Abstract
Individual posttranslational modifications (PTMs) on histones have well established roles in certain biological processes, notably transcriptional programming. Recent genomewide studies describe patterns of covalent modifications, such as H3 methylation and acetylation at promoters of specific target genes, or “bivalent domains,” in stem cells, suggestive of a possible combinatorial interplay between PTMs on the same histone. However, detection of long-range PTM associations is often problematic in antibody-based or traditional mass spectrometric-based analyses. Here, histone H3 from a ciliate model was analyzed as an enriched source of transcriptionally active chromatin. Using a recently developed mass spectrometric approach, combinatorial modification states on single, long N-terminal H3 fragments (residues 1–50) were determined. The entire modification status of intact N termini was obtained and indicated correlations between K4 methylation and H3 acetylation. In addition, K4 and K27 methylation were identified concurrently on one H3 species. This methodology is applicable to other histones and larger polypeptides and will likely be a valuable tool in understanding the roles of combinatorial patterns of PTMs.
Keywords: bivalent domain, electron transfer dissociation, mass spectrometry, posttranslational modifications, Tetrahymena
In eukaryotes, DNA is wrapped around octameric cores of the histone proteins H3, H4, H2A, and H2B, forming nucleosomes, the fundamental subunit of chromatin (1, 2). Flexible “histone tails” that extend from nucleosomes are subject to a wealth of posttranslational modifications (PTMs) including methylation, acetylation, and ubiquitination on lysines, methylation on arginines, and phosphorylation on serine or threonine residues (1–4). The complexity and nonrandom distribution of known PTMs on the N terminus of histone H3 alone (Fig. 1A) suggests functional implications for unique combinations of modifications, several of which are under active investigation (5–7).
Fig. 1.
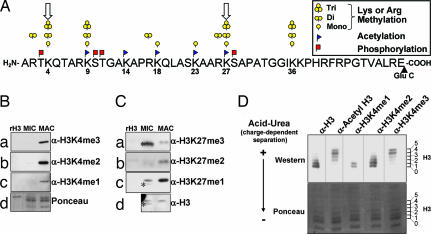
Analysis of somatic chromatin suggests long-range connectivity among modifications on histone H3. (A) Schematic of PTMs observed on the Glu-C generated N-terminal peptide of histone H3 [for the entire sequence of Tetrahymena H3.2, see supporting information (SI) Fig. 4]. Conventional mass spectrometry protocols often require digestion into much smaller pieces such that connectivity between modifications spaced by many amino acids as may occur in bivalent domains (open arrows on K4 and K27) is poorly understood. (B) Highly purified Tetrahymena MIC and MAC histones were resolved by SDS/PAGE and detected by Western blot analysis with antibodies recognizing monomethylated, dimethylated, or trimethylated species of H3K4. (C) Histones were purified and resolved as in B; however, Western blot analysis was performed with antibodies recognizing monomethylated, dimethylated, or trimethylated species of H3K27. A MIC-specific H3 isoform that is proteolytically processed endogenously is indicated by ∗. (D) MAC histones were acid-extracted and resolved by acid-urea gel to separate positively and negatively charged histone H3 species into six rungs representing H3 isoforms that range from relatively hypoacetylated (0) to hyperacetylated (5). Equivalent Ponceau staining was used as a loading control.
Emerging studies suggest that neighboring histone PTMs may function together on the same histone tail in a combinatorial fashion, sometimes reducing or enhancing the ability for the targeted residue to become modified in what is referred to as modification “cross-talk” (8–10). Furthermore, immunogen-based approaches such as ChIP or ChIP on Chip suggest unique groupings of histone PTMs are involved in demarcating chromatin domains as either active or repressed, in an epigenetically stable fashion. In somatic cell lineages, for example, H3K4me3 and H3 hyperacetylation are consistently colocalized at 5′ regions of transcriptionally active genes in euchromatin (11, 12). In contrast, transcriptionally silent heterochromatin, such as the inactive X chromosome (Xi) in mammalian females, is often characterized by H3K27me, H3K9me, and H3 hypoacetylation (13). Interestingly, undifferentiated embryonic stem cells were recently suggested to have “bivalent domains,” regions of the genome enriched for both H3K4me3 and H3K27me3 on a nucleosomal scale (see ref. 14 and downward arrows in Fig. 1A), which, during differentiation, might be resolved into either a repressive H3K27me3-enriched environment or a transcriptionally favorable H3K4me3-enriched region. However, while these reports are suggestive of functional interplay among histone PTMs, direct determination of their interdependence remains poorly understood, in part because the analytical techniques available for the study of long-distance combinatorial patterns on the same histone tail (or protein) are limited.
