man-PTS, we expressed the man-PTS genes from Lb. sakei 23K in the lactococcal clone B464 (ptn deletion mutant). Indeed, heterologous expression of the complete Lactobacillus man-PTS operon (manLMN) (clone B630) or only the gene pair manMN coding for the membrane-located IIC and IID (clone B634) rendered the L. lactis ptn deletion mutant sensitive to these bacteriocins (data not shown).
Furthermore, by using the flag-tagging approach for purification and detection of the immunity proteins (EntiP and SaiA) specific for enterocin P and sakacin A, we observed that these immunity proteins also formed complex with the man-PTS components in a bacteriocin-dependent manner as similarly found for LciA (Fig. 4). (The identity of the man-PTS components was verified by MS; data not shown). Taken together, these results demonstrate that several class II bacteriocins can use components of the man-PTS as a receptor to target susceptible cells and that their immunity proteins probably involve a common mechanism in self-protection of the bacteriocin producing cell.
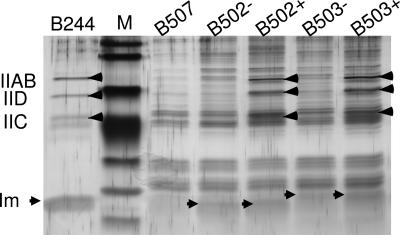
Fig. 4.
Silver-stained SDS/PAGE gel showing that the immunity proteins specific for enterocin P and sakacin A form complex with the man-PTS components only in the presence of their cognate bacteriocins. L. sakei clones: B502 expressing fentiP, B503 expressing fsaiA, B507 expressing empty vector; B244 is a lactococcal strain expressing lcnA and flciA. Protein samples were purified from cultures grown in the absence (−) or presence (+) of added bacteriocins (≈50 BU/ml for enterocin P in B502+ and 25 BU/ml for sakacin A in B503+). Arrowheads signify the man-PTS components (IIAB, IIC, and IID) and the immunity proteins (Im: fLciA in B244, fEntiP in B502, and fSaiA in B503); M; molecule weight protein markers. Equal volumes of the eluted fractions (each 5 μl of total 100 μl) were applied to wells for direct comparison.
Discussion
By using the fusion immunity protein fLciA as bait, we were able to fish out proteins that formed complex with fLciA when it conferred immunity against lactococcin A. Through mass spectrometric analysis, three of these proteins were identified as components of the man-PTS, namely IIAB, IIC, and IID. The interaction between these proteins and fLciA was shown to be dynamic, involving an on–off type mechanism that depended on the presence or absence of lactococcin A. The role of the man-PTS components was subsequently assigned as target/receptor for lactococcin A based on the observation that the deletion mutant lacking the ptn operon became resistant to lactococcin A. This finding was further corroborated by two additional functional studies: (i) reintroduction of the ptn operon into the deletion mutant reestablished sensitivity to lactococcin A and (ii) a lactococcin A-resistant Lactobacillus strain (Lb790) became sensitive to lactococcin A by heterologously expressing the lactococcal ptn operon. The latter observation is of particular importance because lactobacilli are known to be resistant to lactococcin A despite the fact that they also harbor a man-PTS highly similar to the lactococcal man-PTS. Attempts to complement the lactococcal ptn deletion mutant with the corresponding genes from L. sakei 23K (sharing over 57% identity and 72% similarity at the amino acid level) failed to establish sensitivity to lactococcin A (data not shown). The specific requirement of a lactococcal man-PTS for lactococcin A-induced sensitivity can thus account for the previous observation that this bacteriocin acts primarily toward lactococcal strains but poorly toward species outside the Lactococcus genus (20, 21).
Detailed genetic analysis combined with immunocoprecipitation studies demonstrated that the expression of ptnC and ptnD (without ptnAB) was sufficient for lactococcin A-induced sensitivity and also for complex formation between the encoded proteins and fLciA. This finding is surprising because IIAB (encoded by ptnAB) was associated with the protein complex when the complete ptn operon was expressed (e.g., in the wild-type strain). These results might suggest that the membrane-located proteins, IIC and IID, together could form a functional receptor for lactococcin A and that IIAB was associated with the bacteriocin–fLciA–protein complex as a mere cargo, probably because of its ability to form reversible contacts with a membrane-located part (19).
