Abstract
We have developed a two-component system involving reconstituted caspase (recCaspase) for selective and/or conditional ablation of targeted cells. Caspases, the executioners of programmed cell death, are normally synthesized as inactive zymogens and are activated by proteolytic processing of their subunits. We show here, using two different caspases, Caenorhabditis elegans CED-3 and human Caspase-3, that coexpression of the subunits generates constitutively active caspase activity that leads to cell death. This recCaspase activity, however, occurred only when the subunits associated through binding of linked antiparallel leucine-zipper domains. We exploited the dual-component nature of recCaspases by expressing the individual subunits from combinations of promoters either to target selectively the subset of cells for apoptosis or induce cell death in specific cells at specific times during development. The high degree of target specificity and tight regulation of induction of recCaspase would be advantageous in creating animal models that are ablated for specific cells and in other targeted cell killings.
Keywords: apoptosis, Caenorhabditis elegans, dual-component system, genetic ablation
Targeted cell killing is a powerful method to study cell function and has been suggested as a possible strategy to remove unwanted cells, e.g., tumor cells. Laser ablation has been extensively used in model organisms to study the role of specific cells on the development and behavior of the animals (1–3). Genetic ablation is potentially a much more efficient method of removing cells, because large numbers of animals lacking specific cells could be generated. Genetically ablated lines can be maintained indefinitely, shared within the research community, and used for population assays and genetic screens. Genetically ablated animals can be made by several methods, which use cell-specific promoters to express cytotoxic products (4–10). However, cell-specific promoters are rare, and even when available, are associated with some leakiness that may be unsuitable for expressing highly toxic gene products. To overcome the limitations of single-component systems to genetically ablate cells, we have developed a two-component system based on the cell killing activity of caspases.
Caspases are cysteine proteases that play a central role in the process of programmed cell death (11). All caspases are synthesized as inactive procaspase zymogens that comprise of three domains, a prodomain and the two catalytic subunits. Proteolytic cleavage at the conserved aspartate residue(s) present in the linker region connecting the two catalytic subunits and between the prodomain and the large subunit leads to generation of the active heterotetrameric caspase consisting of two copies of their large and small subunits (12–15).
The apoptotic pathway is conserved in metazoans from nematodes to humans (16, 17). Genetic analysis of the programmed cell death pathway in the nematode Caenorhabditis elegans has identified a single gene, ced-3 that encodes the apoptotic caspase responsible for the majority of the programmed cell death (18–20). CED-3 activation requires the adaptor protein CED-4, an ortholog of mammalian Apaf-1 (21–23). CED-3 contains a large prodomain that interacts with CED-4, and the CED-4-mediated oligomerization of CED-3 procaspase leads to autoproteolytic activation (24).
Mammals have 11 caspases, seven of which are known to form a caspase cascade that executes programmed cell death. Based on their role in the caspase cascade, the apoptotic caspases are broadly classified into two groups: the upstream or the initiator caspases, such as caspases-2, -8, -9, and -10, and the effector or the executioner caspases, such as caspases-3, -6, and -7 (14, 15). The initiator caspases, once activated, cleave and activate the downstream caspases, which cleave various cellular proteins leading to cell death.
Despite differences in the mechanism of activation of various caspases, the basic structure of all activated caspases is essentially the same: The activated caspases are heterotetramers consisting of two active sites each formed by a pair of large and small subunits. In this article, we demonstrate reconstitution of caspase activity from individually expressed subunits by assisted oligomerization, forming what we have termed reconstituted caspase (recCaspase). In addition, we have used the dual-component nature of the recCaspase to selectively target specific cells for apoptosis by expressing the two subunits from appropriate combination of promoters that show overlapping expression only in those specific cells. Further, using a combination of a heat-shock promoter and a cell-specific promoter, we show that cell death can be induced at specific times during development.
Results
In Vivo Reconstitution of CED-3 Activity from Its Subunits.
The ced-3 gene encodes the only apoptotic caspase in C. elegans (18–20). The CED-4 protein mediates oligomerization of CED-3 procaspase, which leads to autoproteolytic activation (24) to produce the large (p17) and the small (p15) subunits (Fig. 1A). To test whether CED-3 caspase activity resulted from the expression of the individual subunits in the same cell, we expressed the small (p15) and the large (p17) subunits of CED-3 (20) (Fig. 1A) separately from the mec-18 promoter, which is active only in six touch receptor neurons of C. elegans (G. Gu and M.C., unpublished data). The DNAs for both subunits were injected into a strain whose touch receptor neurons were labeled with GFP, so that the loss of the cells could be monitored by the loss of GFP fluorescence. No cell loss was detected in transformed animals (three stable lines; 25 animals per line), suggesting that the expression of the caspase subunits by themselves cannot cause death of the touch receptor neurons (Fig. 1G). Because C. elegans transformation usually results in the production of extrachromosomal arrays that can be lost during cell division, we created two lines in which the transformed DNA is integrated into one of the C. elegans chromosomes. No touch receptor loss was detected in either of the integrated lines (Fig. 1 D and G; 50 animals per line).