Mass spectrometry has become an enabling method for mapping PTMs on histones (reviewed in refs. 15 and 16), without some of the caveats of antibody-based analyses that include antibody production, unwanted cross-reactivity, epitope occlusion, and the inability to detect multiple modifications (patterns) or novel modification sites. However, a drawback of conventional mass spectrometry approaches is that histones are typically digested into short peptides to make sequence and PTM analysis possible (17, 18), referred to as “bottom-up” analysis. With short peptides, it becomes difficult to determine which peptides originated from the same molecule, and information about interdependence of modifications on a given histone molecule is often lost (see ref. 19).
A newer mass spectrometry technique, electron capture dissociation (ECD), allows sequencing of intact proteins (20). This “top-down” analysis (21) has been successfully used to determine histone PTMs. Using ECD, Kelleher and coworkers (22–25) characterized PTMs on all four human core histones, Burlingame and colleagues (26) characterized Tetrahymena H2B variants, and Zhang and Freitas (27) characterized histone H4 from calf thymus. Recently, an ion/ion analogue of ECD, termed electron transfer dissociation (ETD) (28), was developed. This technique can sequence highly charged, longer peptides (>20 aa), and in combination with a second ion/ion reaction, termed proton transfer charge reduction (PTR) (29, 30), can determine the N- and C-terminal sequence of intact proteins on a chromatographic time scale (31, 32). Using this technique, we examined the PTM status of the N-terminal H3 peptide residues 1–50 (H31–50) from macronuclei isolated from the ciliated protozoan Tetrahymena thermophila to determine long-range combinations of histone PTMs (i.e., PTMs separated by 20 or more residues) associated with a transcriptionally active state.
The genome of T. thermophila is functionally separated into two nuclei that can be easily purified: a “somatic” macronucleus (MAC) from which all transcription occurs, and a transcriptionally silent “germ-line” micronucleus (MIC) (33). To determine PTM correlations on H31–50, we purified MAC H3 based on charge and modification state and used the combination of ETD/PTR and accurate mass measurements to determine patterns of histone PTMs. We observed a strong correlation on the same histone molecule between H3K4me3 and increased acetylation occupancy at distinct lysine residues, as well as coexistence of H3K4me1 with H3K27me1 or H3K27me2. These long-range histone PTM associations point to an underlying hierarchical mechanism of establishing histone modification patterns and suggest a unique configuration of transcriptionally poised H3 modifications that resemble the bivalent domains recently reported for embryonic stem cells (14).
Results
Relative Enrichment of Methylation at H3K4 and H3K27 Suggests Bivalent-Like Domains in Transcriptionally Active Nuclei from Tetrahymena.
Although properties of H3K4me3/H3K27me3 bivalent domains in embryonic stem cells are poorly defined, their discovery suggests functional overlap between these methylation sites at distinct genomic regions (14). Thus, we sought to determine whether methylation at lysines H3K4 and H3K27 could be detected in a transcriptionally active nucleus, where H3K4 methylation and, to a much lesser extent, H3K27 methylation, have been reported using microsequencing (34). In the present work, histone acid extracts from MACs were resolved by SDS or acid-urea gel electrophoresis and analyzed with antibodies specific for distinct methylated lysine states (Fig. 1 B–D). Equivalent staining or detection of general H3 by Western analyses served as loading controls.
As expected for transcriptionally active chromatin (34), analysis with K4 methylation state-specific antibodies indicated that MAC histones are highly enriched in H3K4me1, H3K4me2, and H3K4me3 modifications (Fig. 1B a–c, MAC). In contrast, H3K4 methylation was not detected on H3 isoforms present in transcriptionally silent, germ-line MICs (Fig. 1C a–c, MIC). Unexpectedly, however, strong enrichment of H3K27me1 and H3K27me2 was observed with macronuclear H3 (Fig. 1C b and c, MAC). In contrast, H3K27me3 was localized almost exclusively to micronuclear H3 (Fig. 1Ca, MIC), and essentially all of this reactivity was associated with the proteolytically processed form of H3 in that nucleus (see ref. 35 for details and asterisks in Fig. 1C). The finding that certain methylation states of H3K27me (monomethylation and dimethylation) are enriched along with essentially all H3K4 methylation states in MACs suggested a potential functional overlap among these marks, at least at an H3 population level. Whether these marks coexist on the same H3 tail is not clear from these analyses, prompting further investigation.
Increased Acetylation Occupancy Correlates with Trimethylation Status on Histone H3 from Macronuclei in Tetrahymena.
