Abstract
The active sites of intramembrane proteases are positioned in the lipid bilayer to facilitate peptide bond hydrolysis in the membrane. Previous crystallographic analysis of Escherichia coli GlpG, an intramembrane protease of the rhomboid family, has revealed an internal and hydrophilic active site in an apparently closed conformation. Here we describe the crystal structure of GlpG in a more open conformation, where the capping loop L5 has been lifted, exposing the previously buried and catalytically essential Ser-201 to outside aqueous solution. A water molecule now moves into the putative oxyanion hole that is constituted of a main-chain amide (Ser-201) and two conserved side chains (His-150 and Asn-154). The loop movement also destabilizes a hydrophobic side chain (Phe-245) previously buried between transmembrane helices S2 and S5 and creates a side portal from the lipid to protease active site. These results provide insights into the conformational plasticity of GlpG to accommodate substrate binding and catalysis and into the chirality of the reaction intermediate.
Keywords: intramembrane proteolysis, membrane protein, rhomboid protease, x-ray crystallography
Intramembrane proteases represent a unique set of membrane proteins (for a review, see refs. 1 and 2). Their activities are involved in numerous important biological processes, such as cholesterol metabolism, development, growth factor signaling, amyloid β-peptide synthesis, and many others. There are three known classes of intramembrane proteases that are thought to be homologous in their catalytic mechanisms to the soluble metallo-, aspartyl, and serine proteases, respectively (1, 2). All intramembrane proteases are polytopic integral membrane proteins and cleave peptide bonds that are normally embedded in cell membranes. It has been proposed that to facilitate intramembrane proteolysis, their active sites are also positioned in the lipid bilayer (3–5). This important prediction was confirmed experimentally recently when the first crystal structure of an intramembrane protease, Escherichia coli GlpG from the rhomboid family, became solved (6).
The rhomboid proteases belong to the “serine class” (5, 7–9). The crystal structure of GlpG established two important spatial relationships (6). First, the catalytically essential serine (Ser-201) is positioned below the membrane surface in a central and hydrophilic cavity of the intramembrane protease (Fig. 1A). Second, the other catalytically essential histidine (His-254) is immediately nearby and forms a hydrogen bond with the serine, probably priming it for nucleophilic attack (Fig. 1B). Understanding the enzymatic mechanism of GlpG not only has theoretical value, but it is also of practical importance, especially in the design of inhibitors that target similar proteases in human parasites such as Toxoplasma and Plasmodium (10, 11).
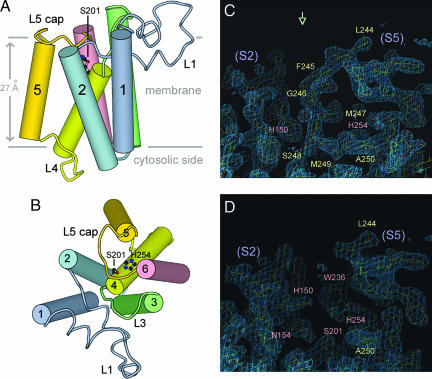
Fig. 1.
The L5 cap has become disordered. (A and B) Side and top views of GlpG, respectively. The transmembrane helices, shown as cylinders, are labeled 1–6. The two horizontal lines indicate boundaries for the hydrophobic region of the lipid bilayer. (C and D) Electron density features around L5 for the closed and open-cap structures (top view). The 2Fo − Fc maps are contoured at the 1.5σ level.
