Abstract
Proteorhodopsin (PR) is a light-powered proton pump identified by community sequencing of ocean samples. Previous studies have established the ecological distribution and enzymatic activity of PR, but its role in powering cells and participation in ocean energy fluxes remains unclear. Here, we show that when cellular respiration is inhibited by depleting oxygen or by the respiratory poison azide, Escherichia coli cells expressing PR become light-powered. Illumination of these cells with light coinciding with PR's absorption spectrum creates a proton motive force (pmf) that turns the flagellar motor, yielding cells that swim when illuminated with green light. By measuring the pmf of individual illuminated cells, we quantify the coupling between light-driven and respiratory proton currents, estimate the Michaelis–Menten constant (Km) of PR (103 photons per second/nm2), and show that light-driven pumping by PR can fully replace respiration as a cellular energy source in some environmental conditions. Moreover, sunlight-illuminated PR+ cells are less sensitive to azide than PR− cells, consistent with PR+ cells possessing an alternative means of maintaining cellular pmf and, thus, viability. Proteorhodopsin allows Escherichia coli cells to withstand environmental respiration challenges by harvesting light energy.
Keywords: light-driven proton pumps, oceanic bacteria
Aquatic ecosystems play a major role in the conversion of light energy into chemical energy, principally via chlorophyll-based photosynthesis (1, 2). Other light-harvesting mechanisms include the light-driven proton pump bacteriorhodopsin, used by halobacteria living in salt ponds to supplement respiration (3). In 2000, a novel light-driven proton pump, proteorhodopsin (PR), was discovered (4). The world's oceans contain an estimated 1028 PR-expressing bacteria, placing them among the most prevalent organisms on Earth (5, 6). Proteorhodopsins are widely distributed and spectrally tuned to their oceanic environments (5, 7–12). PR is distinguished from bacteriorhodopsin by its high sequence homology to sensory rhodopsins (postulated to have evolved from a different origin than bacteriorhodopsin; ref. 4), its presence in members of Eubacteria (as opposed to Archaea), and significant differences in the critical amino acids that eject transported protons out of the cell (4).
Abundant evidence exists for PR's function as a transmembrane proton pump, including light-mediated transport of protons in right-side-out PR+ vesicles and the electrogenic extrusion of protons by illuminated PR+ Escherichia coli cells (4, 7, 13). Despite the ability of PR to pump protons, green light illumination did not increase SAR11 growth rates or cell yield in cell culture experiments (14), leaving proteorhodopsin's in vivo contribution unclear. Moreover, the extent to which light-harvesting by PR can complement or replace other cellular energy sources remains to be quantified.
The proton motive force (pmf) is the electrochemical potential of protons across the membrane, maintained under aerobic conditions by oxidative phosphorylation. Bacteria use the pmf to synthesize ATP, drive chemiosmotic reactions, and power the rotary flagellar motor (15, 16) In 1974, Racker and Stoeckenius (17) demonstrated that the pmf generated by light-driven proton pumping by bacteriorhodopsin could be used to produce ATP. We reasoned that measuring the pmf of illuminated PR+ cells could identify a role of PR in cellular energy fluxes. The speed of the E. coli flagellar motor is proportional to the pmf over a wide range of speeds (18, 19), therefore, proton extrusion by PR should increase the flagellar motor's rotation rate when the cells are illuminated. We used E. coli as the heterologous host in our PR experiments because it is the primary model organism for studying Gram-negative bacteria and techniques for pmf measurement and PR expression in E. coli have been refined (4, 5, 7–11, 13, 18, 19).
Results and Discussion
We tracked swimming PR+ E. coli in two dimensions by using dark-field microscopy, periodically illuminating the cells with green light at PR's absorption maximum. We observed single cells to characterize rapid responses of the cellular pmf to light. No detectable increase in cell swimming velocity occurred upon illumination with green light. We surmised that light-driven proton pumping may benefit the cell only under certain environmental conditions, as suggested by Giovannoni et al. (14).
To test the possibility that light-driven proton pumping is most beneficial to aerobically grown cells when their ability to respire is suddenly impaired, we energy-depleted the cells. Because E. coli is difficult to energy-deplete by nutrient limitation because of its endogenous energy stores (20, 21), we additionally used the respiratory poison azide, which has multiple cellular effects (22, 23) but primarily inhibits cytochrome oxidase and, thus, proton extrusion by the respiratory chain, stopping the flagellar motor (18).