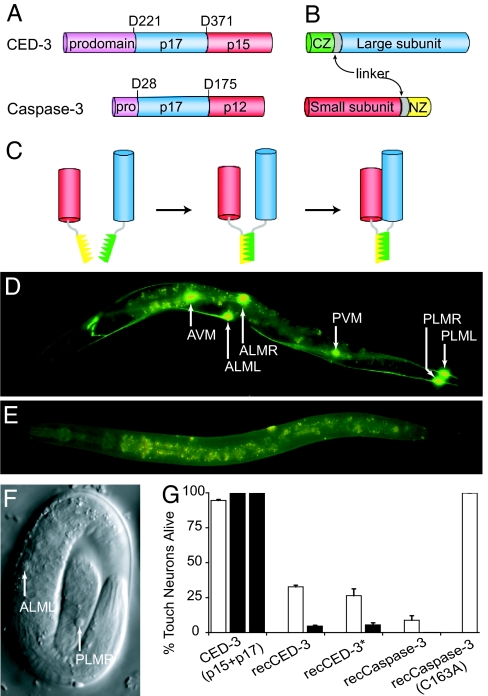
Fig. 1.
Cell killing activity of recCaspases in C. elegans. (A–C) Schematics of reconstitution of activated caspase from its subunits. (A) Subunit organization in the CED-3 and caspase-3 procaspases. The aspartate residues that are cleaved for activation are shown. (B) recCaspase components. Antiparallel leucine-zipper domains NZ (yellow) and CZ (green) are attached to the small (red) and large (blue) caspase subunits through a linker sequence (gray). (C) Formation of recCaspase. Interaction between the leucine-zipper domains leads to oligomerization of the caspase subunits that, in turn, results in the association and activation of caspase subunits. The precise subunit composition of the final recCaspase is not known. (D–G) Cell killing by recCaspases in C. elegans. Fluorescent images of adult worms expressing either the CED-3 subunits without the leucine-zipper sequences (p17 + p15) (D) or the recCED-3 subunits from touch neuron-specific mec-18 promoter (E). The GFP-positive touch receptor neurons are indicated by arrows. (F) Differential interference contrast (DIC) image of an embryo showing recCED-3 mediated apoptosis of an ALM and a PLM neuron. (G) Percentage of touch receptor neurons remaining in adults expressing the CED-3 subunits (p15 + p17) and recCaspases. GFP-positive cells were counted in 1-day-old adults expressing recCaspase from stable line with extragenic arrays of the constructs (open bars) or from the chromosomally integrated arrays (filled bars). Three stable lines or three different experiments in case of integrated lines were used to calculate mean and SEM. recCED-3* contains an activating mutation (45).
Effective cell killing was observed when we included interacting antiparallel leucine-zipper domains to promote the association of the subunits. These domains can reconstitute fluorescent GFP from split polypeptides (25, 26). We added the antiparallel leucine-zipper domains to the N terminus of the large and C terminus of the small subunits (Fig. 1B) based on the x-ray crystallographic structures of caspase-3 and caspase-7 (27–29) and the production of activated “reverse” caspase-3 and caspase-6 (30). Expression of the leucine-zipper-caspase subunits from the mec-18 promoter caused the death of >60% of the touch sensory neurons (Fig. 1G; four stable lines; 20 animals per line). Because the association of the leucine-zipper domains reconstituted caspase activity (Fig. 1C), we refer to the resulting active protein as recCED-3. Chromosomal integration of the transformed DNA resulted in death of >95% of the touch receptor neurons (Fig. 1 E and G). This loss of GFP expression was due to apoptotic death of the touch neurons, because cell corpses with the distinct flat, refractile disk-like appearance that is characteristic of apoptosis in C. elegans (31) were clearly visible in the positions of touch neuron cell bodies in late embryos or early L1 larvae (Fig. 1F). No cells other than the targeted cells were killed in these animals; none of the 311 cells in the head of hermaphrodite L1 larvae expressing recCED-3 showed apoptotic morphology (n = 35 animals and three independent experiments). In contrast, one-third of the PLM touch receptor neurons (31 ± 5%) in the tail of these animals showed apoptotic morphology at this stage; the other two-thirds already had died by this stage as indicated by the absence of GFP. These results demonstrate the high target specificity of the recCaspase.