By immunodetection, we observed a significant pool of the fLciA in the membrane-associated fraction in addition to the cytoplasmic pool, regardless of the presence or absence of the bacteriocin (data not shown). This membrane-associated pool has been estimated to constitute 50–70% of the total amount of the protein in cells (31, 22). The finding that fLciA-complex formation involves an on–off-type immunity mechanism suggests that the membrane-associated pool of the protein can be divided further into two forms of complexes: one non- or loosely associated with the receptor proteins in the absence of the bacteriocin and the second tightly associated with the receptor proteins in the presence of the bacteriocin. Whether lactococcin A can form direct contacts with its cognate immunity protein or not has been addressed previously. It was shown that LciA did not affect the antimicrobial activity of lactococcin A in an in vitro experiment, implying little or no contact between lactococcin A and its immunity protein (31). By using an antibody recognizing the C-terminal half of LciA, Venema et al. (22) observed that the antibody could react with right-side-out vesicles from immune (LciA-positive) cells but failed to bind inside-out vesicles or protease-K-treated right-side-out vesicles from immune cells. Based on these results, these authors suggest that the antibody-reacting part of LciA is located on the outside of the membrane and that this part presumably forms contacts with the bacteriocin, because the latter blocks antibody binding. The results presented here seem to unify these apparently conflicting results into one mechanism: the interaction between lactococcin A and its immunity protein requires the presence of, and is mediated by, the receptor proteins (IIC and IID); otherwise, no or poor interaction occurs between them. Taken together, our proposed model (Fig. 5) is that LciA confers immunity, not by preventing the bacteriocin from binding to its receptor (in a competitive manner), but, rather, by direct interaction with the receptor–bacteriocin complex (in a bind-and-lock manner) thereby blocking the subsequent steps leading to membrane permeabilization and cell death.
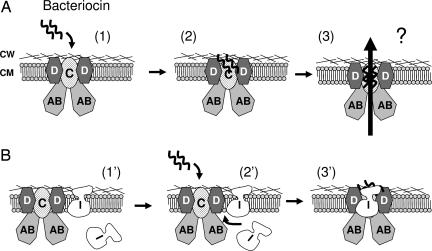
Fig. 5.
Model of target cell killing and immunity for lactococcin A and probably also for lactococcin B and some pediocin-like bacteriocins. (A) The bacteriocin employs IIC and IID of the man-PTS as a receptor on target cells (states 1 and 2). After binding, the bound bacteriocin somehow triggers permeabilization of the membrane (state 3), causing leakage of cellular components and, eventually, cell death. Although the exact nature of the pores is unknown, it is conceivable that they are formed either by oligomerized bacteriocin molecules or by disruption of the man-PTS. (B) In immune cells without bacteriocin production (e.g., the clone B268), the immunity protein (I) is not tightly associated with the man-PTS (state 1′). Only when exogenous bacteriocin is added to the culture medium (state 2′), the immunity protein approaches and binds strongly the target proteins (IIC and IID). The cells are thereby protected, probably through the blocking of the bound bacteriocin molecules, from advancing to the subsequent steps that lead to cell death (state 3′). In bacteriocin-producing cells (e.g., B244), the cognate immunity protein is tightly associated with the bacteriocin-bound receptor proteins to protect cells from being killed (state 3′). In all cases, the cytoplasmic component IIAB may form contact(s) with its membrane-located partners without being involved in the receptor-function. CW, cell wall; CM, cytoplasmic membrane. The man-PTS unit is shown with a stoichiometric ratio of 2:1:2 for IIAB/IIC/IID, based on the analogy to the E. coli counterpart (42).