Strikingly, with respiration inhibited by azide, PR+ cells responded to green light. PR+ cells in 30 mM azide swam slowly in red illumination, but they showed a marked velocity increase with green illumination (Fig. 1b). Upon removal of the green light, they slowed to their previous velocity. To increase the accuracy of our flagellar rotation measurements, we subsequently used a tethered cell geometry (Fig. 2a), permitting extended observation of the same bacterium in different illumination conditions. PR+ cells were allowed to stick to the coverslip via a flagellum, and we then monitored the angular rotation rate of cells (Fig. 2a). A typical tethered cell rotated at a mean rate of 0.2–1 Hz, depending on its length and the position of the stuck flagellum along its body. To facilitate data interpretation, we deleted the cheY gene (24, 25), yielding smooth-swimming mutants whose flagellar motors do not reverse.
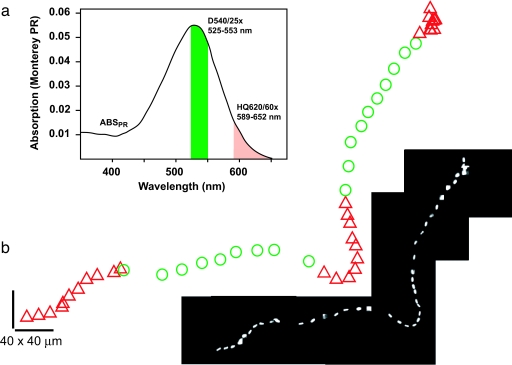
Fig. 1.
Light-powering of PR+ E. coli. (a) Overview of spectra and spectral overlaps. The PR absorption spectrum [redrawn from Beja (4)] is shown in black, with green and red bars indicating illumination wavelengths. (b) Single PR+ bacterium swims faster when illuminated with green light. The cell's position is recorded at a rate of 0.5 Hz via constant red dark-field illumination throughout the entire track. Illumination with green light at the absorption peak is periodic (20 s on, 20 s off) and occurs only during green circles of the path. Velocity during periods of green illumination (10.6 ± 0.9 μm/s) is 96% higher than during periods of red illumination alone (5.4 ± 0.4 μm/s; P < 0.005, one-tailed t test). Respiration has been inhibited with 30 mM azide in motility buffer. (Inset) Raw tiled movie frames of swimming cell.
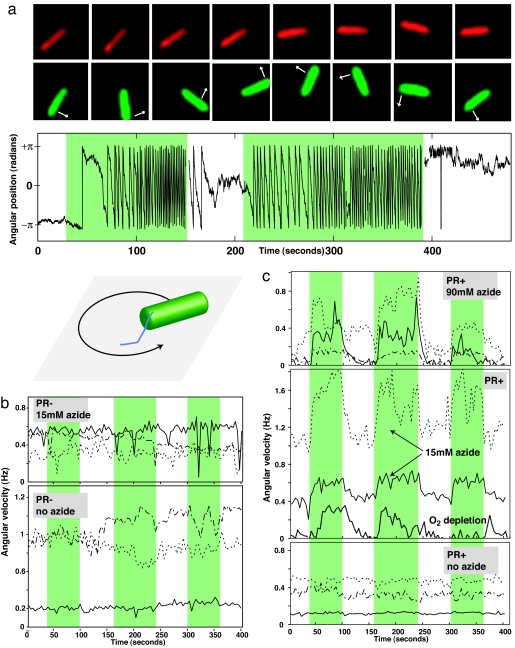
Fig. 2.
Flagellar response to green light measured in PR+ and PR− bacteria observed at different levels of respiratory inhibition. (a) Movie frames showing a tethered cell in red light and green light (full movie is available at SI Movie 1), a plot of the angular position versus time of a cell in 60 mM azide, and a schematic of tethered cell geometry. In red light, the cell rotationally diffuses about its attachment point. Green light (shaded area) leads to counter clockwise rotation of the cell. (b and c) Cells without (b) and with (c) PR. Solid, dotted, and dashed lines show three representative cells per condition. Note the considerable cell-to-cell variation in absolute rotation rates due to variation of cell length and tethering geometry. With or without azide, cells lacking PR show no response to green light. Removal of oxygen also leads to light-responsiveness. At high azide concentrations (c Top), angular velocity is nearly zero until cells are illuminated with green light.
