Abstract
The development of antimicrobials is critical in this time of increasing antibiotic resistance of most clinically relevant bacteria. To date, all current antibiotics focus on inhibiting crucial enzymatic activities of their protein targets (i.e., trimethoprim for dihydrofolate reductase), thus disrupting in vitro essential gene functions. In contrast, we have previously reported the identification of virstatin, a small molecule that inhibits virulence regulation in Vibrio cholerae, thereby preventing intestinal colonization in an infant mouse model for cholera. Virstatin prevents expression of the two major V. cholerae virulence factors, cholera toxin (CT) and the toxin coregulated pilus, by inhibiting the virulence transcriptional activator ToxT. It has previously been described that the N-terminal domain of ToxT has the ability to form homodimers. We now demonstrate that virstatin inhibits ToxT dimerization, thus demonstrating that it further falls into a unique class of inhibitors that works by disrupting protein-protein interactions, particularly homodimerization. Using virstatin, truncation mutants of ToxT, and a virstatin-resistant mutant, we show that dimerization is required for ToxT activation of the ctx promoter. In contrast, ToxT dimerization does not appear to be required at all of the other ToxT-regulated promoters, suggesting multiple mechanisms may exist for its transcriptional activity.
Keywords: antibiotics, cholera, pharmacology, regulation, virulence
Over the past several decades, much effort has been invested in the elucidation of virulence mechanisms used by many bacterial species to cause disease. These efforts have laid the groundwork for a new class of antibiotics that target regulation of these virulence mechanisms. Several recent reports have described the identification of many such drugs, including inhibitors of type III secretion, quorum sensing, and toxin activity (1–7).
We previously reported the identification of virstatin, a small molecule that inhibits Vibrio cholerae virulence regulation (2). V. cholerae is a Gram-negative, facultatively anaerobic pathogen that causes the diarrheal disease cholera by elaboration of two major virulence factors, cholera toxin (CT) and the toxin-coregulated pilus (8). In response to unknown stimuli in the host, a transcriptional cascade results in the expression of both virulence factors. The transcriptional factor ToxT is the most downstream regulator, directly activating ctxAB and the tcp genes. We previously identified ToxT to be the target of virstatin and isolated a virstatin-resistant mutant that contains a single leucine to proline point mutation at position 113 in the N-terminal domain of ToxT.
ToxT is a member of the large AraC/XylS family of transcriptional regulators defined by two conserved helix–turn–helix DNA-binding domains (9). In ToxT, these domains are located in the C-terminal portion of the protein and have been defined as the domains critical for both binding and activation of transcription by ToxT (10). The N-terminal portion of AraC/XylS transcriptional regulators displays a large amount of sequence diversity; however, it functions in oligomerization and binding of cofactors in some members of the family (11, 12). By analogy, it has long been speculated that the N-terminal domain of ToxT may result in dimerization as a prerequisite to activity. Prouty et al. (13) recently demonstrated the ability of the N-terminal domain to dimerize in heterologous systems; however, Withey et al. (14) raised the possibility that ToxT can bind and activate transcription as a monomer based on mapping of putative ToxT-binding sites at various ToxT-regulated promoters.
Based on the location of the mutation in the virstatin-resistant mutant of ToxT (L113P), we hypothesized that virstatin may inhibit dimerization of ToxT. In this study, we demonstrate that there is a direct correlation between the activity of ToxT at the ctx promoter and its ability to dimerize as determined by studies with ToxT truncation mutants. Furthermore, we show that virstatin inhibits dimerization of the N-terminal domains in a bacterial-2-hybrid system and by gel filtration chromatography. Finally, we use virstatin as a tool to regulate dimerization of ToxT to identify differences in the mechanism of ToxT at the various promoters that cannot be explained solely by the location and orientation of the ToxT-binding sites.
Results
The Transcriptional Activity of ToxT at the ctx Promoter Requires Amino Acids 10–276.
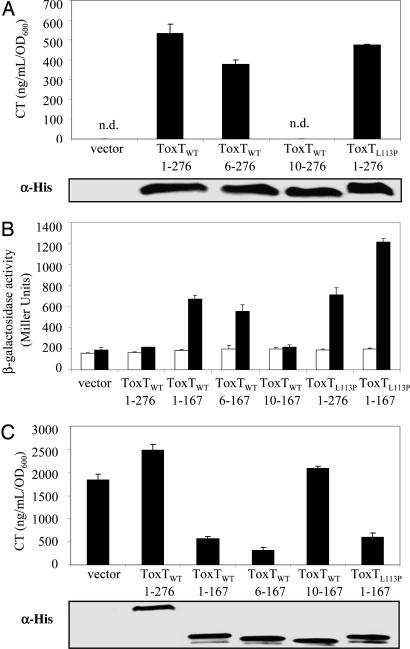
To characterize the N-terminal domain of ToxT with respect to activity, we determined the minimal sequence necessary for activation of the ctx promoter. We constructed truncation mutants of the ToxT N terminus, deleting the first five amino acids as well as the first nine amino acids and examined the ability of these mutants to complement a toxT deletion in V. cholerae classical strain O395. We induced expression of ToxT variants from a plasmid as C-terminal His6-tag fusions in strain O395ΔtoxT and assayed CT production by CT ELISA.