Our results also suggest that the man-PTS in immune (LciA-positive) cells may exist in two different forms: (i) one non- or loosely associated with the immunity protein and (ii) one tightly associated with both the bacteriocin and immunity protein (state 1′ versus state 3′ in Fig. 5B). With regard to its involvement in sugar transport, it is reasonable to ask whether the structure or functionality of the man-PTS is affected when it is converted from one form to another. If adversely affected, one would expect bacterial growth on mannose or glucose as sole carbohydrate source to be perturbed, because the man-PTS is a key pathway for uptake of these sugars. Our growth study indicates that there is, indeed, a fitness cost to compensate immunity (see SI Fig. 8). Immune cells (e.g., fLciA-positive B268) growing in a medium containing lactococcin A [200–250 bacteriocin unit (BU)/ml], showed an almost doubling in generation time when mannose or glucose was used as sole carbohydrate. Such an impaired growth rate was not observed when galactose, which uses an uptake pathway (probably a galactose permease; 32) different from the man-PTS, was used as sole carbohydrate. The man-PTS is known to be involved in a hierarchical regulatory network, so-called catabolite repression (18); consequently, it will be of great interest in future work to employ transcriptome analysis to examine whether immune cells show an altered metabolic pattern when exposed to bacteriocin as a consequence of fitness cost. This approach also has great potential to be a useful means of identifying regulated receptors targeted by other antimicrobials.
In addition to lactococcin A, we demonstrated that lactococcin B as well as some Listeria-active pediocin-like bacteriocins could also use components (IIC and IID) of the man-PTS as a receptor in cell targeting and that the immunity proteins for these one-peptide bacteriocins seem to share a common mechanism in producer-cell self-protection. Interestingly, we failed to demonstrate such an involvement of the man-PTS system in the killing of susceptible cells by the two-peptide nonlantibiotic bacteriocins (subclass IIb), plantarcin EF from L. plantarum C11 (33), and lactococcin G from L. lactis LMG2081 (34) (data not shown), suggesting that these two-peptide bacteriocins might apply a different mechanism than the one-peptide bacteriocins in cell targeting.
The PTSs are ubiquitous in bacteria but do not occur in eukaryotes. This uniqueness makes them a potential target for developing new antimicrobials. This study demonstrates that the man-PTS can, indeed, serve as such a target for a number of membrane-permeabilizing peptides. Our current view is that the man-PTSs from different species/genera may serve as receptors for several different bacteriocins and that each bacteriocin recognizes specifically just one or a limited number of man-PTSs. The identification of the recognition sequences that specifically define such a discrimination will be of key importance in further work. The information from such studies will be valuable for constructing efficient antimicrobial peptides that might circumvent resistance as has been with the antilisterial peptides (16, 35).
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains and their derivatives used in this study are listed in SI Table 1. Lactococcal strains were grown in M17 medium (Oxoid, Hampshire, U.K.) supplemented with 0.5% (wt/vol) glucose at 30°C, whereas Lactobacillus and Enterococcus strains were propagated in MRS medium (Oxoid) at 30°C. Where appropriate, erythromycin and/or chloramphenicol (each at 5 μg/ml) was added to the growth medium for selection of transformants.
Bacteriocin Concentration and Assays.
Bacteriocin was concentrated from supernatants of overnight cultures, by ammonium sulfate precipitation (30% saturation), and pellets were dissolved in water. Bacteriocin concentration was determined by a microtiter plate assay as described (20). One BU is defined as the minimum amount of bacteriocin required to produce 50% growth inhibition of a 200 μl-culture of the indicator IL1403 (for lactococcin A) or L. sakei 23K (for enterocin P and sakacin A).
Plasmid Construction.
Strategies and PCR primers used to construct the various plasmids are detailed in SI Tables 2 and 3.
Gene Expression, Protein Purification, and Western Blot Analysis.