Without bound substrate, the intramembrane protease has to adopt a closed conformation so that its internal active site and water molecules present in the active site are physically separated from the surrounding lipid hydrocarbon tail groups. In the solved crystal structure, the active site of GlpG is located near the end of a short and central helix S4, and it is completely encircled by five other transmembrane helices and an unusual membrane-embedded loop L1 (Fig. 1 A and B). An extracellular loop (L5) tightly caps the Ser–His dyad from above and prevents substrate binding. It is reasonable to believe that this structure faithfully represents the conformation of the free enzyme in its native membrane environment (6) and is not an experimental artifact. However, based on the solved structure alone, it was not obvious how transmembrane substrates gain access to the internal and closed active site of the membrane protease. Because the natural substrate for GlpG is not known at this time, a structure of GlpG in complex with substrate, which should provide a straightforward answer to this question, will probably take some time to emerge. In this report, we describe unexpected experimental results that have shed light on the conformational flexibility of the protease, and we discuss the implications of these results in substrate binding and catalysis.
Results
Open-Cap Movement.
While trying to modify the active-site serine covalently with a class-specific inhibitor 3,4-dichloroisocoumarin (DCI) (5, 9, 12) by directly soaking the inhibitor into preformed GlpG crystals, we noticed a strong negative peak in the difference Fourier map that covered a surface loop (L5), previously depicted as the “cap” (6) [Fig. 1B and supporting information (SI) Fig. 6]. There was no continuous positive density nearby, suggesting that L5 had become disordered. The difference was not caused by the inhibitor because similar changes were observed in crystals soaked with a blank DMSO solution that was used to dissolve DCI. The difference map did not show any major positive peaks near Ser-201, suggesting that the inhibitor had failed to react with it under the present experimental conditions.
The refined crystal structure at 2.5 Å resolution confirmed that L5, from residue 245 to 249, had become disordered (Fig. 1 C and D and Table 1). Structural changes in the membrane protein were restricted to areas around L5, where several side chains also shifted. As a result of the movement of L5, two hydrophobic side chains (Met-247 and Met-249) were lifted from the immediate vicinity of Ser-201 (Fig. 2A), thereby fully exposing it to the aqueous environment outside of the membrane (Fig. 2B). The void left behind was occupied by new water molecules. A neighboring histidine, His-150 from transmembrane helix S2, moved in here through simple rotations of its Cα–Cβ and Cβ–Cγ bonds (Fig. 1 C and D and SI Fig. 7). Because the new location of His-150 partially overlaps with the original path of L5 main chain, it is apparent that the disordered loop can no longer be present in this general area. As a result, an open trough appears on the top of the membrane-embedded protease, which could be where substrate binds (Fig. 2B).
Table 1.
Data collection and refinement statistics
| Measurement | Value |
|---|---|
| Data collection | |
| Space group | R32 |
| Cell dimensions, Å | a = 111.4, c = 128.6 |
| Wavelength, Å | 0.9795 |
| Resolution, Å* | 40.0–2.5 (2.59–2.50) |
| Observed reflections | 110,141 |
| Unique reflections | 10,493 |
| Redundancy | 10.5 |
| Completeness, %* | 96.8 (97.9) |
| <I/σ>* | 11.7 (4.3) |
| Rmerge*,† | 0.091 (0.347) |
| Refinement | |
| Resolution, Å | 40.0–2.5 |
| Rwork/Rfree‡ | 0.240/0.285 |
| No. of atoms | |
| Protein | 1,395 |
| Detergent | 21 |
| Water | 40 |
| B factors | |
| Protein | 41.3 |
| Detergent | 69.2 |
| Water | 47.8 |
| rms deviations | |
| Bond length, Å | 0.009 |
| Bond angle, ° | 1.33 |
*Highest resolution shell is shown in parentheses.
†Rmerge = Σ|Ii − 〈I〉|/Σ Ii.
‡Rwork = Σ|Fo − Fc |/ΣFo. Rfree is the cross-validation R factor for the test set of reflections (10% of the total) omitted in model refinement.
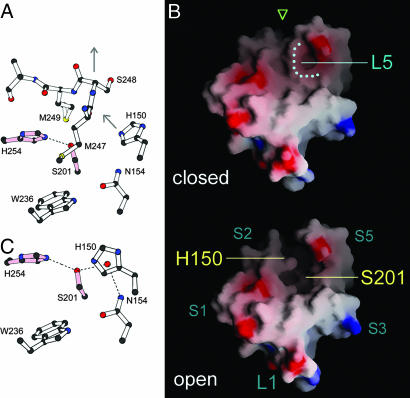
Fig. 2.