Transformation of either leucine-zipper domain-caspase subunit by itself did not result in touch receptor loss (data not shown). The position of the leucine-zipper domains was important, because no cell loss was seen when they were placed in the opposite orientation, i.e., at the end of C terminus of large subunit and N terminus of the small subunit (96 ± 4% GFP positive cells; four stable lines; 20 animals per line). These results suggest that the subunits in CED-3 are similarly oriented as they are in other caspases. Our observation that the individually synthesized caspase subunits are active when brought together in the correct orientation indicates that the zymogen structure is not only important to regulate the activity of the caspases but also for assembly of the subunits.
Reconstitution of Caspase-3 Activity in C. elegans.
We produced a similar activated form of the human caspase-3 enzyme in C. elegans. Caspase-3 is one of the main executioner caspases (32–34). ProCaspase-3 exists as a homodimer, does not undergo autoproteolytic activation even when present at very high concentrations, and remains inactive until cleaved by an upstream initiator caspase (33, 34). Because C. elegans does not have a caspase cascade and lacks initiator caspases that are necessary to activate caspase-3, we first tested whether caspase-3 activity could be reconstituted in C. elegans by expressing the leucine-zipper-caspase subunits (Fig. 1A). Expression of the recCaspase-3 subunits from the mec-18 promoter caused loss of ≈80% of the touch sensory neurons (Fig. 1G; three lines; 30 animals per line). Chromosomal integration of the transformed DNA resulted in death of nearly 100% of the touch receptor neurons (Fig. 1G). Because C. elegans lacks the initiator caspases necessary for activation of caspase-3, our results indicate that the reconstituted caspase-3 is constitutively active. RecCaspase-3-mediated and recCED-3-mediated touch receptor neuron cell deaths were indistinguishable from each other in terms of the apoptotic morphology of the dying cells. Because recCaspase-3 was equally efficient as recCED-3 in causing cell death in C. elegans, it is likely to act on the same set of downstream targets. Introduction of the active site mutation C163A into the large subunit of the recCaspase-3 abolished the cell-killing activity of recCaspase-3, indicating that overexpression does not lead to nonspecific cell killing (Fig. 1G).
Cell death by the recCaspases in C. elegans did not require either endogenous CED-3 or CED-4, components that are needed for programmed cell death [see supporting information (SI) Text]. Thus, the recCaspases appear to be constitutively active.
Reconstitution of Caspase-3 Activity in HeLa Cells.
To show that the recCaspase activity is not restricted to C. elegans, we expressed the subunits of human recCaspase-3 under the Tet-inducible promoter in transiently transfected HeLa cells. A cotransfected pCaspase3-sensor vector was used to monitor caspase activity. This vector produces an enhanced yellow fluorescent protein (EYFP) fusion protein with an N-terminal nuclear export signal, a caspase-3-specific cleavage site, EYFP, and a C-terminal nuclear localization signal. The nuclear export signal is dominant over the nuclear localization signal, keeping the uncleaved EYFP protein out of the nucleus (Fig. 2A) and allowing cleaved protein, which lacks the nuclear export signal, to go to the nucleus (Fig. 2B). This change in the localization can be detected at very early stages of apoptosis, even before any cell morphological changes become visible. However, as the cells progress through apoptosis, characteristic morphological changes associated with apoptosis like cell shrinkage, membrane blebbing and nuclear fragmentation can be observed (Fig. 2C). In uninduced cells, <2% of the EYFP-positive cells had protein localized to the nucleus (Fig. 2D), about the same as cells transfected with the empty vector. Induction of recCaspase expression with doxycycline increased this number to 30–40% within 8–12 h (Fig. 2D). Induction of caspase-3 subunits without the leucine-zipper domains did not show any significant increase in apoptotic activity (Fig. 2D). Similar recCaspase-3 activity also was observed in CHO cells (data not shown). Our results demonstrate the reconstitution of human caspase-3 in the mammalian cell lines and show that tight regulation of such caspase activity can be achieved by the use of appropriate promoters.