The NICE expression system has been described previously (36, 37). Lactococcal cells or Lactobacillus cells from overnight cultures (50–250 ml) expressing cloned gene(s) were harvested by centrifugation, washed twice with ice-cold Tris-buffer saline (TBS) before cells were mechanically lysed either by French press or fast-prep (Bio101 Savant Instruments Inc, Holbrook, NY). After centrifugation for the removal of unlysed cells, the flag-tagged protein fLciA from each cell extract (10 mg) was immunoprecipitated with anti-flag M2-affinity gel (10 μl), washed, and eluted following the manufacturer's protocol (A 2220; Sigma, St. Louis, MO). The resulting samples were analyzed by SDS/PAGE (38). For immunoblotting, proteins from SDS/PAGE gels were transferred onto nitrocellulose membranes, and blots were then sequentially incubated with the mouse antibody M2 (Sigma) and horseradish peroxidase-conjugated anti-mouse antibody (Amersham, Piscataway, NJ). Blots were developed by enhanced chemiluminesence (ECL) according to the manufacturer's instructions (Amersham).
Peptide Preparation and Mass Spectrometry.
Proteins were excised from the SDS/PAGE gel, digested, and extracted following the protocol of Jensen et al. (39). The extracted peptide samples were dried in a Speed-Vac, rehydrated, and desalted by using C18 STAGE tips (40). The desalted samples were eluted in 2–5 μl of 50% ACN/0.1% TFA, and 0.5–1 μl of the eluate was mixed 1:1 with matrix solution and applied on a stainless steel MALDI target (Bruker Daltonics, Billerica, MA). Acquisition of MS data was performed on an Ultraflex MALDI-TOF/TOF (Bruker Daltonics) instrument operated in reflectron mode with delayed extraction. Positively charged ions in the m/z range of 200–4,000 were analyzed by using an acceleration voltage of 25 kV and a delayed extraction (PIE) setting of 40 ns. The sample spectra were calibrated externally with a calibration standard covering the m/z range 700–3,100 (Bruker Daltonics). MS/MS spectra of selected peptides were recorded by using the LIFT ion optics of the mass spectrometer (41). After acquisition and mass axis recalibration, database searches were performed by using the public Mascot server (www.matrixscience.com), with the mass error tolerance set to 50 ppm. The database selected was NCBInr, and the taxonomy was set to “Other firmicutes.”
Supplementary Material
Acknowledgments
This work was supported by the Research Council of Norway.
Abbreviations
- BU
bacteriocin unit
- man-PTS
mannose phosphotransferase system.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608775104/DC1.
References
- 1.Hughes D. Nat Rev Genet. 2003;4:432–441. doi: 10.1038/nrg1084. [DOI] [PubMed] [Google Scholar]
- 2.Levy SB, Marshall B. Nat Med Sup. 2004;10:122–129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Hill C, Ross RP. Nat Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 4.Peschel A, Sahl HG. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 5.Drider D, Fimland G, Hechard Y, McMullen LM, Prevost H. Microbiol Mol Biol Rev. 2006;70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hechard Y, Sahl HG. Biochimie. 2002;84:545–557. doi: 10.1016/s0300-9084(02)01417-7. [DOI] [PubMed] [Google Scholar]
- 7.Moll GN, Konings WN, Driessen AJ. Antonie Leeuwenhoek. 1999;76:185–198. [PubMed] [Google Scholar]
- 8.Nes IF, Diep DB, Havarstein LS, Brurberg MB, Eijsink V, Holo H. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 9.Eijsink VG, Axelsson L, Diep DB, Havarstein LS, Holo H, Nes IF. Antonie Leeuwenhoek. 2002;81:639–654. doi: 10.1023/a:1020582211262. [DOI] [PubMed] [Google Scholar]