The active site is exposed in the open-cap conformation. (A) Detailed picture of the closed cap and the catalytic dyad viewed from the back of GlpG, as indicated by the green arrow in Fig. 1C. (B) Comparison of the surface features of the closed and open-cap GlpG structures viewed from the top. Blue areas are positively charged, and red areas are negatively charged. The dotted line marks the path of L5 in the closed structure. (C) With the cap lifted, a water molecule (red dot) moves into the putative oxyanion hole.
Putative Oxyanion Hole.
For any given protease, the oxyanion-binding site has the important function of stabilizing the negative charge developed on the carbonyl oxygen of the scissile bond during catalysis. The location of this site was not obvious in the closed state of GlpG because of the two methionine side chains plugged into the active site. Now with the capping loop L5 removed, water molecules immobilized in the active site slightly adjusted their positions. One water, previously bound between Ser-201, His-150, and Gly-198, had moved by ≈1.3 Å to a new location, where it broke off the hydrogen bond with the backbone carbonyl of Gly-198 and formed a new bond with the side chain amide of Asn-154 (Fig. 2C). This new location corresponds roughly to where the oxyanion hole for a classic serine protease is (13) and raises the possibility that not only the activation mechanism of the catalytic serine by a histidine is conserved, but also the relative position of the oxyanion hole is similar between the two types of serine protease. We noted previously that the backbone amide of Gly-199 (of the conserved GXSG motif) pointed away from the Ser–His dyad and could not contribute to oxyanion binding. Now it seems that Asn-154, or even the nearby flexible His-150, could play a role in this crucial function, compensating for the loss of the backbone amide. Like that of the soluble serine protease, the backbone amide group of GlpG Ser-201 is well positioned to form another hydrogen bond with the bound oxyanion.
The hypothesis that His-150 and Asn-154 may participate in oxyanion binding is consistent with the fact that both of these residues are highly conserved in the rhomboid protease family (14, 15). Mutagenesis studies have shown that substitution of Asn-154 by alanine greatly reduced protease activity for most rhomboids studied (5, 7), which would also be consistent with the current proposal. However, it should be noted that for E. coli GlpG and Bacillus subtilis rhomboid YqgP, mutation of Asn-154 did not appear to affect protease activity significantly (8, 9), suggesting that the contribution of this asparagine to the overall stabilization of the oxyanion reaction intermediate may vary among different proteases and that multiple functional groups (e.g., His-150) could be involved in this function. Based on these findings, in Fig. 3 we present a hypothetical model for a bound substrate dipeptide fragment where its carbonyl oxygen is hydrogen-bonded to the backbone amide of Ser-201 and the side chains of both His-150 and Asn-154.
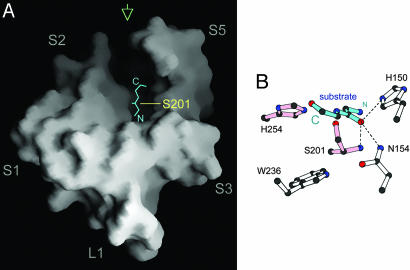
Fig. 3.
A hypothetical model of bound substrate peptide. (A) The peptide (cyan) is positioned in the open trough at the top of the membrane protease, with its carboxyl terminus pointing toward the gap between transmembrane helices S2 and S5. (B) A detailed view of the hypothetical substrate (cyan) bound to the GlpG active site. This picture corresponds to a view from the back of GlpG as indicated by the green arrow in A. The carbonyl carbon atom of the scissile bond is positioned just above the hydroxyl group of Ser-201, and the carbonyl oxygen atom occupies the proposed oxyanion-binding site defined by the main chain of Ser-201 and the side chains of His-150 and Asn-154.