Expression of full-length ToxTWT (aa1–276), virstatin-resistant mutant ToxTL113P (aa1–276), and truncation mutant ToxTWT6 (aa6–276) produced similar quantities of CT, whereas expression of truncation mutant ToxTWT10 (aa10–276) did not complement the toxT deletion (Fig. 1A). Expression of all constructs was confirmed to be equal by Western blot analysis by using an anti-His antibody. These results suggest that the N-terminal amino acids 6–9 are necessary for ToxT activation of the ctx promoter.
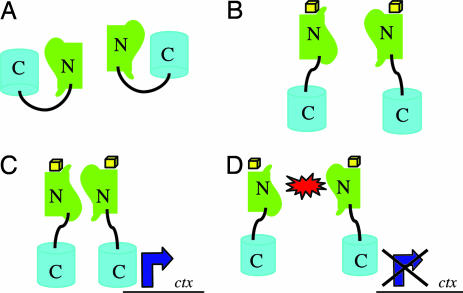
Fig. 1.
The N-terminal domain of ToxT dimerizes. (A) (Upper) Activity of ToxT at the ctx promoter when it is expressed from a plasmid in O395ΔtoxT as measured by CT ELISA. ToxTWT6 (aa6–276) is active, whereas ToxTWT10 (aa10–267) does not complement the toxT deletion. Full-length ToxT is 276 aa. (Lower) Western blot with α-His demonstrates that all portions of ToxT were expressed to equal levels. (B) Portions of, but not full-length, ToxT drive a protein–protein interaction in a bacterial two-hybrid system in E. coli when induced with IPTG as measured by β-galactosidase assay. Truncation of the first five amino acids allows lacZ transcription, whereas truncation of the first nine abolishes activity. Full-length and N-terminal portion of ToxTL113P point mutant (2) are able to dimerize in this system. Vector control is a transformant containing empty pACTrAp-Zif and pBRGpω plasmids. White, no IPTG; black, 10 μM IPTG. (C) (Upper) Only N-terminal portions of ToxT that can induce a protein–protein interaction can exert a dominant negative effect on CT production. ToxT truncations were expressed from a plasmid in wild-type O395. (Lower) Western blot with α-His demonstrates that all N-terminal portions were expressed to equal levels.
The N-Terminal Domain of ToxT Is Able to Dimerize.
On the basis of an analogy to other AraC/XylS family transcriptional regulators, it has been proposed that dimeric ToxT is required to activate transcription and that the capacity to form dimers is encoded in the N-terminal domain (13). Using a bacterial two-hybrid system in Escherichia coli, we confirmed the ability of the N-terminal domain to dimerize. We constructed various translational fusions of ToxT with a zinc-finger DNA-binding protein (Zif) and with the ω subunit of the E. coli RNA polymerase. The ability of the various ToxT variants to recruit RNA polymerase to Zif was assayed by using the transcriptional lacZ reporter gene (15).
The N-terminal domain of ToxT (aa1–167, chosen based on an analogy to the N-terminal domain of AraC), when fused to both Zif and the ω subunit of the E. coli RNA polymerase, activated lacZ transcription strongly, thus demonstrating its ability to homodimerize (Fig. 1B). We also examined ToxTL113P, previously identified in a screen for virstatin-resistant ToxT mutants (2), in the two-hybrid system and discovered that its N-terminal domain (aa1–167) also homodimerized and, in fact, resulted in increased β-galactosidase relative to wild-type. Translational fusions of full-length ToxTWT were unable to dimerize, whereas the full-length ToxTL113P dimerized effectively although not as well as its N-terminal domain alone. The fact that full-length ToxTL113P dimerizes but not ToxTWT suggests that the conformations of the fusion proteins differ. It seems less likely that the inability of the full-length wild-type fusion to dimerize is an artifact of the two-hybrid system. It is possible that a conformational change may be required for full-length ToxT to dimerize after some activation step such as binding a small molecule activator. Even in the absence of an inducer, ToxTL113P may be in a conformation more conducive to dimerization than ToxTWT, although the increased β-galactosidase activity of the ToxTL113P N-terminal domain over full-length suggests that a small conformational change may still be necessary.