- 10.Bauer R, Dicks LM. Int J Food Microbiol. 2005;101:201–216. doi: 10.1016/j.ijfoodmicro.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Fimland G, Johnsen L, Dalhus B, Nissen-Meyer J. J Pept Sci. 2005;11:688–696. doi: 10.1002/psc.699. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee C, Paul M, Xie L, van der Donk WA. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 13.Breukink E, de Kruijff B. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 14.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. J Biol Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 15.Ramnath M, Beukes M, Tamura K, Hastings JW. Appl Environ Microbiol. 2000;66:3098–3101. doi: 10.1128/aem.66.7.3098-3101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jansch L, Hechard Y, Hastings JW, Knochel S. Microbiology. 2002;148:2361–2369. doi: 10.1099/00221287-148-8-2361. [DOI] [PubMed] [Google Scholar]
- 17.Ramnath M, Arous S, Gravesen A, Hastings JW, Hechard Y. Microbiology. 2004;150:2663–2668. doi: 10.1099/mic.0.27002-0. [DOI] [PubMed] [Google Scholar]
- 18.Postma PW, Lengeler JW, Jacobson GR. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Q, Schunk T, Flukiger K, Erni B. J Biol Chem. 1995;270:5258–5265. doi: 10.1074/jbc.270.10.5258. [DOI] [PubMed] [Google Scholar]
- 20.Holo H, Nilssen O, Nes IF. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum MJ, Kok J, Venema G, Holo H, Nes IF, Konings WN, Abee T. J Bacteriol. 1991;173:7934–7941. doi: 10.1128/jb.173.24.7934-7941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venema K, Haverkort RE, Abee T, Haandrikman AJ, Leenhouts KJ, de Leij L, Venema G, Kok J. Mol Microbiol. 1994;14:521–532. doi: 10.1111/j.1365-2958.1994.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 23.Stoddard GW, Petzel JP, van Belkum MJ, Kok J, McKay LL. Appl Environ Microbiol. 1992;58:1952–1961. doi: 10.1128/aem.58.6.1952-1961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Havarstein LS, Diep DB, Nes IF. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 25.Venema K, Dost MH, Beun PA, Haandrikman AJ, Venema G, Kok J. Appl Environ Microbiol. 1996;62:1689–1692. doi: 10.1128/aem.62.5.1689-1692.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venema K, Abee T, Haandrikman AJ, Leenhouts KJ, Kok J, Konings WN, Venema G. Appl Environ Microbiol. 1993;59:1041–1048. doi: 10.1128/aem.59.4.1041-1048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cintas LM, Casaus P, Herranz C, Havarstein LS, Holo H, Hernandez PE, Nes IF. J Bacteriol. 2000;182:6806–6814. doi: 10.1128/jb.182.23.6806-6814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Axelsson L, Holck A, Birkeland SE, Aukrust T, Blom H. Appl Environ Microbiol. 1993;59:2868–2875. doi: 10.1128/aem.59.9.2868-2875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marugg JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, Toonen MY, Walker SA, Zoetmulder LC, Vandenbergh PA. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diep DB, Godager L, Brede D, Nes IF. Microbiology. 2006;152:1649–1659. doi: 10.1099/mic.0.28794-0. [DOI] [PubMed] [Google Scholar]
- 31.Nissen-Meyer J, Havarstein LS, Holo H, Sletten K, Nes IF. J Gen Microbiol. 1993;139:1503–1509. doi: 10.1099/00221287-139-7-1503. [DOI] [PubMed] [Google Scholar]
- 32.Grossiord BP, Luesink EJ, Vaughan EE, Arnaud A, de Vos WM. J Bacteriol. 2003;185:870–878. doi: 10.1128/JB.185.3.870-878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diep DB, Havarstein LS, Nes IF. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissen-Meyer J, Holo H, Havarstein LS, Sletten K, Nes IF. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vadyvaloo V, Snoep JL, Hastings JW, Rautenbach M. Microbiology. 2004;150:335–340. doi: 10.1099/mic.0.26731-0. [DOI] [PubMed] [Google Scholar]
- 36.Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Ruyter PG, Kuipers OP, de Vos WM. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Jensen ON, Larsen MR, Roepstorff P. Proteins. 1998;2:74–89. doi: 10.1002/(sici)1097-0134(1998)33:2+<74::aid-prot9>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber J, Ishihama Y, Mann M. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 41.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. Anal Bioanal Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 42.Rhiel E, Flukiger K, Wehrli C, Erni B. Biol Chem Hoppe Seyler. 1994;375:551–559. doi: 10.1515/bchm3.1994.375.8.551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.