Side Portal.
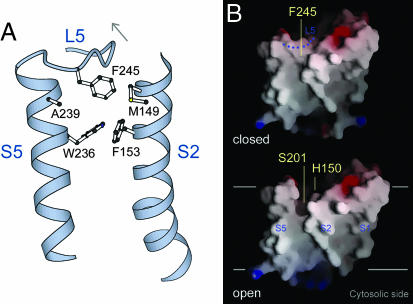
The loop movement also affected Phe-245. In the closed structure, the side chain of Phe-245 is inserted into a gap between transmembrane helices S2 and S5, roughly at the same level of the internal active site (Fig. 4A). By virtue of its size and snug fit with Met-149, Phe-153, Trp-236, and Ala-239, Phe-245 physically separates the membrane-embedded active site from the lipid. In the open-cap structure, Phe-245 had surprisingly become disordered (SI Fig. 8). Although the side chain of Met-149 from S2 moved inward slightly to adjust for the change (SI Fig. 7), it was not sufficient to bridge the gap. Therefore, an opening was left in the wall of protein structures that surrounded the active site, exposing internal hydrophilic residues unfavorably to lipid (Fig. 4B).
Fig. 4.
Cap movement creates a lateral opening. (A) Phe-245 is normally inserted between S2 and S5, blocking an entrance to the active site (back view). (B) Molecular surface of GlpG viewed from the back. Blue areas are positively charged, and red are negatively charged. The two horizontal lines mark the hydrophobic region of lipid bilayer. The dotted line depicts the path of L5 in the closed structure.
Discussion
The structural changes described in this report were unexpected because they have apparently occurred without substrate or inhibitor bound to the protease. We have analyzed many soaked crystals, and they all yielded similar results. The changes must be caused by soaking, resulting either directly from the new solvent or indirectly from changes in the lattice force. Regardless of the source, the perturbation appears small because most parts of the protein are not affected. The dramatic change specifically involving only L5 could not have been anticipated. In the closed structure, L5 is well defined by electron density (Fig. 1C), and its B factor (<40 Å2) is only slightly higher than average (≈34 Å2) and much lower than some part of the protein, e.g., L4 (≈60 Å2). L5 is not involved in crystal packing. Therefore, the observed movement seems to reflect an intrinsic propensity of this loop to switch to the open conformation and indicates that the free-energy difference between the closed and open states is relatively small.
Despite the unusual circumstance from which the new crystal structure was derived, a comparison of the two solved structures provides important insights regarding the membrane protease plasticity and its relationship to substrate binding. In the previous structure, the active site is completely buried inside the membrane protein (6) (Fig. 1 A and B). To bind substrate, the protein structure must become open, but which parts of the protein are movable, and in what sequence, could not be determined. The proximity of L5 to the catalytic dyad already hinted that this loop had to move at some point, but it could not be decided, based on the previous structure alone, whether the L5 movement occurs early or at a later stage after changes in other parts of the protein. These data, showing that L5 is intrinsically flexible, are consistent with the expectation that this loop will eventually become displaced by substrate, and indicate that a similar conformational end point (i.e., the “open-cap” state) may be achievable with a minimal input of free energy by different means, e.g., substrate binding, or artificial forces generated during soaking. The observation that L5 is able to open, apparently in the absence of other changes in the protein, strongly suggests that structural rearrangements around L5 are independent and possibly early events to accommodate substrate binding. This sequence of action is again reminiscent of some soluble proteases whose active sites are also capped by flexible loops (16).