Finally, to examine the sequence requirements for transcriptional activity in V. cholerae compared with those for dimerization, we tested the ability of the truncation mutants in both the N-terminal and C-terminal ends to activate transcription in the two-hybrid system. Truncation of ToxT from the C-terminal end before amino acid 167 results in a gradual loss of dimerization ability (data not shown). Although truncating the first five amino acids at the N terminus (aa6–167) led to only a slight decrease in β-galactosidase activity, truncating the first nine amino acids (aa10–167) completely abolished the interaction (Fig. 1B). These results directly correlate with those obtained in the CT ELISA for ToxT activity. Thus, the ability of the N-terminal domain of ToxT to dimerize in the two-hybrid system requires the same amino acids (6–9) that are required for ToxT activation of the ctx promoter in V. cholerae.
The N-Terminal Domain of ToxT Behaves as a Dominant Negative for Cholera Toxin Expression.
Having demonstrated the ability of the N-terminal domain of ToxT to form homodimers in a heterologous system, we examined the ability of the N-terminal domain of ToxT to behave as a dominant negative in V. cholerae strain O395. We expressed various ToxT mutants from a plasmid in O395 and measured activity by CT ELISA. Although expression of the empty plasmid or full-length ToxT resulted in normal CT production, expression of the N-terminal domains of both ToxTWT and the ToxTL113P mutant significantly reduced CT production (Fig. 1C). This result demonstrates that the N-terminal domain is able to form a heterodimer with and sequester the wild-type, full-length ToxT expressed by O395 and thus prevent normal transcriptional activation.
In concordance with the observations made in the bacterial-2-hybrid system, expression of the aa6–167 fragment of ToxT produced the dominant negative effect, whereas expression of the aa10–167 fragment had no effect on CT production. Again, the same amino acids (6–9) that were required for dimerization in our heterologous E. coli system and the activation of ctx transcription in V. cholerae are also required for the dominant negative effect in V. cholerae. We confirmed that all N-terminal constructs were expressed equally by performing Western blot analysis against a C-terminal His6-tag (Fig. 1C).
Virstatin Inhibits the Ability of ToxT to Form Dimers.
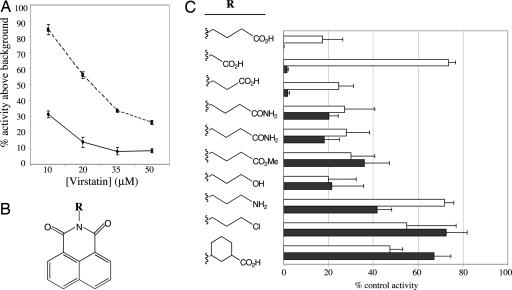
As a result of the position of the virstatin-resistant mutation at position 113 in the N-terminal domain, we hypothesized that virstatin may inhibit the dimerization of ToxT. We adapted the bacterial-2-hybrid system to test the effect of virstatin on dimerization by deleting tolC in the E. coli reporter strain KDZif1Z to increase the cellular permeability of virstatin. We compared the effects of virstatin on dimerization of the N-terminal domains of ToxTWT and ToxTL113P. KDZif1ZΔtolC carrying plasmids that express N-terminal domain fusions of either ToxTWT or the ToxTL113P mutant were grown overnight in the presence of varying concentrations of virstatin. These samples were then subcultured into media containing virstatin and IPTG to induce expression of the chimeric proteins and assayed for ability to induce expression of the β-galactosidase reporter.
Virstatin inhibited dimerization of the ToxTWT N-terminal domain fusion even at 10 μM, and at higher concentrations (20, 35, and 50 μM), it reduced transcriptional activity to baseline levels (Fig. 2A). In contrast, the ToxTL113P N-terminal domain fusion was relatively more resistant to virstatin than wild-type. At 10 μM, virstatin had no effect on the amount of β-galactosidase activity of the ToxTL113P mutant. Even at higher concentrations, virstatin only partially reduced the β-galactosidase activity of the mutant. Because the mutant also appears to dimerize more efficiently than wild-type, this stronger interaction could compete out virstatin binding, thus resulting in its relative resistance to virstatin. These results further demonstrate a strong correlation between the ability to dimerize and the previously reported transcriptional activity of ToxTWT and ToxTL113P in the presence and absence of virstatin.
Fig. 2.