Fig. 2.
recCaspase-3 activity in HeLa cells. (A–C) Induction of recCaspase-3 activity with 1 μg/ml doxycycline. (A–C Left) Fluorescent micrographs. (A–C Right) DIC photomicrograph of the same field. Without doxycycline (A), the caspase3-sensor EYFP is found throughout cells; after induction (B and C), EYFP fluorescence localizes to the nucleus. (C) Dying cells (arrows) exhibiting the characteristic apoptotic morphology. (D) recCaspase-3, but not caspase-3, subunits are active in HeLa cells. Percentages of nuclear-localized GFP cells out of the total GFP-positive cells are given. Observations from three independent experiments were used to calculate mean and SEM.
Targeted Cell Killing by recCaspases.
Our laboratory has described the use of the two-component recGFP system to selectively label subsets of cells (26). To demonstrate the usefulness of the two-component recCaspases in targeting subset of cells for killing, we have constructed animals in which only the AVD interneurons die in the head of C. elegans. The touch receptor neurons in the anterior of the animal form gap junctions onto the AVD interneurons and chemical synapses onto the AVB interneuron (35). These interneuronal connections are redundant (35), so a mutant affecting one set of connections cannot be easily identified by the loss of touch sensitivity. Unfortunately, AVD cell-specific promoter is not yet identified, so that only these cells could be eliminated. We were able, however, to generate animals lacking the AVD cells by expressing the two parts of recCED-3 from the cfi-1 promoter, which is expressed in the IL2, URAD, URAV, AVD, PVC, and LUA neurons and some head muscles (36), and from the nmr-1 promoter, which is expressed in the AVA, AVD, AVE, RIM, AVG, and PVC neurons (37) (Fig. 3B). Our cfi-1 promoter fusion constructs, which lacked the introns and the coding sequences present in the cfi-1 protein fusion constructs of Shaham and Bargmann (36), never showed expression in PVC and, hence, did not cause the death of PVC neurons (Fig. 3 A, C, and D) in many stable lines examined. Similar strains that are deleted for specific neurons will be useful not only for further genetic analysis, but also for analysis of neuronal circuits.
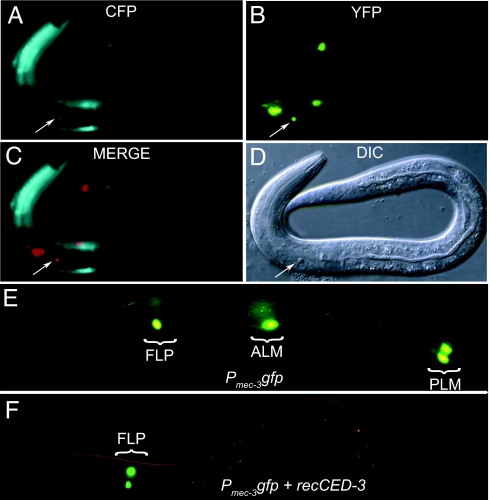
Fig. 3.
Targeted cell killing by recCaspase expressed from a combination of promoters with overlapping expression patterns. (A–D) Targeting the AVD interneuron (indicated by arrow) for cell death by recCaspase3 expressed from a combination of promoters Pnmr-1cz::caspase-3(p17) and Pcfi-1caspase-3(p12)::nz. Although the cfi-1 promoter expresses CFP in many neurons and head muscle (A) and the nmr-1 promoter expresses yellow fluorescent protein in multiple neurons (B), they are both expressed only in the AVD cells (C; the yellow fluorescent protein image was artificially colored red). (D) The death of an AVD neuron (arrow) as seen by DIC microscopy. (E and F) Restriction of GFP to FLP neurons by recCED-3 mediated cell death. (E) Without recCED-3, Pmec-3gfp is expressed in the ALM and PLM touch neurons and the FLP neurons of a newly hatched larva. (F) When recCED-3 is expressed by using Pmec-3 cz::ced-3(p17) and Pmec-18ced-3(p15)::nz, the touch neurons, which express both the promoters, die, and GFP fluorescence is found only in the FLP neurons.
The recCaspases can be used with GFP and recGFP expressed from other promoters to further restrict fluorescent protein expression in multiple component systems by eliminating unwanted cells from a set of labeled cells. For example, no promoter has been identified in C. elegans that uniquely labels the two FLP neurons. A mec-3::gfp fusion, however, is expressed in the FLP neurons and touch neurons in embryos and newly hatched animals (38) (Fig. 3E). By expressing the two subunits of recCED-3 from the mec-3 and mec-18 promoters, we were able to kill the touch receptor neurons, which express both promoters, but not the FLP neurons in animals expressing the mec-3::gfp fusion (Fig. 3F). Only FLP cells are tagged with GFP in embryos and early larval stages of the resulting animals.