The loop movement unexpectedly pulls Phe-245 from its buried location between S2 and S5 and creates a gap in the tightly sealed portion of the protein structure that is embedded in the hydrophobic region of the bilayer (Fig. 4B). The depth of the gap roughly matches that of the internal active site. This observation, combined with surface features of the open-cap GlpG from the extracellular side (Fig. 2B), raises the possibility where transmembrane substrate may enter the protease active site through the gap between S2 and S5. This model is simpler than the one we proposed previously (6) because it does not require much more conformational changes in the protease other than those already described here. Besides, because S5 appears less extensively involved in packing with other membrane-spanning helices and it is flanked by two potentially mobile loops (SI Fig. 9), it even seems possible that S5 could move slightly to influence the opening of the cap. Given the known membrane topology of rhomboid substrates (amino terminus outside), according to this model and the assumption about the oxyanion-binding site (Fig. 2C), a mechanistic prediction can thus be made in which Ser-201 attacks substrate from the si-face of the peptide bond (Fig. 3B), in a fashion similar to E. coli signal peptidase (17) but different from most other serine proteases that make the nucleophilic attack on the re-face. This prediction should be particularly relevant to the design of compounds that mimic the transition state and, therefore, are capable of specifically inhibiting the rhomboid proteases.
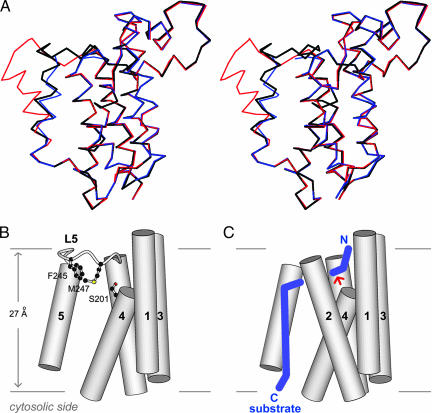
Two new rhomboid structures have just been solved and are available from the protein data bank: PDB entry 2IRV, by Bibi et al. (27), describes the structure of E. coli GlpG with a lipid bound to its active site; PDB entry 2NR9, by James et al. (28), describes the structure of a related rhomboid protease from Haemophilus influenzae. After this paper was submitted, an on-line publication appeared that described the structure of the same E. coli GlpG free enzyme in a different crystal form (18), which was solved with the help of the published GlpG coordinates (6). As expected, these structures were virtually identical (black and blue in Fig. 5A), with the exception that in the crystal form described by the online publication (18), crystal packing had caused transmembrane helix S5 in one of the two GlpG molecules in the asymmetric unit to bend away from the main body of the protease (red in Fig. 5A). Although the position of the “tilted” S5 observed by these authors (18) is obviously an artifact of crystallization because, in this conformation, the entire hydrophilic interior of the free membrane protease would be in direct and unfavorable contact with the hydrocarbon region of the lipid bilayer, the interesting fact that the position of S5 could be influenced by a neighboring molecule in the crystal is consistent with our hypothesis regarding conformational plasticity in this general region of the membrane protein (SI Fig. 9).
Fig. 5.
The relationship between protease plasticity and substrate binding. (A) Stereo diagram comparing the GlpG structure (Cα trace) described in ref. 6 (black) and those in ref. 18 (blue and red). (B) Schematic diagram illustrating the positions of the cap (Phe-245 and Met-247) and the internal active site (Ser-201) relative to the hydrophobic region of the membrane (horizontal lines). The transmembrane helices are shown as cylinders. Transmembrane helix S2 and loops L1–L4 were omitted for the purpose of clarity. (C) Diagram illustrating the approximate path of bound transmembrane substrate (blue). The red arrow points to the cleavage site. The portion of substrate entering the protease likely has an extended conformation, whereas the portion remaining in the membrane outside of the protease is probably helical.