The restriction of H3K4 methylation to macronuclei (somatic) (Fig. 1B), with regard to previously reported high levels of H3 acetylation in MACs, is consistent with ChIP experiments colocalizing these PTMs to active regions of chromatin (11, 34, 36). To this end, we asked whether monomethylation, dimethylation, or trimethylation at H3K4 correlated with acetylation in a manner suggestive of potential long-distance interactions. Acid-urea gel electrophoresis was chosen to resolve MAC H3 on the basis of charge (and size) and probed with PTM-specific antibodies. Because one acetylation (or phosphorylation) decreases positive charge by one, histones can be resolved as “ladders” with each slower-migrating “rung” representing an increased phospho or acetyl occupancy. Tetrahymena H3 resolves into six detectable rungs (37) when visualized with general H3 antibodies (Fig. 1D, α-H3): 0 (unacetylated H3) to 5 (species of H3 with five acetylations). These rungs are acetylation-dependent because phosphatase treatment does not disrupt resolution of this H3 ladder (data not shown), and previous analyses show that MAC H3 has no detectable phosphorylation (37, 38).
As expected, slower-migrating species of H3 were detected with an α-acetyl H3 antibody (Fig. 1D, rungs 2–5). Strikingly, H3K4me3 was predominately localized to the slowest-migrating isoforms (Fig. 1D, rungs 4–5), suggestive of H3 with high acetyllysine occupancy, supporting ChIP assays showing a link between H3K4me3 and hyperacetylated H3 in other models (see ref. 11 and references within). H3K4me2 comigrated with unacetylated or intermediately acetylated H3 species (Fig. 1D, rungs 0–3), and H3K4me1 was exclusively localized to either unacetylated or singly acetylated H3 species (Fig. 1D, rungs 0–1). These data suggest a long-distance coupling between acetyl occupancy of H3 and distinct methylation states of H3K4, but suffer from the fact that these analyses are carried out on a population level.
Acetylations and Methylations on H3 Are Predominantly Localized at the N Terminus.
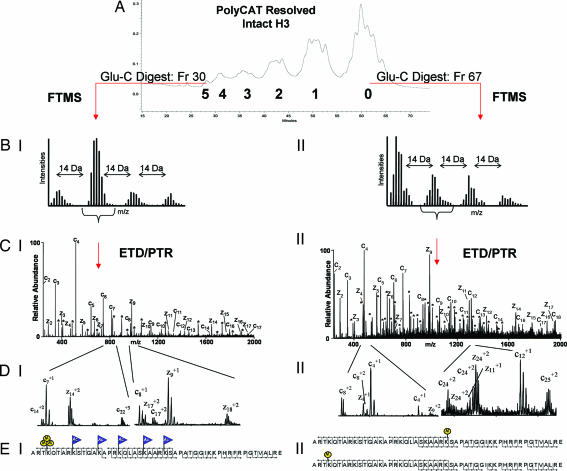
Although the above data suggested a link between H3K4 methylation and high acetylation occupancy on H3, it remained unclear whether these PTMs reside on the same histone. To better examine potential PTM linkage, MAC H3 was purified by RP-HPLC, separated into fractions enriched for H3 isoforms differing in charge by using PolyCAT A chromatography, and subjected to mass spectrometry analysis (Fig. 2).
Fig. 2.
Schematic of the mass spectrometry approach. (A) PolyCAT A separation of intact histone H3. (B) FTMS mass measurements of the N-terminal H3 Glu-C peptide (H31–50). Shown are the +9 charge states of the species present in fraction 30 of group 5 (I) and the +10 charge states of fraction 67 of group 0 (II). (C) The resulting ETD/PTR zoom scan spectrum of the indicated species from groups 5 (I) and 0 (II) are shown. Singly charged c-type (containing the N terminus) and z-type fragment ions (containing the C terminus) are labeled in the spectra. Doubly charged fragment ions of type c and z are indicated by ∗. (D) Selected m/z range of the corresponding spectra displayed in C. Note the double ion series in CII. Two c4 fragment ions are observed corresponding to two H3 species; one is unmodified and one is monomethylated at K4. This ion series is traceable up to the +2 fragment ion series as shown by the two c8+2 and c24+2 fragment ions. The H3 species in this spectrum that is not methylated at K4 affords a methylation on a different residue to satisfy the FTMS measurements. The second H3 species is methylated at K27 as two z24+2 fragment ions corresponding to K27 and differing by nominal 14 Da are observed. The spectra in CII and DII correspond to two species, one monomethylated on K4 and one monomethylated on K27. (E) Sequence of H31–50. Modified residues are indicated by blue flags for acetylation (Ac) and by yellow circles for methylation (M). Peptide backbone cleavages observed in the spectra are indicated. EI shows a species trimethylated on K4 and acetylated on K9, K14, K18, K23, and K27, as interpreted from CI and DI. EII shows two species, one monomethylated on K4 and one monomethylated on K27, as interpreted from CII and DII.