As expected, there was no effect of green light on the cell's rotation rate in the absence of azide (Fig. 2b). However, as we inhibited respiration by adding azide, the cells again became light-responsive. PR+ cells sped up upon illumination with green light [Fig. 2 a and c, see also supporting information (SI) Movie 1]. The PR+ cells were converting light energy into an electrochemical potential used to do mechanical work. PR+ cells stopped moving or slowed considerably when the green illumination light was removed. For example, at low concentrations of azide (5–15 mM), angular velocity dropped by one-fourth when the green light was removed. At higher azide concentrations (80–110 mM), angular velocity dropped further (50–60%) because more of the cells' cytochromes were bound by azide (26).
To determine the extent to which light can replace respiration as an energy source, we varied the azide concentration. Increasing the azide concentration from 0 to 110 mM caused cellular pmf levels, and therefore average cellular angular velocity, to decrease (SI Fig. 4a). The pmf dropped to 50% of its original value at an azide concentration of 55 mM, in agreement with inhibition studies of isolated bo cytochrome (26). When the cells were illuminated with intense green light, their rotation rate was restored to the speed of cells with an unimpaired respiratory system. As proton extrusion by the respiratory system dropped with increasing azide, PR provided an ever larger fraction of the proton efflux needed to maintain the cellular pmf at the original, no-azide value (Fig. 3a). At the highest azide concentration studied, the average cell velocity increased 70% upon green light illumination. Cells lacking PR exhibited no increasing angular velocity in green light, indicating that PR expression is required.
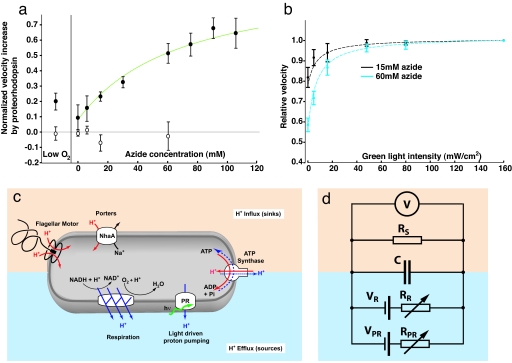
Fig. 3.
The response of PR+ bacteria to green light increases with respiratory inhibition and light intensity. (a) Benefit of illumination versus degree of inhibition of the respiratory system. The difference between angular velocity in green light and red light (ωG–ωR) becomes pronounced in PR+ bacteria (filled circles) as respiration is inhibited by low oxygen or sodium azide. PR− cells (open circles) show no change between red and green illumination. To facilitate comparison between cells, the angular velocities are normalized by each bacterium's maximum velocity. n = 5–14 cells per condition. Green line, fit to model described in d. (b) The rotation speed of PR+ cells depends on the intensity of green illumination. Individual PR+ spinner cells were exposed to six intensities of green light. The mean angular velocity at each intensity is plotted (n = 5–6 cells for each intensity), normalized by the velocity at maximum illumination. Dashed lines, fits to model described in d. (c) Overview of transmembrane fluxes and proton pumping in PR+ E. coli. Sources of proton motive force include respiration and PR. Sinks include rotation of the flagellar motor and ATP synthesis. (d) Model including sources of pmf (respiration and proteorhodopsin), sinks (such as the flagellar motor), and the membrane capacitance. The variable resistors RR and RPR model the effect of azide and light on proton extrusion by respiration and PR, respectively. The voltmeter (top-most circuit element) measures the potential difference across the membrane (equivalent to the pmf).
We subsequently varied the intensity of the green light illumination. The rotation rate was clearly stimulated even at the lowest light intensity studied (≈5 mW/cm2) (Fig. 3b). The rate increased rapidly with intensity up to 10 mW/cm2 (15 mM azide) or 20 mW/cm2 (60 mM azide) (Fig. 3b), where the effect saturated. At >50 mW/cm2, there was no detectable benefit of increased illumination. The energy absorbed by PR through our filter (Fig. 1a) at intensities of 10–20 mW/cm2 is equivalent to the energy absorbed by PR from the sun at sea level, taking into account the absorption cross-section of PR and the solar irradiation spectrum (detailed information is published as SI Text).