The schematic diagrams in Fig. 5 B and C summarize our present thoughts on the role of the “cap” (L5), particularly Phe-245 and Met-247, in controlling substrate entry into the buried active site. We propose that docking of substrate transmembrane domain near the S2/S5 gap could destabilize the L5 cap, causing it to open, so that the top portion of the substrate could unwind and bend into the active site (Fig. 5 B and C). In the online publication mentioned above (18), it has been proposed that substrate entry was “gated” by some sort of lateral movement of transmembrane helix S5, in a mechanism “similar” to that of protein-conducting channel (19). This comparison may be inappropriate because opening of lateral gate in the channel protein probably allows a complete transmembrane segment of the translocating peptide to move out from the channel into the lipid bilayer. There is no evidence at this time suggesting that the transmembrane domain of a rhomboid substrate ever becomes fully engulfed by the protease (a movement in the reverse direction of channel “substrate”). Therefore, the need for a lateral gate involving a full transmembrane helix (S5) is questionable. Indeed, features of GlpG crystal structure suggest that only the top portion of substrate membrane-spanning sequence bends into the protease (Fig. 5C), which is consistent with earlier biochemical studies showing that this region is critical for cleavability (20).
Methods
The core domain of GlpG was purified and crystallized as decribed previously (6). The inhibitor DCI powder (Calbiochem, San Diego, CA) was dissolved in DMSO as 100 mM solutions. The membrane protein crystals were transferred stepwise to a final cryoprotecting solution that contained 25% (vol/vol) glycerol, 0.6% NG, 3.0 M NaCl, 100 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (pH 7.0). The inhibitor stock solution was added at the last soaking step to the cryoprotecting solution to achieve a final concentration of 2.5 mM DCI and 2% (vol/vol) DMSO. The crystals were soaked for 5 days in the presence of DCI/DMSO at room temperature before flash freezing in liquid nitrogen. For the control, the crystals were treated identically except that no inhibitor was present in the DMSO solution.
The x-ray diffraction data were collected at 100 K on beamline X25 at the National Synchrotron Light Source (NSLS; Upton, NY). Many data sets were collected, and the one presented in Table 1 was the best among them. All diffraction images were processed by HKL2000 (21). A model of GlpG (PDB ID code 2IC8), stripped of detergent and solvent molecules, was rigid-body-refined, and a difference Fourier map was calculated (SI Fig. 6). After confirming the disorder of residues 245–249 by a 2Fo − Fc map, this part of GlpG was omitted from the model in subsequent refinement steps. The model was improved by iterative rounds of minor adjustments by using O (22) and conjugate gradient minimization and B factor refinement by CNS (23). At the last step of the refinement, a single detergent and 40 water molecules were added to the protein model.
The illustrations in Figs. 1 A and B, 2 A and C, 3B, 4A, and Fig. 5 were generated by MOLSCRIPT and Raster3D (24, 25). Those in Figs. 2B, 3A, and 4B were generated by GRASP (26).
Supplementary Material
Acknowledgments
We thank A. Héroux at the synchrotron beamline for assistance and S. C. Harrison for insightful discussions. Diffraction data for this work were measured at beamline X25 of NSLS. This work was supported by a New Scholar Award in Aging from the Ellison Medical Foundation (to Y.H.) and a gift from the Neuroscience Education and Research Foundation (to Y.H.).
Abbreviation
- DCI
3,4-dichloroisocoumarin.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2O7L).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611080104/DC1.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe MS, Kopan R. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 3.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 5.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Zhang Y, Ha Y. Nature. 2006;444:179–183. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 7.Urban S, Schlieper D, Freeman M. Curr Biol. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 8.Maegawa S, Ito K, Akiyama Y. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 9.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brossier F, Jewett TJ, Sibley LD, Urban S. Proc Natl Acad Sci USA. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urban S, Wolfe MS. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. pp. 40–43. [Google Scholar]
- 14.Wasserman JD, Urban S, Freeman M. Genes Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS, Rogozin IB, Laetitia D, Letellier MC, Pellegrini L. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott WR, Schiffer CA. Structure (London) 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 17.Paetzel M, Dalbey RE, Strynadka NC. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Nat Struct Mol Biol. 2006;12:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 19.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 20.Urban S, Freeman M. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Merritt EA, Murphy ME. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls A, Sharp K, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 27.Ben-shem A, Fass D, Bibi E. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. Proc Natl Acad Sci USA. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.