(A) Virstatin inhibits ToxT dimerization in bacterial two-hybrid system in E. coli. The addition of increasing concentrations of virstatin inhibited the protein–protein interaction of the ToxT N terminus with itself as measured by β-galactosidase assay. Point mutant ToxTL113P was more resistant to virstatin. Activity is presented as a percentage of reporter activity above background in the presence of virstatin compared with no virstatin. Solid line, ToxT; dashed line, ToxTL113P. (B) Chemical structure of virstatin base with the R group labeled in bold. (C) Inhibition of ToxT dimerization in the bacterial two-hybrid system and CT production in V. cholerae by analogs of virstatin is correlated. Each compound is designated by its R group and inhibition is graphed as the percentage of control activity. Virstatin is the first compound listed. White, β-galactosidase production; black, CT.
To further define the relationship among virstatin, ToxT dimerization, and ToxT transcriptional activity, we analyzed the ability of several virstatin structural variants to inhibit dimerization and ToxT activity at the ctx promoter. Reporter strains KDZif1ZΔtolC containing wild-type N-terminal domain fusions were grown in the presence or absence of 50 μM virstatin or its structural variants and assayed for β-galactosidase activity. The amount of N-terminal domain dimerization in the presence and absence of the structural variants correlates for the most part with the transcriptional activity of ToxT in the presence of 20 μM virstatin and its variants as assayed by CT ELISA (Fig. 2B). A few virstatin variants (e.g., the carboxylate series) showed more activity in inhibiting ToxT-mediated transcription in V. cholerae than in blocking ToxT dimerization in the E. coli-based two-hybrid system; however, these exceptions may be the result of differences in the permeability or metabolism of molecules with different chemical structures in the two different bacterial species.
Virstatin Favors the Monomeric Form of ToxT.
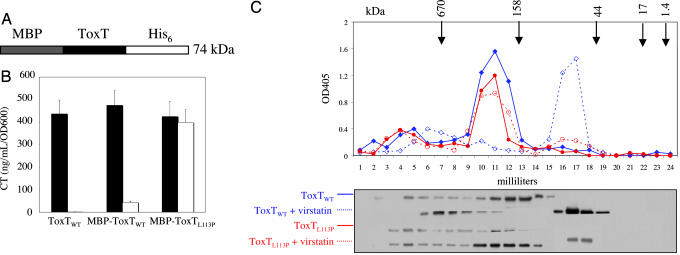
We determined the oligomeric state of ToxT in the presence and absence of virstatin by gel filtration chromatography. We constructed a 74-kDa fusion protein of an N-terminal maltose-binding protein (MBP) and a C-terminally His6-tagged ToxTWT or ToxTL113P (Fig. 3A). The fusion was more soluble than ToxT alone. Both the ToxTWT and ToxTL113P fusions complemented a toxT deletion in V. cholerae (O395ΔtoxT) as well as ToxT wild-type and could be inhibited by virstatin as shown by CT ELISA (Fig. 3B).
Fig. 3.
Virstatin favors ToxT monomers. (A) MBP–ToxT–His6 fusion construct. (B) The MBP–ToxT fusions complement a toxT deletion in O395 as well as wild-type ToxT. MBP–ToxTWT is inhibited by virstatin, whereas MBP–ToxTL113P is resistant to virstatin. No virstatin, black; virstatin, white. (C) (Upper) ToxTWT and ToxTL113P amounts in the absence and presence of virstatin in FPLC fractions are quantified by ELISA using Ni+2-coated plates and an anti-MBP antibody (molecular mass standards are indicated with arrows above the graph). (Lower) Western blot analysis of each of the fractions was performed by using an α-MBP antibody, demonstrating that monomeric ToxTWT can be isolated only in the presence of virstatin (bands shown are all 74 kDa).
The wild-type and mutant fusions were expressed in O395 from a plasmid driven by a Ptac-inducible promoter and induced with IPTG in the presence or absence of virstatin. All subsequent purification steps were carried out in the presence or absence of virstatin (100 μM) corresponding to their respective expression conditions. The ToxT fusions were purified from O395 on a Ni2+ column and protein concentrations of the eluted proteins were normalized by Bradford assay. The Ni2+ purified fusion proteins were then run on a Superdex 75 10/300 column (Amersham, Biosciences, Piscataway, NJ). The amount of the MBP–ToxT–His6 fusion in each fraction was assayed by sandwich ELISA with a Ni2+-coated plate and an α-MBP antibody. We also performed Western blot analysis of individual fractions by using an α-MBP antibody that demonstrated an isolated 74-kDa band at varying molecular weights suggestive of varying oligomerization states.