We determined the amount of cell death of both targeted cells (the PLM neurons) and nontargeted cells (FLP neurons and other cell in the head region) in an integrated line expressing recCaspase-3 from the combination of mec-3 and mec-18 promoters. Apoptotic cells [65 ± 2% (SEM)], as observed by DIC microscopy, were readily seen in the position of PLM neurons in the tail of newly hatched larvae (1 h after hatching). The dying cells were confirmed to be PLM by their faint GFP fluorescence. In contrast, no apoptotic cells were observed in the position of FLP neurons and other cells in the head region of these animals. Furthermore, absence of cell death in nontargeted FLP neurons was confirmed by the presence of GFP-positive FLP cells (98 ± 1%), which was very similar to the number of FLPs observed in an isogenic strain without the recCaspase-3 (96 ± 1%). Although 12% of PLMs showed wild-type GFP fluorescence at this stage, they completely disappeared by the time the animals reached the fourth larval stage (n = 30 animals). These results again confirm that recCaspase-mediated cell death is very efficient and highly target specific.
Temporal Induction of recCaspase-3 Activity.
The dual component nature of the recCaspase also could be exploited to induce cell death in specific cells at specific developmental stages. We used the inducible heat shock promoter (hsp-16) in combination with the touch neuron-specific promoter (mec-18) to express recCaspase-3 at specific times. Heat shocking the animals that expressed both subunits of recCaspase-3 in embryos leads to very efficient killing of the embryonically derived ALM and PLM touch neurons (>90%), whereas virtually none of the AVM and PVM, which are generated postembryonically in the mid-L1 larval stage, were affected (SI Fig. 5). Although death could be specifically induced in touch neurons at any stage of development, induction of cell death was progressively less efficient after the L1 stage (SI Fig. 5). Expression of only the large subunit (p17) of recCaspase-3 from hsp-16 promoter at any stage of the animal's life cycle did not cause any death of the touch receptor neurons as observed by loss of GFP expression (SI Fig. 5). These results demonstrate that recCaspase activity can be very tightly regulated and induced in a stage-specific manner.
To test the general usefulness of the conditional expression of recCaspase, we expressed it in muscle cells. A 2-h heat shock of animals that expressed only the large subunit (p17) of recCaspase-3 from the hsp-16 promoter did not cause any changes in the morphology of the muscle cells or paralysis of the animals (Fig. 4 A and E). Heat shocking the animals that expressed both subunits of recCaspase-3 from the combination of hsp-16 and myo-3 promoters resulted in rounding up of the large muscle cells, which quickly accumulated a large number of vacuoles (Fig. 4 B and C). Absence of any kind of movement defect or paralysis in >99% of the non-heat-shocked animals grown at 15°C (data not shown) indicates that the hsp-16 promoter is very tightly regulated and the expression of the small subunit (p12) of recCaspase-3 from the strong myo-3 promoter under non-heat-shock conditions is not toxic to the cells.
Fig. 4.
Conditional induction of recCaspase-3 activity in muscle cells. (A) Expression of only the p17 subunit of the recCaspase-3 from the heat shock promoter [(Phsp-16 cz::caspase-3(p17)] had no effect on survival of body wall muscle. (A and B) GFP fluorescence from the muscle-specific myo-3 promoter (A and B Upper); a DIC image of the same animal (A and B Lower). For both animals, newly hatched L1 larvae were heat shocked for two hours and photographed 24 h later. (B) Expression of Pmyo-3 caspase-3(p12)::nz (causing the p12 subunit of recCaspase to be expressed in muscles) in combination with Phsp-16 cz::caspase-3(p17) resulted in death of the body wall muscles. (C) Enlargement of the DIC image of the animal in B showing the vacuolated deaths of muscle cells. (D) Superimposed DIC and fluorescence (Pmyo-3gfp) images of an embryo heat shocked for 30 min and photographed 8 h later. Arrows indicate some of the apoptotic nuclei, which show the characteristic refractile, flat disk-like morphology. (E) Occurrence of muscle cell death with induction of recCaspase activity at different times. Animals were heat shocked at the indicated times and scored 48 h later for paralysis caused by the death of muscle cells. Data from the expression of the single subunit (from Phsp-16; green line) as well as both the subunits (from Phsp-16 and Pmyo-3; blue line) of recCaspase are shown. Data from at least two stable lines (50 animals per line) and three independent experiments were used to calculate mean and SEM. (F) Time course of induction of recCaspase activity. First-stage larvae expressing both the subunits of the recCaspase were collected 12 h after hatching and subjected to a 2-h heat shock. At the indicated times after heat shock, animals were scored as completely paralyzed (blue), partially paralyzed (red), or wild type. Data from three stable lines and at least 100 animals per line were used to calculate mean and SEM. The mean values of partially and completely paralyzed animals were added to obtain the total number of animals affected (black) at each time point.