It was now essential to confirm the postulated mechanism of action. Reduced proton pumping by the respiratory system causes the pmf to drop. Light-based proton pumping by proteorhodopsin can then increase the pmf. Removal of oxygen from the cell culture also should lead to light-responsive bacteria with none of the possible confounding effects of azide. We needed to reduce O2 levels substantially, because at 0.3 ppm, the E. coli respiratory system still generates ≈50% of the normal pmf (27). We gently bubbled nitrogen through the cell culture for 15 min and then used a nitrogen-filled glovebox to prepare sealed imaging chambers containing 2 μl of cells. Just as we had seen upon addition of azide, PR+ cells became light-responsive upon oxygen depletion. Illumination increased the cells' angular velocity by 45 ± 25% (Fig. 2c; P < 0.005, one-sided t test). Addition of the protonophore carbonyl cyanide 3-chlorophenylhydrazone (100 μM) to low oxygen cultures eliminated all bacterial motility, and movement was not restored with 50 seconds of green illumination (n = 300), demonstrating that cells unable to maintain a pmf cannot be revived.
To clarify the relationships between pmf, azide, and light, we constructed a highly simplified model of E. coli membrane fluxes. Our cells have multiple proton pumps that can contribute to the pmf (28), including PR, the respiratory chain, and the ATPase (Fig. 3c). Pmf is consumed by the flagellar motor and numerous transporters. In addition, the bacterial membrane has a basal permeability to protons (29). In a simpler system, Rotterdam et al. (30) showed that lipid vesicles with only one kind of ion pump reach a steady-state pmf whose magnitude is well approximated by a simple RC circuit.
Hypothesizing that an analogous circuit might be able to capture the functional relationship between pmf, azide, and light, we constructed the model shown in Fig. 3d (see also SI Fig. 5) and parameterized it by fitting to the data shown in Fig. 3 a and b and in SI Fig. 4. This model describes in vivo time-dependent dynamics between light-driven proton pumping and respiration and is described in detail in SI Text. Despite the model's simplicity, it suggests why no effect of PR on growth rates has been reported. The model indicates that the maximum potential PR can generate by using the free energy from photon absorption (VPR) is similar to the potential generated by E. coli respiration. Thus, in E. coli grown at neutral pH in rich or minimal media, or in E. coli respiring aerobically by using endogenous energy stores, PR cannot pump protons. Only when the pmf falls below the maximum potential (VPR) during respiratory stress does PR begin to pump, and the proton flux through PR increases as the pmf falls. PR is able to maintain E. coli cellular pmf near this maximum potential (VPR ≈ −0.2V) with sufficiently bright illumination (KM ≈ 60mW/cm2).
The cell motility studies together with the results of the pmf modeling raised the possibility that PR could pump sufficient protons to increase cell viability in addition to powering the flagellar motor. To clarify a possible relationship between viability and proton pumping by PR, we plated cell cultures after their exposure to 30 mM azide for 30 min in sunlight. The cells lacking retinal were most sensitive to azide; no colonies were recovered after plating the azide exposed cells. The cells lacking PR but having retinal were slightly more azide resistant; 1% of cells survived (SI Table 1). The cells with both PR and retinal were significantly more azide resistant than in all three other conditions (11% of cells survived; P < 0.005, Mann–Whitney U test). Consistent with the motility studies and the pmf model, PR is able to sustain cellular pmf at a level that increases viability. These findings directly illustrate the survival benefit of PR-based proton pumping under natural illumination conditions.
Our data demonstrate that during respiratory challenges, light-driven proton pumping by PR can augment the cellular pmf to the extent that it powers cell motility and increases cell survival, yielding cells that can withstand respiratory poisons and oxygen depletion via light harvesting. Conservation of the basic features of energy metabolism in proteobacteria such as SAR86 and SAR11 makes it likely that PR expression will confer similar benefits to other members of this class and, perhaps, even to more distantly related Gram-negative bacteria. Therefore, it may be possible to synthesize a diverse array of light-powered bacteria for a variety of purposes via PR expression combined with modulation of the cell's natural pmf-generating mechanisms.
