Abstract
Darwin claimed that a unique inclusively hierarchical pattern of relationships between all organisms based on their similarities and differences [the Tree of Life (TOL)] was a fact of nature, for which evolution, and in particular a branching process of descent with modification, was the explanation. However, there is no independent evidence that the natural order is an inclusive hierarchy, and incorporation of prokaryotes into the TOL is especially problematic. The only data sets from which we might construct a universal hierarchy including prokaryotes, the sequences of genes, often disagree and can seldom be proven to agree. Hierarchical structure can always be imposed on or extracted from such data sets by algorithms designed to do so, but at its base the universal TOL rests on an unproven assumption about pattern that, given what we know about process, is unlikely to be broadly true. This is not to say that similarities and differences between organisms are not to be accounted for by evolutionary mechanisms, but descent with modification is only one of these mechanisms, and a single tree-like pattern is not the necessary (or expected) result of their collective operation. Pattern pluralism (the recognition that different evolutionary models and representations of relationships will be appropriate, and true, for different taxa or at different scales or for different purposes) is an attractive alternative to the quixotic pursuit of a single true TOL.
Keywords: lateral gene transfer, phylogeny
The meaning, role in biology, and support in evidence of the universal “Tree of Life” (TOL) are currently in dispute (1–15). Some evolutionists believe (i) that a single rooted and dichotomously branching representation of the relationships between all life forms is appropriate (at all levels above species), because it best represents their history; (ii) that we can with available data and methods reconstruct this tree quite accurately; and (iii) that we have in fact done so, at least for the major groups of organisms. Other evolutionists question the second and third of these beliefs, holding that data are as yet insufficiently numerous and phylogenetic models as yet insufficiently accurate to allow reconstruction of life's earliest divisions, although they do not doubt that some rooted and dichotomously branching tree can in principle represent the history of all life. Still other evolutionists, ourselves included, question even this most fundamental belief, that there is a single true tree. All sides express confidence in their positions, and the debate often seems to be at an impasse (see for instance refs. 9–12).
This situation has its roots deep in the history of phylogenetics and indeed in the pre-Darwinian philosophical and systematic tradition. Our purpose here is to show that the debate owes its intensity and protracted nature to an unresolved and largely unrecognized difference in what phylogeneticists think the TOL is supposed to represent. For many of its supporters, the TOL is a biological fact (a reality outside of our own minds), first established nearly 150 years ago by Darwin, and needing only elaboration (16). For those who question it, the TOL is a scientific hypothesis (a heuristic epistemological model), forcefully and eloquently articulated by Darwin but not yet proven to be true (17, 18). We develop this second position here, suggesting a formulation of the TOL hypothesis that might generally be accepted as being faithful to Darwin's original intent, and discussing its testability and status in the context of prokaryotic data.
This exercise has implications for phylogenetic practice and many areas of biological theory. Questions about the structure of the TOL are, after all, secondary to questions about whether such a branching pattern actually corresponds to anything in nature (rather than being imposed on nature by the habits of systematists), and if so, whether a branching evolutionary process is its underlying cause.
Darwin's “Hidden Bond”
Classification is an important practice in the management of knowledge of all sorts. It has long been central to biology, as has been the notion that some classification schemes are more natural than others because they more closely map to an underlying natural structure or principle, which could be linear (the great chain of being), partially circular (the Quinarian systematics of the early 19th century), or tree-like in character (17, 19, 20). Darwin accepted as his explanandum (that which is to be explained) that a natural classification does exist and that it is tree-like (an inclusive hierarchy). He sought in his theory of evolution the explanans (the explanation), the cause of that structure in nature.
Darwin makes this agenda perfectly clear in several passages in On the Origin of Species (16). At the beginning of Chapter 13, for instance, he suggests that classification is not an arbitrary practice.
From the first dawn of life, all organic beings are found to resemble each other in descending degrees, so that they can be classed in groups under groups. This classification is evidently not arbitrary like the grouping of the stars in constellations.
And then, a dozen paragraphs later, he tells us why it is not arbitrary, because a natural process (evolution) is the root cause of the hierarchical patterns long recognized by systematists.
All of the foregoing rules and aids and difficulties in classification are explained, if I do not greatly deceive myself, on the view that the natural system is founded on descent with modification; that the characters which naturalists consider as showing true affinity between any two or more species, are those which have been inherited from a common parent, and, in so far, all true classification is genealogical; that community of descent is the hidden bond which naturalists have been unconsciously seeking, and not some unknown plan of creation, or the enunciation of general propositions, and the mere putting together and separating objects more or less alike.
Any classification of groups under groups is of course an inclusive hierarchy, representable as a tree, a model endorsed by Darwin in Chapter 4.