We were surprised that the flattened disk-like, refractile appearance of the nuclei that is characteristic of C. elegans apoptotic cells (31) rarely was seen in animals expressing active recCaspase in their muscles. We hypothesized that strong expression of the recCaspase from the combination of myo-3 and hsp-16 promoters overwhelmed the cells so that the refractile nuclei were not seen readily. When we reduced the duration of the heat shock from 2 h to 30 min, however, the characteristic morphology of the apoptotic nuclei was seen more often (Fig. 4D).
Upon heat shock, >90% of the animals that expressed both subunits of recCaspase were paralyzed (Fig. 4E). Induction of recCaspase in muscle cells, in contrast to its expression in touch receptor neurons, was almost as efficient in adult animals as in L1 larvae, reflecting the strong and consistent expression of the myo-3 promoter throughout development. Our observations point out that the strength of individual promoters and their temporal expression pattern are key factors in regulation of the activity of dual component recCaspases.
Time Course of Induction of recCaspase Activity.
For recCaspase to be effective as a cell-killing tool, it has to be very efficient and act fast. We looked at the activity of heat-shock-induced recCaspase-3 (expressed from the combination of myo-3 and hsp-16 promoters) in muscle cells of L1 larvae (12 h after hatching at 15°C) subjected to a 2-h heat shock. The number of animals that were either completely or partially paralyzed was counted at different times. The effects of recCaspase were seen very quickly (Fig. 4F). More than 50% of the animals were affected within 3 h after heat shock; nearly all of the animals were completely paralyzed 12 h after heat shock.
Discussion
We have developed a previously undescribed way of generating constitutively active caspases: Individually expressed caspase subunits are brought together in the correct orientation by using the antiparallel leucine-zipper sequences to generate the constitutively active recCaspase. Because recCaspase are constitutively active, they do not depend on any upstream components for their activation. As we have demonstrated, our reconstitution strategy works for both long-prodomain caspases (CED-3) as well as short-domain caspases (caspase-3) that are activated by different mechanisms. Hence, this mode of reconstitution should apply to all caspases because they all have similar basic structure and are activated by similar cleavage reactions from the procaspase polypeptides. Furthermore, the finding that recCaspases are active in mammalian cells and C. elegans indicates that these dual-component systems will be effective in a broad range of organisms.
Previous attempts to establish lines that are genetically ablated for specific cells by ectopically expressing toxic gene products from cell-specific promoters in C. elegans (8, 9) had limited success because few cell-specific promoters have been identified. Only 12% of the 118 neuronal groups of White et al. (39) can be killed by using cell-specific promoters (SI Table 1). As we have shown for the touch receptor neurons, expressing the two subunits of a recCaspase from the same promoter can ablate these cells. Seventy percent more neuronal groups, however, could be killed selectively if two promoters with overlapping expression pattern were used to express recCaspase (SI Table 1). In addition, expression of recCaspases from a combination of inducible and cell-specific promoters allows for killing of specific cells at particular times, making recCaspase a very useful tool for developmental studies. Thus, the recCaspase system provides a highly selective means of creating genetically ablated animals.
Caspases are being considered for targeted cell killing in cancer therapy (10, 40, 41). Use of two-component caspases would have a potential advantage in targeted gene therapy of tumor and cancer cells. The recCaspases can provide a dual control mechanism to tightly regulate the induction of caspase activity and cell death in a given set of cells. By using a combination of promoters, one of which is expressed in specific cells or tissue and the other whose expression can be induced, one should be able to target the killing of specific cells at specific times. Alternatively, using two promoters that target the same cells or tissues could ensure tight regulation of caspase activity or cell death.
We envision additional layers of regulation imposed over the recCaspases. A third promoter could be added to express an additional component that could either induce or further restrict the expression of recCaspases. For example, Tet-inducible system could be used to tightly control the induction of expression: one of the recCaspase subunits can be expressed from the tet-responsive promoter, whereas the other recCaspase subunit and the tet-transactivator could be expressed from two different cell-specific promoters that show overlapping expression pattern. The recCaspase expression can be restricted further by expressing an inhibitor of caspase activity, e.g., p35 (42) or a fold-back RNAi construct for one of the subunits, from a third promoter. Finally, recCaspases could be used both in vivo and in vitro to identify new substrates and inhibitors of caspases and will be especially useful in the case of the caspases that have not been well characterized.