Individual PolyCAT A fractions of MAC H3 were digested with Glu-C and analyzed on the Finnigan LTQ-Fourier transform mass spectrometer (Thermo Electron, San Jose, CA) to obtain mass measurements, accurate to within 2 ppm (SI Tables 1 and 2). Comparison of the mass differences between the modified peptides and their unmodified counterparts allowed the determination of the number and character of PTMs. The only PTMs detected were acetylation and methylation, and these were confined to H31–50 (SI Tables 1 and 2).
Because intact H3, resolved by the PolyCAT A column (Fig. 2A, groups 0–5), eluted in six groups, we reasoned that peaks contained within each group were clustered by total acetylation occupancy (see ref. 19), in agreement with our acid-urea analysis that indicated five distinct acetylated H3 isoforms plus one unmodified species (Fig. 1D). Furthermore, Fourier transform mass spectrometry (FTMS) on H31–50 showed that the earliest eluting fractions were comprised of H3 molecules with an addition of five acetyl groups, followed by fractions that harbored four, three, two, one, and zero acetyl groups (Figs. 2 and 3 and SI Table 2). Thus, at least six total distinct acetylation occupancy states on intact H3 (including unmodified) were resolved by PolyCAT A chromatography.
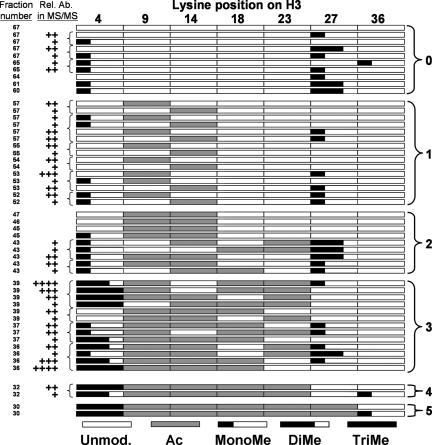
Fig. 3.
Summary of H3 species identified in the mass spectrometry analysis. Shown are the H31–50 modifications states identified in this study, categorized by fraction number. Species identified within the same spectrum are indicated with a left bracket, and the relative abundance of each species within this spectrum is indicated by the number of + symbols. The species with the most + symbols is most abundant, and the species with the least number of + symbols is of lowest abundance. Where no + symbol is noted next to a fraction number, only one species per spectrum was identified. The numbers on the right indicate grouping by acetylation occupancy.
Sequencing of Macronuclear Histone H3 Using ETD/PTR.
To determine the location, nature, and linkage of H3 PTMs, H31–50 was sequenced by ETD/PTR using an ion/ion reaction-enabled LTQ ion trap mass spectrometer (28). We operated the LTQ in high-resolution mode (zoom scan) over the entire mass range (250–2,000 Da) to resolve multiply charged fragment ions, which extends the mass range to 4,000 Da (i.e., +2 ion m/z of mass 2,000 = 4,000 Da). This combination of PolyCATA chromatography and ETD/PTR mass spectrometry is necessary as previous attempts to sequence H31–50 with on-line chromatography and normal scan mode, but no prior separation of H3 isoforms, resulted in complicated mixed spectra (31). The spectra were mixtures of several isobaric species (i.e., methylations on different residues or different modifications with identical mass, i.e., me3 versus me1 + me2) that only permitted the use of singly charged fragment ions, providing sequence information of the first 15–16 N- and C-terminal residues. Here, the prior separation of H3 isoforms and the acquisition of high-resolution scan mode enabled us to use doubly charged fragment ions, allowing complete sequence coverage for H31–50.
Fig. 2CI shows the ETD/PTR fragmentation spectrum recorded on the peptide indicated in Fig. 2BI from the early eluting fraction, group 5. Accurate mass measurement from the FTMS analysis reveals five acetyl and three methyl groups (Fig. 2BI). The fragmentation spectrum in Fig. 2CI shows the addition of 42 Da to K4, K9, K14, K18, K23, and K27. However, the mass accuracy of the linear ion trap does not permit differentiation between trimethylation (42.0469 Da) and acetylation (42.0106 Da). Therefore, H31–50 was further digested with trypsin and analyzed with LTQ-FTMS (39) to determine the site of trimethylation. All fractions of groups 2, 3, 4, and 5, respectively, were combined, and each group was subjected to this bottom-up experiment. The peptide H33–8 containing the K4 residue was exclusively methylated in all fractions (data not shown). Consequently, the spectrum in Fig. 2CI was interpreted as trimethylation on H3K4 and acetylations on H3K9, H3K14, H3K18, H3K23, and H3K27, as diagramed in Fig. 2EI, confirming the coexistence of H3K4me3 and H3 hyperacetylation on a single peptide species.