Materials and Methods
Cell Culture.
We expressed the SAR86 γ-proteobacterial PR-variant (Geneart, Toronto, ON, Canada) in E. coli cells (RP437 DE3 ΔcheY CmR) by using a T7-based expression system (pET200, KanR; Invitrogen, Carlsbad, CA). Cells were grown in T broth (1% tryptone/0.5% NaCl) supplemented with kanamycin (25 μg/ml). In mid-log phase, PR expression was induced with 1 mM isopropyl β-d-thiogalactoside and the medium was supplemented with 10 μM ethanolic all-trans-retinal. Cells were collected in late log phase by gentle centrifugation (4,500 × g for 5min) and carefully resuspended in motility medium (1 mM PBS (Ambion, Foster City, CA)/0.1 mM EDTA, pH 7.4). Throughout the article, PR+ denotes cells expressing proteorhodopsin, and PR− denotes cells (RP437 DE3 ΔcheY CmR) without the PR plasmid. PR− cells, unless otherwise noted, also were induced with isopropyl β-d-thiogalactoside, supplemented with all-trans-retinal and grown in T broth with chloramphenicol (25 μg/ml). Unless otherwise noted, all experiments were done in glucose-free motility medium at room temperature.
Instrumentation.
Throughout the article, power density values for “green light” refer to the power density passed by a D540/25× filter (Chroma, Rockingham, VT) and originating at a 175 W Xenon bulb (Lambda light source; Sutter Instruments, Novato, CA), or, for “red light,” to the power density passed by a HQ620/60× filter and originating at a 100 W Quartz Halogen bulb (Nikon). To visualize the cells, the sample chamber was illuminated continuously with faint red light (0.09 mW/cm2) at the tail of PR's absorption spectrum (4) (Fig. 1a). We periodically illuminated the sample chamber with bright green light (160 mW/cm2) coinciding with the maximum of PR's absorption spectrum, ≈525 nm (4). Cells were imaged at a framerate of 5 Hz by using an Andor (South Windsor, CT) iXon camera mounted to a Nikon TE2000 microscope. The Nikon microscope was modified for dark-field work by attaching a Zeiss 1.2–1.4 N.A. oil immersion dark-field condenser to the Nikon condenser turret by using a custom adapter. Custom software written in C++ was used to control the Andor camera, and Matlab was used to process the images. All errors and error bars represent standard errors of the mean.
Cell Viability Experiments.
PR+ and PR− cells were grown in LB plus Kanamycin (PR+) or LB plus Chloramphenicol (PR−) to an OD600 of 0.25–0.3, induced with 1 mM isopropyl β-d-thiogalactoside and supplemented with 10 μM all-trans-retinal (if retinal+), grown to an OD600 of 0.5–0.6, spun down (4,500 × g for 5 min), resuspended in motility buffer (1 mM PBS/0.1 mM EDTA) to an OD600 of 0.1–0.2. One to two milliliters of each culture was placed in a clear plastic culture tube, sodium azide was added to a concentration of 30 mM, and tubes were placed outside in the sun. Samples were taken before addition of azide (0 min) and after 30 min of exposure to sunlight. Fifty microliters of each sample was plated at a dilution of 1/104 (0 min) or 1/103 (30 min) onto LB with the appropriate antibiotic. Cell density at 0 min (before the addition of azide) was ≈3 × 106 colonies in 50 μl, a density that matches well with the measured OD600 of 0.1–0.2. Colonies were counted manually after overnight incubation. The one-sided Mann–Whitney U test (31) was used to test the null hypothesis that the ability to pump protons does not increase cell survival.
Supplementary Material
Acknowledgments
We thank Howard Berg (Harvard University) for suggesting the oxygen depletion experiment, Ed Delong (Massachusetts Institute of Technology) for encouragement and feedback, Sydney Kustu (University of California, Berkeley, CA) for her keen microbiological eye, and Andy Spakowitz (Stanford University). We thank the Zusman laboratory (University of California, Berkeley) for the gift of strain RP437. We also thank Ann McEvoy, Adam Politzer, and Elizabeth Kremen. J.M.W. was supported by the National Science Foundation for Graduate Research Support. This work was supported by the University of California, Berkeley (J.L.), the Hellman Faculty Fund (J.L.), the Sloan and Searle foundations (J.L.), and the Department of Energy Office of Science, Energy Biosciences Program (J.L. and C.B.).