Materials and Methods
Nematode Protocols.
Animals were maintained, unless otherwise mentioned, at 20°C as described (43). Transgenic animals were generated by microinjection into wild type (N2) or TU2769 (uIs31) (this strain contains integrated insertion of mec-17::gfp, which expresses GFP specifically in the touch neurons; ref. 44). The expression plasmids (50 μg/ml if injected alone or 25 μg/ml if two were injected) were injected with the dominant roller plasmid, pRF4 (50 μg/ml) that serves as the transformation marker (45). For heat-shock constructs, the expression plasmids were injected at a reduced concentration of 10 μg/ml (for each construct) along with 80 μg/ml of the roller plasmid. At least three stable lines were obtained for each genotype. The extrachromosomal array was integrated into the chromosome by following the slightly modified integration protocol of I. Greenwald and O. Hobert (personal communication). Animals were irradiated with gamma rays (4,800 rads), and lines that inherited the transformation marker 100% of the times in the subsequent generations were selected.
Expression Constructs.
The sequences corresponding to the antiparallel leucine-zipper domains NZ (ALKKELQANKKELAQLKWELQALKKELAQ) and CZ (AQLEKKLQALEKKLAQLEWKNQALEKKLAQ) along with the linker sequence “GGSG” were amplified from bacterial expression plasmids pET11a-NZGFP and pET11a-CZGFP (Ghosh et al. (25); a gift from Lynne Regan, Yale University, New Haven, CT). The sequences corresponding to p17 and p15 subunits of CED-3 (Fig. 1A) were amplified either from genomic DNA or from Pmec-7ac-ced-3 (ac-ced-3 was a gift from Ding Xue, University of Colorado, Boulder, CO), which contains a mutation that results in a constitutive ced-3 activity (46). The sequences for p17 and p12 subunits of Caspase-3 (Fig. 1A) were amplified from a human Caspase-3 cDNA (Mammalian Gene Collection full-length cDNA clone ID 4419175). All of the constructs used for C. elegans expression were derived from the promoter-less GFP plasmid pPD95.75 (a gift of Andy Fire, Stanford University, Stanford, CA; Fire laboratory Vector kit; ftp://www.ciwemb.edu/pub/FireLabInfo/FireLabVectors). Plasmid constructs used for expression in the HeLa Tet-ON cell line (Clontech, Mountain View, CA) were derived from pTRE-Tight (Clontech), which contains a tetracycline-responsive promoter. Details regarding the cloning of expression constructs are provided in SI Table 2.
Cell Death Assay.
The death of the C. elegans touch receptor neurons, which were labeled with GFP, was monitored by the loss of GFP fluorescence in adult worms under a Leica stereo dissection microscope equipped for fluorescence microscopy. L1 larvae (collected 1–2 h after hatching) were observed by using a Zeiss Axioscope 2 microscope. The percent of surviving cells were calculated by dividing the number of GFP-positive cells by the total number of touch receptor neurons (number of animals times 6 for adults and times 4 for the L1 larvae). The death of body wall muscle cells, which resulted in the paralysis of the animals, was scored as a percentage of animals that were paralyzed 2 days after heat shock. The change in the morphology of the dying body wall muscle cells, which were also labeled by Pmyo-3::gfp in transgenic animals, was observed by using a Zeiss Axioscope 2 microscope.
Heat-Shock Experiments.
The animals, grown at least for two generations at 15°C, were heat-shocked at the specified stages by incubating them at 34°C for 2 h and immediately transferred back to 15°C after the heat shock. Unless indicated otherwise, the death of touch receptor neurons or the muscle cells were scored 48 h after heat shock.
Caspase Activity in HeLa Cells.
HeLa Tet-On cells, maintained in DMEM with 10% Tet system approved FBS (BD Biosciences, San Jose, CA), were transfected transiently by using Lipofectamine2000 (Invitrogen). The DNA mix contained pCaspase3-sensor vector (Clontech), tetracycline transactivator rtTA2-M2 plasmid (47), and plasmids for caspase-3 subunits with or without leucine zippers or the control vector (pTRE-Tight). Cells were split 8 h after transfection, plated on coverslips, and after another 12 h of growth, 1 μg/ml doxycycline was added to induce expression. Cells were fixed at 12 h after induction, and the percentage of cells with caspase activity was determined by the number of cells with nuclear localized yellow fluorescent protein divided by the total number of fluorescent cells. More than 300 fluorescent cells were counted from randomly chosen fields in each experiment; data from three independent experiments were used to calculate mean and SEM.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM30997 (to M.C.).