The affinities of all of the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth. The green and budding twigs may represent existing species; and those produced during former years may represent the long succession of extinct species […] The limbs, divided into great branches, and these into lesser and lesser branches, were themselves once, when the tree was young, budding twigs, and this connection of the former and present buds by ramifying branches may well represent the classification of all extinct and living species in groups subordinate to groups.
In Darwin's theory, similarities between species reveal their common descent, whereas differences that result in or from speciation reveal species' modifications that are driven by natural selection. In directing readers' attentions to the only figure in On the Origin of Species (his tree diagram), he stressed the essential role of competition and selection in diversification, without which the TOL might resemble the poplars of Lombardy more than the live oaks of Louisiana.
I attempted also to show that there is a constant tendency in the forms that are increasing in number and diverging in character, to supplant and exterminate the less divergent, the less improved, and preceding forms. I request the reader to turn to the diagram illustrating the action, as formerly explained, of these several principles; and he will see that the inevitable result is that the modified descendants proceeding from one progenitor become broken up into groups subordinate to groups. [Our italics.]
The TOL Hypothesis
We paraphrase Darwin's reasoning as “the TOL hypothesis,” which we take as half of his larger theory (the other half being concerned with the operation of natural selection), as follows.
The pattern of groups subordinate to groups embraced by a unique inclusively hierarchical classification based on homologies (true affinities in Darwin's language) is indeed not arbitrary. It reflects an underlying natural reality with a natural cause, rather than “some unknown plan of creation, or the enunciation of general propositions” in Aristotelian logic, embedded in the practices of systematists.
That natural cause is historical, and in particular, it is direct descent with modification, a branching process whose branches will be recaptured in the most truly natural and correct classification, which might in principle be extended to include the last common ancestor (or ancestors) of all extant forms.
Modification is driven by natural selection.
The TOL hypothesis could be falsified by substantial failure of any of these propositions. First, and most fundamentally, the pattern of groups subordinate to groups might be illusory or artifactual, “the mere putting together and separating objects more or less alike” (16) in accordance to expectation. In this case there would be no explanandum, no all-embracing pattern or fact existing in nature and independent of our desire to impose order. Second, similarities between species used to erect the TOL (or any natural scheme) might not predominantly reflect common descent. Patterns of resemblance recognized by systematists could after all result from some natural cause other than direct (branching) descent with modification, such as environmental constraint and convergence, parallelism, or reticulation. Third, selection and branching species divergence might not be inevitably connected. Sometimes selection will drive reticulation [as with lateral gene transfer (LGT) of novel adaptations], whereas sometimes divergence will be produced by stochastic processes (drift).
As to this third possibility, modern evolutionists accept the uncoupling of selection from divergence, not only at the molecular level (the neutral theory) but in certain models for speciation, without seeing the Darwinian (or at least the neo-Darwinian) theory as refuted (21, 22). We have come to appreciate the plurality of evolutionary processes of lineage diversification. But most of us hold on to the first two tenets, that there is a real and universal natural hierarchy, and that descent with modification explains it, in much the same way as Darwin did. We may be process pluralists, but we remain pattern monists.
Of course, a trivial case for falsification could be made on the basis of a single reticulation event, but biologists in general have long accepted that theories about pattern and process need be true only in general. We will argue that inclusive hierarchical classifications do not emerge naturally and consistently from the relevant prokaryotic data considered in general (in their entirety). Instead, they have been imposed on them by selective analyses that are based on the assumption that a tree must be the real natural pattern, even if only certain of the data can be trusted to reveal it. Furthermore, we propose that the underlying historical processes affecting prokaryotes are more complex and various than those imagined by Darwin (or by neo-Darwinists), and not of necessity expected to give rise to a natural hierarchy.
Problematically, Darwin depended on the notion that the true pattern of natural relationships is a tree in the construction of his theory of the responsible process and, as Panchen (17) notes, his explanandum was subsequently considered by him as a part of the proof that his theory (explanans) was right. That classifications should be constructed as hierarchies because evolution is a branching process and that hierarchical classification is a proof of branching evolution is the mixed message many of us took from our early education as biologists. But we now have ample other evidence supporting the reality of evolution. We could thus dispense with the tree (and such semicircular reasoning), should this particular historical premise about branching fall short, without weakening the solid edifice of evolutionary biology.
Classification, Evolution, and the Nature of Biology
The body of data (the explanandum) for which a hypothesis (the explanans) proposes to account cannot at the same time constitute proof for that hypothesis (17, 23), nor can further data of the same kind. We might construct a hierarchical taxonomy of Drosophila based on certain morphological characters and claim that its branching pattern reflects an evolutionary branching process. Adding more taxa would bush out the tree but not strengthen this fundamental claim about process, nor would adding more characters, necessarily, if there were reason to believe that by functional constraint these characters were correlated with the first set. Much of what has happened in post-Darwinian phylogenetics has been an enormous expansion of the explanandum (accepted from the outset by Darwin) by the addition of new taxa or characters (24–32). Moreover, this expansion has for the most part used algorithmic tools that are constrained to produce trees.