Both ToxTWT and ToxTL113P fusion proteins expressed and isolated in the absence of virstatin ran at relatively high molecular weights by gel filtration, likely representing higher oligomers. SDS/PAGE and Western analysis confirmed that these higher molecular weight fractions in fact consisted of the 74-kDa ToxT fusion. In contrast, the ToxTWT fusion protein in the presence of virstatin (100 μM) resulted in an ≈74-kDa peak corresponding to the monomer. Only a very minor peak at ≈74 kDa was produced with the ToxTL113P fusion protein in the presence of virstatin (Fig. 3C). Because the oligomerization state may be altered through the isolation steps, caution must be taken in extrapolating these results to the state of ToxT within a cell. Nevertheless, these results demonstrate that virstatin allows the ToxTWT fusion to remain as a monomer compared with the higher oligomers isolated in the absence of virstatin. Furthermore, the inability to isolate a significant amount of the monomer of the ToxTL113P fusion is consistent with the mutant's relative resistance to virstatin and the mechanism of virstatin inhibiting ToxT dimerization.
The Oligomerization State of ToxT Required for Transcriptional Activation Varies at Different Promoters.
Recently, there have been several reports published describing the ToxT-binding sites at various promoters (10, 14, 16). It has been suggested that dimerization of ToxT may not be required to activate all of these promoters or that ToxT may at least have the capacity of binding to some of them as a monomer. We used virstatin as a tool to prevent dimerization and determined the ability of monomeric ToxT to activate various transcriptional reporters.
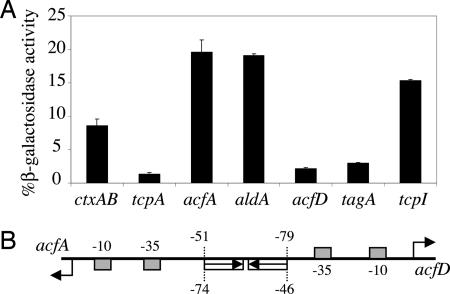
Transcriptional reporters fusing the promoters of ctxAB, tcpA, acfA, aldA, acfD, tagA, and tcpI to the lacZ were introduced into V. cholerae strain O395ΔlacZ. These reporter strains were grown under virulence-inducing conditions (Luria broth at pH 6.5, 30°C) in the presence or absence of virstatin (100 μM) and assayed for β-galactosidase activity. We discovered a variation in virstatin-mediated repression between several of the promoters tested. Two classes of promoters were identified. Virstatin-sensitive promoters included tcpA, acfD, ctxAB, and tagA that were repressed the most efficiently to 3% to 8% of the control activity in the presence of virstatin. A second class of promoters included tcpI, acfA, and aldA, which were repressed to a lesser extent, ≈15–20% of control activity (Fig. 4A). This latter class may be characterized by aldA, which has been shown to contain only one ToxT-binding site (14, 17), thus consistent with the possibility that monomeric ToxT is able to activate this promoter. In contrast, the canonic ToxT-binding motif in the acfA and tcpI promoters do not differ significantly in sequence from the promoters in the first class and yet they show relative virstatin indifference to the same extent as aldA.
Fig. 4.
ToxT function varies at different promoters. (A) Virstatin inhibited activity of ToxT at the ctxAB, tcpA, acfD, and tagA promoters to a greater extent than at the acfA, aldA, and tcpI promoters. Activity at each of the promoters was assayed by measuring β-galactosidase activity by using a lacZ transcriptional reporter in O395ΔlacZ. Data are presented as the percentage of activity in the presence of 100 μM virstatin compared with control activity. (B) Map of ToxT-binding sites at acfA and acfD promoters.
Most surprisingly, the sensitivity of the acfA and acfD promoters to virstatin appeared to differ, despite the previous assumption that these two promoters share two common ToxT-binding sites located between them and thus were likely regulated in the same manner by ToxT (Fig. 4B). This variation in virstatin sensitivity raises the possibility that the mechanism of ToxT may vary at the different promoters and that differences in orientation and location of the binding sites alone are not sufficient to distinguish these promoters.
Discussion
In the more than 20 years since the discovery of cholera toxin and its role in causing cAMP-mediated secretory diarrhea, much attention has focused on understanding the regulatory cascade that governs virulence expression in V. cholerae. Many transcriptional regulators have been identified, including ToxT, which is the direct activator of ctx and tcp expression. However, many details about ToxT's mechanism of transcriptional activation remain unclear. Recent research has focused on understanding the details of the binding of ToxT and activation of its responsive promoters (10, 13, 14).
We made a series of N-terminal truncation mutants of ToxT and measured their ability to activate ctx transcription as determined by CT ELISA. We found that deletion of the first five residues is tolerated but that deletion of the first nine amino acids abrogates activity. This finding is reminiscent of AraC, in which the N terminus is required to bind arabinose to induce a conformational change resulting in transcriptional activation (18) and raises the consideration that the N terminus of ToxT may similarly bind a small molecule effector.