Abbreviations
- DIC
differential interference contrast
- EYFP
enhanced yellow fluorescent protein
- recCaspase
reconstituted caspase.
Footnotes
Conflict of interest statement: D.S.C. has submitted a patent based on this work.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610877104/DC1.
References
- 1.Sulston JE, White JG. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt RR, Nguyen N, Kuwada JY. Neuron. 1992;8:869–882. doi: 10.1016/0896-6273(92)90201-n. [DOI] [PubMed] [Google Scholar]
- 3.Shah EM, Jay DG. Curr Opin Neurobiol. 1993;3:738–742. doi: 10.1016/0959-4388(93)90146-p. [DOI] [PubMed] [Google Scholar]
- 4.Palmiter RD, Behringer RR, Quaife CJ, Maxwell F, Maxwell IH, Brinster RL. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 5.Borrelli E, Heyman R, Hsi M, Evans RM. Proc Natl Acad Sci USA. 1988;85:7572–7576. doi: 10.1073/pnas.85.20.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunes S, Steller H. Genes Dev. 1991;5:970–983. doi: 10.1101/gad.5.6.970. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi K, Morita S, Sawada H, Mizuguchi T, Yamada K, Nagatsu I, Fujita K, Kreitman RJ, Pastan I, Nagatsu T. Proc Natl Acad Sci USA. 1995;92:1132–1136. doi: 10.1073/pnas.92.4.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaham S, Horvitz HR. Genes Dev. 1996;10:578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 9.Harbinder S, Tavernarakis N, Herndon LA, Kinnell M, Xu SQ, Fire A, Driscoll M. Proc Natl Acad Sci USA. 1997;94:13128–13133. doi: 10.1073/pnas.94.24.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacCorkle RA, Freeman KW, Spencer DM. Proc Natl Acad Sci USA. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornberry NA, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 12.Cohen GM. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengartner MO. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 14.Degterev A, Boyce M, Yuan J. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 15.Riedl SJ, Shi Y. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 16.Horvitz HR. ChemBioChem. 2003;4:697–711. doi: 10.1002/cbic.200300614. [DOI] [PubMed] [Google Scholar]
- 17.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 18.Ellis HM, Horvitz HR. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 20.Xue D, Shaham S, Horvitz HR. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 21.Chinnaiyan AM, Chaudhary D, O'Rourke K, Koonin EV, Dixit VM. Nature. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 22.Seshagiri S, Miller LK. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 23.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Chang HY, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh I, Hamilton AD, Regan L. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 26.Zhang S, Ma C, Chalfie M. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Rotonda J, Nicholson DW, Fazil KM, Gallant M, Gareau Y, Labelle M, Peterson EP, Rasper DM, Ruel R, Vaillancourt JP, et al. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 28.Mittl PR, Di Marco S, Krebs JF, Bai X, Karanewsky DS, Priestle JP, Tomaselli KJ, Grutter MG. J Biol Chem. 1997;272:6539–6547. doi: 10.1074/jbc.272.10.6539. [DOI] [PubMed] [Google Scholar]
- 29.Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasula SM, Ahmad M, MacFarlane M, Luo Z, Huang Z, Fernandes-Alnemri T, Alnemri ES. J Biol Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 31.Sulston JE, Horvitz HR. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes-Alnemri T, Litwack G, Alnemri ES. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 33.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 34.Tewari M, Quan LT, O'Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 35.Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaham S, Bargmann CI. Genes Dev. 2002;16:972–983. doi: 10.1101/gad.976002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. Neuron. 2001;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 38.Way JC, Chalfie M. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 39.White JG, Southgate E, Thomson JN, Brenner S. Philos Trans R Soc London B. 1986;341:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 40.Shariat SF, Desai S, Song W, Khan T, Zhao J, Nguyen C, Foster BA, Greenberg N, Spencer DM, Slawin KM. Cancer Res. 2001;61:2562–2571. [PubMed] [Google Scholar]
- 41.Shinoura N, Hamada H. Curr Gene Ther. 2003;3:147–153. doi: 10.2174/1566523034578410. [DOI] [PubMed] [Google Scholar]
- 42.Xue D, Horvitz HR. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 43.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Hagan R, Chalfie M, Goodman MB. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 45.Mello CC, Kramer JM, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 47.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.