Abbreviations
- pmf
proton motive force
- PR
proteorhodopsin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611035104/DC1.
References
- 1.Behrenfeld MJ, Falkowski PG. Limnol Oceanogr. 1997;42:1–20. [Google Scholar]
- 2.del Giorgio PA, Duarte CM. Nature. 2002;420:379–384. doi: 10.1038/nature01165. [DOI] [PubMed] [Google Scholar]
- 3.Danon A, Stoeckenius W. Proc Natl Acad Sci USA. 1974;71:1234–1238. doi: 10.1073/pnas.71.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, et al. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. Nature. 2002;420:806. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 7.Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 8.Sabehi G, Beja O, Suzuki MT, Preston CM, DeLong EF. Environ Microbiol. 2004;6:903–910. doi: 10.1111/j.1462-2920.2004.00676.x. [DOI] [PubMed] [Google Scholar]
- 9.Sabehi G, Massana R, Bielawski JP, Rosenberg M, Delong EF, Beja O. Environ Microbiol. 2003;5:842–849. doi: 10.1046/j.1462-2920.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 10.Man-Aharonovich D, Sabehi G, Sineshchekov OA, Spudich EN, Spudich JL, Beja O. Photochem Photobiol Sci. 2004;3:459–462. doi: 10.1039/b316071h. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre JR, Christianson LM, Beja O, Suzuki MT, Karl DM, Heidelberg J, DeLong EF. Proc Natl Acad Sci USA. 2003;100:12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, Isaacson T, Hirschberg J, Wagner M, Beja O. PLoS Biol. 2005;3:1409–1417. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang WW, Sineshchekov OA, Spudich EN, Spudich JL. J Biol Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, Rappe MS, Laney S, Wilhelm LJ, Tripp HJ, et al. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- 15.Macnab RM. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt FC, editor. Washington, DC: Am Soc Microbiol; 2005. Chap 10. [Google Scholar]
- 16.Schuster SC, Khan S. Annu Rev Biophys Biomol Struct. 1994;23:509–539. doi: 10.1146/annurev.bb.23.060194.002453. [DOI] [PubMed] [Google Scholar]
- 17.Racker E, Stoeckenius W. J Biol Chem. 1974;249:662–663. [PubMed] [Google Scholar]
- 18.Gabel CV, Berg HC. Proc Natl Acad Sci USA. 2003;100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung DC, Berg HC. Nature. 1995;375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 20.Berg HC, Tedesco PM. Proc Natl Acad Sci USA. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler J, Templeton B. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 22.Noumi T, Maeda M, Futai M. FEBS Lett. 1987;213:381–384. doi: 10.1016/0014-5793(87)81526-0. [DOI] [PubMed] [Google Scholar]
- 23.Dioumaev AK, Wang JM, Balint Z, Varo G, Lanyi JK. Biochemistry. 2003;42:6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkinson JS, Houts SE. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kita K, Konishi K, Anraku Y. J Biol Chem. 1984;259:3375–3381. [PubMed] [Google Scholar]
- 27.Setty OH, Hendler RW, Shrager RI. Biophys J. 1983;43:371–381. doi: 10.1016/S0006-3495(83)84360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harold FM, Maloney PC. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt FC, editor. Washington, DC: Am Soc Microbiol; 2005. Chap 19. [Google Scholar]
- 29.Maloney PC. J Bacteriol. 1979;140:197–205. doi: 10.1128/jb.140.1.197-205.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rotterdam BJ, Crielaard W, van Stokkum IH, Hellingwerf KJ, Westerhoff HV. FEBS Lett. 2002;510:105–107. doi: 10.1016/s0014-5793(01)03210-0. [DOI] [PubMed] [Google Scholar]
- 31.Conover WJ. Practical Nonparametric Statistics. 2nd Ed. New York: Wiley; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.