The recent identification of a conserved ToxT DNA-binding sequence, termed the toxbox, has raised the question of whether ToxT requires dimerization for activity in analogy to the other members of the AraC/XylS family of transcriptional regulators (14). The identification of a single toxbox site upstream of aldA as well as differentially spaced and oriented binding sites upstream of ctxAB, tcpA, acfA, acfD, tagA, and tcpI have suggested the possibility that ToxT may be able to either bind DNA or activate transcription simply as a monomer.
To examine the relationship between dimerization and activity, we used a bacterial-2-hybrid system to study ToxT and its truncation mutants. We found that the N-terminal domain (aa1–167) dimerized in this system in contrast to full-length ToxTWT (aa1–267), which did not. Although this may be the result of conformational constraints within this hybrid system, it appears unlikely because the full-length ToxTL113P was able to dimerize effectively, suggesting possible differing conformations of the two full-length fusions. We further found that the same amino acids (6–9) that are critical for activity had a similar effect on dimerization, drawing a strong correlation between the ability to dimerize and the activity of ToxT. The ability of the N-terminal domain to act as a dominant negative in V. cholerae O395 further demonstrates that inhibition of full-length ToxT homodimerization results in loss of activity.
Having previously identified a point mutation conferring resistance to virstatin within the N-terminal, putative dimerization domain of ToxT, we hypothesized that virstatin could be inhibiting dimerization of ToxT, resulting in loss of ToxT activation of the ctx and tcp promoters. We demonstrated that this is indeed the mechanism of virstatin by showing that it inhibits the dimerization of ToxT in the bacterial two-hybrid system, whereas the virstatin-resistant ToxT mutant is able to dimerize regardless of its presence. Using this system, we explored analogs of virstatin that have various levels of inhibitory activity in V. cholerae and, as predicted, found a correlation between inhibition of transcription and dimerization.
A biochemical approach also confirmed this mechanism for virstatin. Using gel filtration chromatography, we showed that ToxT purified in the presence of virstatin favors the monomeric form, whereas ToxT purified in the absence of virstatin aggregates into oligomeric complexes. Although this result must be interpreted with caution because the oligomeric states of ToxT may vary with the purification process, nevertheless, the fact that a monomeric form of ToxT could be observed in the presence but not the absence of virstatin shows that virstatin does inhibit protein–protein association.
On the basis of the evidence correlating the ability of ToxT to dimerize and activate transcription, we propose a model of ToxT activation and inhibition with virstatin at the ctx promoter (Fig. 5). By analogy to AraC, ToxT may require the binding of a small molecule or protein inducer to activate transcription. We posit an inactive state of ToxT in which the C-terminal domain blocks the dimerization site on the N-terminal domain in the absence of inducer. Binding of the hypothetical inducer at either the N or C terminus allows a conformational change exposing the dimerization site. In the absence of virstatin, this “open” conformation of ToxT favors homodimerization and transcriptional activation of the ctx promoter. Although there may be some other type of conformational restraint preventing dimerization of full-length ToxT in the bacterial two-hybrid system, the proposed model explains our result that only the N-terminal domain of wild-type ToxT is able to dimerize; the absence of inducer would favor the inactive conformation of ToxT, thus preventing dimerization and transcriptional activation. Virstatin inhibits the dimerization of the wild-type N-terminal domain and the dimerization of ToxT in its open, activated conformation. The virstatin-resistant mutation L113P may favor an open conformation of full-length ToxT, allowing it to dimerize in the bacterial two-hybrid system.
Fig. 5.
Model of ToxT activation of the ctx promoter and inhibition with virstatin. (A) The C-terminal domain of ToxT blocks the homodimerization site on the N-terminal domain in the absence of a hypothetical small molecule activator. (B) In V. cholerae, this putative activator (cube) binds to either the N- or C- terminal domains, resulting in a conformational change in ToxT, which exposes the dimerization site. (C) Dimerization of ToxT occurs, allowing binding and transcriptional activation of the ctx promoter. (D) Virstatin (red star) prevents dimerization of the N-terminal domains and transcription at the ctx promoter.
Finally, using virstatin as an inhibitor of dimerization, we were able to address differences in promoters activated by ToxT. We discovered that the relationship between the oligomerization state of ToxT and promoter structure is a complex one. The ctx and tcp promoters are ToxT dimerization-dependent based on previous work demonstrating the sensitivity of CT and toxin co-regulated pilus expression to virstatin. In contrast, it has been suggested that the aldA promoter contains a single ToxT-binding site unlike the other ToxT-regulated promoters. Its relative virstatin insensitivity would be consistent with this finding. The lack of correlation, however, between the other promoter structures and their relative virstatin sensitivity, in particular acfA and acfD, which share common ToxT-binding sites, indicate that there is still much to be understood about how ToxT functions.