Fig. 2CII shows an ETD/PTR fragmentation spectrum recorded on a species harboring one methyl group. This methyl group could be located at 14 different residues (Ks and Rs), resulting in a mixed spectrum representing different monomethylated species. The N-terminal fragment ions (c ions) starting from the c4 ion on show a double series differing by 14 Da. The c4 fragment ion of greater intensity corresponds to an unmodified K4 residue, whereas the c4 fragment ion appearing 14 Da higher corresponds to a K4me1 residue (see Fig. 2DII). On the C-terminal ion series (z ions) no modifications are observed until fragment ion z24, corresponding to H3K27. Again, two ion series are observed (differing by 14 Da) corresponding to two peptides, containing either unmodified K27 or K27me1 (see Fig. 2DII). Hence, this fraction contains two distinct species, one with H3K27me1 and one with H3K4me1 (Fig. 2EII). Thus, the combination of ETD/PTR and high-resolution acquisition allows for complete sequence coverage of the entire N–terminal peptide encompassing H3K4 and H3K27 and for unambiguous assignment of the combinatorial interplay of these long-distance modifications, even in a mixed spectrum (i.e. a spectrum containing more than one species).
Combinatorial Patterns of Histone Modifications on a Single H3 Tail.
Our approach, outlined in Fig. 2, allowed us to successfully correlate transcription-associated PTMs within H31–50 that are separated by long distances. Fig. 3 summarizes the histone species identified in a combined schematic and tabular form. Interestingly, the acetylation steady state of histone H3 appears to be following an approximate order or hierarchy. Taking into account the relative abundance of each observed modified form of H31–50 (Fig. 2), the singly acetylated species are predominantly located on H3K9, and to a lesser extent on H3K14 (Fig. 3, group 1). Doubly acetylated histone H31–50 is predominately acetylated on H3K9 and H3K14 (Fig. 3, group 2). Triacetylated histone H31–50 is mostly acetylated on H3K9, H3K14, and H3K18 (Fig. 3, group 3), albeit there are some exceptions. Tetraacetylated histone H31–50 is acetylated on H3K9, H3K14, H3K18, and H3K23 (Fig. 3, group 4), and finally quintuply acetylated histone H31–50 is acetylated on H3K9, H3K14, H3K18, H3K23, and H3K27 (Fig. 3, group 5). Most exceptions to these observations were attributed to species with lower abundance. Methylation on H3K36 has been linked to H3 deacetylation in coding regions and to negative regulation of transcription (47–49); however, H3K36me1 was present on both hyperacetylated and hypoacetylated species (Fig. 3, groups 0, 4, and 5), suggesting a relationship to H3 acetylation occupancy. H3K36me2 and H3K36me3 were not detected in this study.
Overall, our results suggest H3K27 dimethylation and to a lesser extend monomethylation prevail in less acetylated histone H3 fractions and can co-occur with H3K4 monomethylation, whereas H3K4 trimethylation is clearly enriched in histone fractions having an acetylation occupancy of three, four, or five. Although we found H3K4me2 to be clearly enriched in intermediately acetylated species, the complex nature of the spectra precluded their accurate interpretation (see Discussion).
Discussion
Sequencing Histone Complexity by Using ETD/PTR Tandem Mass Spectrometry.
Our mass spectrometric approach permits us to elucidate modifications that are concurrently present on a single histone tail, separated by long distances, thus providing a tool for studying roles of combinatorial histone modifications. We believe that our approach of examining large peptides has advantages, namely, we can provide an exhaustive description of PTMs in the N-terminal region of H3, which contains the majority of H3 PTMs (40). This approach has several important practical consequences. First, restricting the analysis to the N-terminal region allows us to obtain complete sequence coverage and hence unambiguous modification assignment. Complete sequence coverage is rarely achieved with the full length of H3, as not all peptide backbone cleavages are observed and hence some modifications can theoretically be attributed to more than one residue. Second, fragmenting 50 amino acids versus the 135 of intact H3 yields increased sensitivity for the resulting fragment ions, as the ion current is divided among fewer product ions. Third, the size of the N-terminal peptide, ≈5 kDa, requires only singly and doubly charged fragment ions for complete sequence coverage, resolution that is readily achieved operating the modified LTQ in zoom scan mode. In addition, the use of PTR to simplify the fragment ions to singly and doubly charged ions greatly reduced the complexity of the spectra, allowing interpretation of spectra comprised of up to three to four distinctly modified H3 species (Fig. 3). However, a portion of the acquired spectra remained uninterpretable because of a very complicated mixed spectrum, likely a result of our acetylation-biased purification scheme. Nevertheless, we were able to describe the major species of each PolyCAT A fraction. Additional H3 purification steps, for example, allowing methylation status-based separation, should reduce the occurrence of mixed spectra and permit more species to be sequenced in the future, like those enriched with H3K4me2 or species that are less abundant.
Only the intact mass of the N-terminal peptide was measured with high mass accuracy with FTMS. Therefore, bottom-up experiments were required when the number of 42-Da additions exceeded the number of acetylation sites determined by FTMS. In this manner, trimethylation could be differentiated from acetylation.
We previously demonstrated that the speed of ETD/PTR is compatible with conventional liquid chromatography/MS methods (31, 32); however, in this study, the necessity to acquire the data with zoom scan prolonged the analysis time to 3–10 min (see Materials and Methods). In sum, the middle-up analysis described above presents a compromise between a bottom-up analysis that is superior in sensitivity but lacks information about the interdependence of peptides and modifications and a top-down approach that provides an overview of modifications on the intact protein level, but often fails to give unambiguous assignment of modification and generally requires significantly more material than bottom- or middle-up approaches. We believe this methodology will be indispensable for understanding long-range, combinatorial roles of histone PTMs in bivalent domains or transcriptional regulation and PTMs on other protein families.
Distinct Combinations of Histone Modifications and Their Functional Implications.
Our findings that H3K4me3 and H3 hyperacetylation occupancy coexist on the same residue are consistent with reports localizing these PTMs to promoter proximal regions on active genes (see ref. 11 and references within). The identification of numerous chromatin-associated protein motifs, such as chromodomains, PHD fingers, tudor and tandem-tudor domains, WD40 repeats, and bromodomains, as PTM binding modules for methylated or acetylated histone lysines (10, 41) suggests that addition/removal of these modifications may be associated by nonrandom, even hierarchical relationships. Indeed, it was recently shown in vivo that H3 trimethylated at K4 is highly susceptible to acetylation turnover (42). To this end, recent reports suggest acetylation on H3 may be promoted through binding of H3K4me2 by Chd1 (SAGA) (43), binding of H3K4me3 by Yng1 (NuA3) (36, 44), and deacetylation through the H3K4me3/ING2 interaction (mSin3a–HDAC1) (45). Furthermore, because different permutations of histone-binding modules can be found together within multimeric complexes, it seems likely that comprehensive understanding of histone modification combinations will yield insight into how chromatin-templated machineries target diverse biological outputs to specific regions of the genome (10, 46).
Sequence analysis of Tetrahymena MAC H3 also identified a species of H3 with both K4me1 and K27me1 or K27me2 on the same tail (Fig. 3, groups 0–3). Given the recent report of bivalent PTM domains in ES cells (14), perhaps Tetrahymena also uses a variation of K4/K27 bivalent domains to reversibly control the on/off state of genes in the somatic, transcriptionally active nucleus. Our data also document the use of H3K27me3 in a lower, unicellular eukaryote, in this case almost exclusively in the transcriptionally silent, germ-line MIC, possibly serving as a mark to recruit micronuclear-specific silencing functions. In this regard, we look forward to the identification of the “writers” and “readers” of this mark in a model where histone genetics is feasible. The ability to identify unique combinations of histone PTM over relatively long distances of histone proteins should shed insights into how functionally distinct regions of euchromatin and heterochromatin are established and maintained in a wide range of organisms.
Materials and Methods
Tetrahymena Culture and Total Histone Purification.
T. thermophila (strain CU 427) was grown as described (50). MAC and MIC were purified as described (50), except the nucleus isolation buffer contained 1 mM PMSF and 10 mM sodium butyrate, but not spermidine. Total histones were extracted from purified Tetrahymena nuclei in 0.2 M H2SO4 as described (51).
Electrophoresis and Western Blotting.