We have shown that virstatin inhibits virulence expression by inhibiting ToxT dimerization and thus its ability to activate the ctx and tcp promoter. There has been significant interest in finding small molecules that inhibit protein–protein associations because it is such a central tenet of biology. Although there are a growing number of examples of such small molecules (19–21), overall, it has continued to be challenging to find such inhibitors, in part as a result of the large surface areas and the often flat binding surfaces that occur at the interface of such interactions. However, the recognition of “hot spots” on protein interfaces, whereby small molecules that bind at these hot spots are capable of disrupting these interactions, has resulted in the continued appeal of such an approach. The finding that virstatin inhibits dimerization of a transcriptional activator of virulence places it in a very unique class of inhibitors and opens the door to a novel approach to antibiotic discovery by targeting protein–protein interactions critical to virulence.
Materials and Methods
Bacterial Strains and Plasmids.
Classical biotype strain O395 was used in all experiments. Virulence-inducing growth conditions were obtained by 1:1,000 dilution of an overnight culture into LB pH 6.5 and growth at 30°C shaking 260 rpm (Gyratory Shaker; New Brunswick Scientific, Edison, NJ). The method of Skorupski and Taylor (22) was used to construct in-frame deletions of all mutants in V. cholerae. Laboratory stocks of E. coli DH5αλpir and SM10λpir were used for cloning and mating into V. cholerae, respectively.
Bacterial Two-Hybrid System.
Experiments were carried out as described (15). Plasmids and strains were a gift of Dr. Simon Dove. Briefly, identical portions of toxT were cloned between the NdeI and NotI sites of pACTrAp-Zif and pBRGpω. All cloning was done in strain DH5αF'IQ. The constructs were then co-transformed into the E. coli strain KDZif1ZΔtolC. Cultures were grown overnight in the absence or presence of 10 μM IPTG and/or 50 μM virstatin and subcultured into identical conditions in the morning. Activation of the reporter was assayed in triplicate by β-galactosidase assay. A representative experiment is shown.
Detection of Cholera Toxin Expression in O395/O395ΔtoxT Expressing ToxT Variants Under Control of the Heterologous pBAD Promoter.
All portions of toxT were C-terminally His6-tagged and cloned into pBAD18. The resulting plasmids were transformed into O395 (dominant negative experiments) or O395ΔtoxT (complementation experiments) (23). The strains were inoculated into LB pH 6.5 and grown at 30°C shaking in the presence of either 0.01% (dominant negative experiments) or 0.005% (complementation experiments) arabinose and GM1 ganglioside enzyme-linked immunosorbent CT assays (24) were performed as previously described on the resulting supernatants. CT expression was normalized for OD600 of the growing culture and is the result of samples done at least in triplicate. A representative experiment is shown.
α-His Western Detection of ToxT Variants.
Cells were grown in LB at 37°C shaking overnight in the presence of 0.1% arabinose. Cell extracts were subjected to SDS/PAGE, transferred to PVDF membrane, probed with anti-tetra-His antibody (Qiagen, Valencia, CA), and visualized by enhanced chemiluminescence (Amersham Pharmacia).
Virstatin Structural Variant Synthesis.
A typical procedure for the synthesis of the virstatin analogs is as follows (25). A mixture of 1,8-naphthalic anhydride (9.7 mmol) and the functionalized amines (9.7 mmol) in DMF (25 ml) was heated at 120°C for 2–6 h. On completion of the reaction (by TLC), the reaction mixture was cooled to room temperature and a half of DMF was evaporated in vacuo; then Et2O (50 ml) was added, resulting in a white precipitate, which was collected by filtration. The crude product was recrystallized from EtOAc to afford the desired virstatin analogs (47–93% yield).
Gel Filtration Studies.
To construct the MBP–ToxT fusion proteins, full-length (aa1–276) toxTWT and toxTL113P were C-terminally His6-tagged and cloned between XbaI and PstI of pMALc2x (Invitrogen, Carlsbad, CA). The resulting plasmids were transformed into O395ΔtoxT. An overnight culture of the ToxT fusions (wild-type and L113P mutant) was inoculated at a 1:500 dilution into 500 ml of LB in the presence or absence of 100 μM virstatin. The cultures were grown with shaking at 37°C for 4 h and then cooled to 18°C. The cultures were induced with IPTG (0.5 mM) and grown for 12 h at 18°C with shaking. The cultures were pelleted by centrifugation, resuspended in 10 ml of lysis buffer (500 mM NaCl/50 mM Tris, pH 7.5/15 mM imidazole/10% glycerol/40 μg/ml lyzozyme/1 mM PMSF/protease inhibitors/2 mM β-mercaptoethanol) with or without 100 μM virstatin, and flash-frozen at −80°C overnight. Extracts were thawed, sonicated, and pelleted at 12,000 rpm (Sorvall SS34 rotor; Thermo Fisher Scientific, Waltham, MA) for 30 min. The fusion proteins were purified over a nickel column (Invitrogen) and eluted with 250 mM imidazole with or without 100 μM virstatin. The purity of the proteins was confirmed by SDS/PAGE electrophoresis with Coomassie staining and Western blot analysis by using an α-MBP antibody (Invitrogen). The amounts of each fusion were normalized by using the Bradford assay (Bio-Rad, Hercules, CA). The aggregation states of the fusions were analyzed by gel filtration chromatography on a Superdex 75 10/300 column (Amersham).
The amount of fusion protein in each 1-ml fraction was determined by sandwich ELISA by using Ni-coated 96-well plates and an α-MBP primary antibody (Invitrogen), followed by an alkaline phosphatase conjugated goat-anti-rabbit secondary antibody. Western blot analysis was also performed on each of the 1-ml fractions by using an α-MBP antibody.
Transcriptional Reporter Assays.
Transcriptional fusions of tcpA, tcpI, acfD, acfA, tagA, and aldA promoters to lacZ in pTL61T were a gift of J. Withey and V. DiRita (University of Michigan Medical School, Ann Arbor, MI). The ctx reporter was constructed by cloning a 500-bp region upstream of ctxA from O395 between the HindIII and XbaI sites of pTL61T. All plasmids were transformed into O395ΔlacZ. The strains were inoculated into LB (pH 6.5) and grown at 30°C shaking overnight in the presence or absence of 100 μM virstatin. The resulting cultures were assayed in triplicate by β-galactosidase assay. A representative experiment is shown.
Acknowledgments
E.A.S. was supported by a National Science Foundation predoctoral fellowship. D.T.H. was supported by National Institutes of Health (NIH) Grant K08 AI060708-01. This work was supported by NIH Grant AI26289 (to J.J.M.).
Abbreviations
- CT
cholera toxin
- Zif
zinc-finger DNA-binding protein
- MBP
maltose-binding protein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hentzer MWH, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, et al. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 3.Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M, Hagglund U, Uvell H. Infect Immun. 2005;73:3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panchal RG, Hermone AR, Nguyen TL, Wong TY, Schwarzenbacher R, Schmidt J, Lane D, McGrath C, Turk BE, Burnett J, et al. Nat Struct Mol Biol. 2004;11:67–72. doi: 10.1038/nsmb711. [DOI] [PubMed] [Google Scholar]
- 5.Roychoudhury S, Z NA, Ninfa AJ, Allen NE, Jungheim LN, Nicas TI, Chakrabarty AM. Proc Natl Sci Acad USA. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright JJR, III, Novick RP. Proc Natl Sci Acad USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K, Bergstrom S, Elofsson M, Wolf-Watz H, Normark S, Henriques-Normark B. Proc Natl Acad Sci USA. 2006;103:14566–14571. doi: 10.1073/pnas.0606412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldor MM, Mekalanos JJ. In: Enteric Infections and Immunity. Paradise L, editor. New York: Plenum; 1996. pp. 37–55. [Google Scholar]
- 9.Higgins DE, Nazareno E, DiRita VJ. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulbert RR, Taylor RK. J Bacteriol. 2002;184:5533–5544. doi: 10.1128/JB.184.20.5533-5544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin RG, Rosner JL. Curr Opin Microbiol. 2001;4:132–137. doi: 10.1016/s1369-5274(00)00178-8. [DOI] [PubMed] [Google Scholar]
- 13.Prouty MG, Osorio CR, Klose KE. Mol Microbiol. 2005;58:1143–1156. doi: 10.1111/j.1365-2958.2005.04897.x. [DOI] [PubMed] [Google Scholar]
- 14.Withey JH, DiRita VJ. Mol Microbiol. 2006;59:1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 15.Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Proc Natl Acad Sci USA. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu RR, DiRita VJ. Mol Microbiol. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
- 17.Withey JH, Dirita VJ. J Bacteriol. 2005;187:7890–7900. doi: 10.1128/JB.187.23.7890-7900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon KP, Lee NL. Proc Natl Acad Sci USA. 1990;87:3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fry DC. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 20.Arkin MR, Wells JA. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 21.Toogood PL. J Med Chem. 2002;45:1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 22.Skorupski K, Taylor RK. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 23.Hase CC, Mekalanos JJ. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardel CL, Mekalanos JJ. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 25.Adam W, Beck AK, Pichota A, Saha-Moller CR, Seebach D, Vogl N, Zhang R. Tetrahedron Asymmetry. 2003;14:1355–1361. [Google Scholar]