SDS/PAGE was performed as described (52). Acid-urea gel electrophoresis was performed as described (53). Antibodies used to probe membranes included: α-General H3 (1:20,000), α-H3K4me1 (1:11,000; ab8895; Abcam, Cambridge, MA), α-H3K4me2 (1:50,000; ab7766; Abcam); α-H3K4me3 (1:50,000; ab8580; Abcam); α-H3K27me1(1:20,000; 07–448; UBI/Millipore, Billerica, MA), and α-H3K9/14Ac (1: 3,500). The antibodies α-H3K27me2 (1:2,000) and α-H3K27me3 (1:2,000) were gifts from Y. Zhang (University of North Carolina, Chapel Hill, NC) and T. Jenuwein (Research Institute of Molecular Pathology, Vienna, Austria), respectively. Secondary reagents and procedures for Western blot analyses were performed with ECL reagents from Amersham Life Sciences (Piscataway, NJ).
Purification of Histone H3 by Reversed-Phase and Cation Exchange.
H3 was purified from total macronuclear histones by RP-HPLC using a C8 column (220 × 4.6 mm; Aquapore RP-300, Shelton, CT) as described (54). The H3 fractions were pooled and dried under vacuum. Macronuclear H3 was further resolved with cation-exchange chromatography by using a PolyCAT A column (200 × 4.6 mm; Poly LC, Columbia, MD) as described (19). H3 was eluted at 0.4 ml/min with a gradient of 0–100% B in 65 min [solvent A, 8 M urea (deionized), 100 mM NaCl, 23 mM PO4 pH 7, 0.5 mM DTT; solvent B was solvent A with 500 mM NaCl]. Individual PolyCAT A fractions were further purified by RP-HPLC using a C8 column (220 × 2.1 mm; Aquapore RP-300). H3 was eluted at 0.4 ml/min with a gradient of 0–100% B in 25 min (solvent A, 5% acetonitrile in 0.1% TFA; solvent B, 90% acetonitrile in 0.1% TFA), after which fractions containing H3 were pooled, dried under vacuum, and subjected to mass spectrometry analysis.
Mass Spectrometry.
Lyophilized H3 fractions were digested with endoproteinase Glu-C (Roche Diagnostic, Indianapolis, IN) in 100 mM ammonium acetate (pH 4.0) at an enzyme to protein ratio of 1:15 for 4 h at 37°C. Digested aliquots were analyzed on a Finnigan LTQ-FTMS (Thermo Electron) using a linear gradient of 0–100% B in 17 min (A = 0.1 M acetic acid in nanopure water, B = 70% acetonitrile in 0.1 M acetic acid) on a 1100 series binary HPLC (Agilent Technologies, Palo Alto, CA) and a flow rate of 60 nl/ min, and the data were recorded in profile mode with a resolution of 100,000 or 400,000 (at m/z 400). See SI Text for details of chromatography columns.
Glu-C digestion mixtures of representative fractions from groups 0–2 were further fractionated by RP-HPLC on a C18 HAISIL 300 column (Higgins Analytical, Mountain View, CA) as described (39). Fractions containing the 1–50 residue were dried under vacuum and resuspended in 0.1% acetic acid. Before analysis, 0.1% acetic acid in acetonitrile was added to give a mixture of 60% water, 40% acetonitrile, and 0.1% acetic acid. The mixture was infused into the modified Finnigan LTQ (Thermo Electron), using the NanoMate 100 robot (Advion Biosciences, Ithaca, NY). Modifications to the LTQ allowing for ion/ion reactions have been detailed (28). Ion/ion reactions were performed as described (31). In brief, a 10-ms ETD reaction (fluoranthene radical anions) preceded a 100-ms PTR reaction (benzoate anions). The product ions were mass-analyzed by using profile mode and high-resolution scan (zoom scan) over the entire mass range from 250 to 2,000 Da. The acquisition time was on the order of 18 s per zoom scan, and 30–40 scans were averaged, resulting in an average acquisition time of 10 min per spectrum.
Fractions of groups 3–5 were directly loaded onto a nano-HPLC column (see SI Text for details). Peptides were eluted with a linear gradient of 0–20% B in 20 min and 20–100% B in 40 min (A = 0.1 M acetic acid in nanopure water; B = 70% acetonitrile in 0.1 M acetic acid) with a flow rate of 60 nl/min on an Agilent 1100 series binary HPLC. Peak parking (55) was performed by reducing the flow rate to 15 nl/min as the N-terminal peptide eluted. Precursor ions of interest were manually selected on the fly, and the ion/ion reactions were performed as described above. The average scan duration was 11 s, and ≈15 scans were averaged per spectrum, resulting in an average acquisition time of 3 min per spectrum.
Supplementary Material
Acknowledgments
We thank UBI/Millipore (formerly UBI Antibodies) for antibody donations and members of D.F.H.'s and C.D.A.'s laboratories for helpful discussions, especially J. Syka, J. Coon, M. Lachner, J. Tanny, and I. Cristea for critical readings of this manuscript. This work was supported by National Institutes of Health Grants GM53512 (to S.D.T.), GM63959 (to C.D.A.), RR00862 and RR022220 (to B.T.C.), and GM37537 (to D.F.H.) and The Rockefeller University (S.D.T., C.D.A., A.J.T., and B.T.C.).
Abbreviations
- PTM
posttranslational modification
- ETD
electron transfer dissociation
- PTR
proton transfer charge reduction
- MAC
macronucleus
- MIC
micronucleus
- FTMS
Fourier transform mass spectrometry.
Note Added in Proof:
Garcia et al. (56) recently reported additional histone H3 PTMs that are not included in Fig. 1A.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610993104/DC1.
References
- 1.van Holde KE. Chromatin: Springer Series in Molecular Biology. New York: Springer; 1988. [Google Scholar]
- 2.Wolffe AP. Chromatin: Structure and Function. San Diego: Academic; 1998. [Google Scholar]
- 3.Duerre JAB, Buttz HR. In: Protein Methylation. Paik WK, Kim S, editors. Boca Raton, FL: CRC; 1990. pp. 125–138. [Google Scholar]
- 4.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Turner BM. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 9.Ahn SH, Diaz RL, Grunstein M, Allis CD. Mol Cell. 2006;24:1–10. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Seet BT, Dikic I, Zhou MM, Pawson T. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 11.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Millar CB, Grunstein M. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 13.Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Burlingame AL, Zhang X, Chalkley RJ. Methods. 2005;36:383–394. doi: 10.1016/j.ymeth.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Bonaldi T, Regula JT, Imhof A. Methods Enzymol. 2004;377:111–130. doi: 10.1016/S0076-6879(03)77006-2. [DOI] [PubMed] [Google Scholar]
- 17.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 18.Delahunty C, Yates JR., III Methods. 2005;35:248–255. doi: 10.1016/j.ymeth.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Siino JS, Jones PR, Yau PM, Bradbury EM. Proteomics. 2004;4:3765–3775. doi: 10.1002/pmic.200400819. [DOI] [PubMed] [Google Scholar]
- 20.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 21.Kelleher NL. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 22.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. J Am Chem Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. J Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 24.Thomas CE, Kelleher NL, Mizzen CA. J Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 25.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. J Proteome Res. 2006;5:233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 26.Medzihradszky KF, Zhang X, Chalkley RJ, Guan S, McFarland MA, Chalmers MJ, Marshall AG, Diaz RL, Allis CD, Burlingame AL. Mol Cell Proteomics. 2004;3:872–886. doi: 10.1074/mcp.M400041-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Freitas MA. Int J Mass Spectrom. 2004;234:213–225. [Google Scholar]
- 28.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephenson JLJ, McLuckey SA. J Am Chem Soc. 1996;118:7390–7397. [Google Scholar]
- 30.Amunugama R, Hogan JM, Newton KA, McLuckey SA. Anal Chem. 2004;76:720–727. doi: 10.1021/ac034900k. [DOI] [PubMed] [Google Scholar]
- 31.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi A, Bai DL, Geer LY, Shabanowitz J, Hunt DF. Int J Mass Spectrom. 2007;259:197–203. doi: 10.1016/j.ijms.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vavra KJ, Allis CD, Gorovsky MA. J Biol Chem. 1982;257:2591–2598. [PubMed] [Google Scholar]
- 34.Strahl BD, Ohba R, Cook RG, Allis CD. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 36.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allis CD, Glover CV, Bowen JK, Gorovsky MA. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Proc Natl Acad Sci USA. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, et al. J Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 40.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 41.Mellor J. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Hazzalin CA, Mahadevan LC. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 44.Martin DG, Baetz K, Shi X, Walter KL, Macdonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 48.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 49.Joshi AA, Struhl K. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 50.Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 51.Taverna SD, Coyne RS, Allis CD. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 52.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 53.Lennox RW, Cohen LH. Methods Enzymol. 1989;170:532–549. doi: 10.1016/0076-6879(89)70063-x. [DOI] [PubMed] [Google Scholar]
- 54.Garcia BA, Joshi S, Thomas CE, Chitta RK, Diaz RL, Busby SA, Andrews PC, Ogorzalek Loo RR, Shabanowitz J, Kelleher NL, et al. Mol Cell Proteomics. 2006;5:1593–1609. doi: 10.1074/mcp.M600086-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Shabanowitz J, Settlage RE, Marto JA, Christian RE, White FM, Russo PS, Martin SE, Hunt DF. Mass Spectrometry in Biology and Medicine. Clifton, NJ: Humana; 2000. [Google Scholar]
- 56.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. J Biol Chem. 2006 doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.