Alec Panchen elaborates similar and other concerns at much greater philosophical depth in his 1992 Classification, Evolution and the Nature of Biology. He stresses Darwin's acceptance of a natural hierarchy and his theory as an explanation of that “fact.”
Natural selection is one component of evolutionary theory as proposed by Darwin and Wallace, but the other, for which selection is merely an hypothesis of mechanism, is the theory that evolution has occurred […] the theory of evolution states that the apparent relationships of organisms in a systematic classification are real relationships, because “relationship” in such a classification is not a metaphor but is actually to be ascribed to community of descent.
But as Panchen shows, the explanandum of a universal tree-like hierarchy in systematic classification is grounded more in the Western philosophical tradition (the logical division of Plato, Aristotle, and Porphyry) than in observation. Indeed, the Linnaean system, to which we still adhere for taxonomical purposes, is a relatively recent product of that long tradition and was only one of several schema that were popular in the first half of the 19th century. Ironically, it is primarily Darwin's theory, which overlies and obscures the philosophical roots of tree-like representations, that reinforces the current belief in the hierarchy's naturalness, its status as fact. Arguably, our systematic practices today (especially in microbiology) might look quite different had Darwin (or someone else) not formulated a branching theory of evolution.
Importantly, Darwin did not and could not test the reality of the tree pattern. Indeed, one is hard pressed to find some theory-free body of evidence that such a single universal pattern relating all life forms exists independently of our habit of thinking that it should. The notion that a tree pattern is the product of induction, obvious to any intelligent observer, is belied by most of the early history of systematics, during which quite different schemes seemed fully defensible.
In a search for independent evidence of a natural hierarchy, Panchen considers homology, paleontology, and biogeography. The first is problematic in that true (taxic) homologies cannot be distinguished from false ones (homoplasies) without some assumption of hierarchy: homologies are more often deduced from trees than trees are from homologies. Thus, explanans melds with explanandum, and neither is tested. The second and third may offer independent evidence that evolution by descent with modification has occurred but are limited in their relevance and applicability to specific groups, areas or times. They do not justify, except by extrapolation, the expectation that there should be groups under groups at all levels, that there should be a universal TOL, dichotomously branching all of the way down to a single root. Alternatives [extensive reticulation or separate origins from a common inchoate ancestral state (2, 14, 34)] can be entertained.
The possibility that hierarchy is imposed by us rather than already being there in the data is especially relevant to the more recent extension of hierarchical classification into the prokaryotic domains, and of TOL thinking into the ancient unicellular past, with the aid of molecular phylogenetics (33–35), our principal areas of concern in this article. There had been no previous commitment to a single hierarchy by microbial systematists, and little detail provided by the microbial fossil record. Thus, the molecular phylogenetic extrapolation set in train in the mid 1960s has been an exceptionally bold one.
Zuckerkandl and Pauling and the Independence of Molecular Evidence
The essay Evolutionary Divergence and Convergence in Proteins, published in 1965 by Emile Zuckerkandl and Linus Pauling (36), is remarkable for its many insights and since-proven-correct predictions concerning the discipline of molecular phylogenetics. Especially relevant to our purposes here, Zuckerkandl and Pauling articulated the view that molecular phylogenies could be independent from traditional trees based on comparative morphology and other organism-level characters.
There is yet an ultimate reason, of a more philosophical nature, for interest in the paleogenetic approach. Whereas the time dependence of evolutionary transformations at the molecular level can only be established with reference to extraneous sources, the topology of branching of molecular phylogenetic trees should in principle be definable in terms of molecular information alone. It will be determined to what extent the phylogenetic tree, as derived from molecular data in complete independence from the results of organismal biology, coincides with the phylogenetic tree constructed on the basis of organismal biology. If the two phylogenetic trees are mostly in agreement with respect to the topology of branching, the best available single proof of the reality of macroevolution would be furnished.
Zuckerkandl and Pauling's emphasis seems to be largely on epistemological rather than ontological independence of molecular and traditional data. If genotype causes phenotype, then it should not be surprising that trees derived from the former resemble trees based on the later. Although independence can be persuasively argued at a within-species level on the basis of the neutral theory (21) and for certain molecular data for certain purposes at deeper levels [Alu insertions to establish primate phylogenies or gene fusions to root the tree of eukaryotes (13)], it might be going too far to claim that an independent “proof of the reality of macroevolution” could be obtained by the usual sorts of sequence-based phylogenetic analyses.
That said, one could argue that, independent or not, molecular trees are superior to organismal-biology-based trees as indicators of true relationships, because genotype is causally prior to principle phenotype. Zuckerkandl and Pauling asserted this principle forcefully, and most molecular biologists who embrace “the central dogma” would surely agree. Furthermore, from what we know (and Darwin could not) about the nature and replication of genetic information, branchings in molecular trees of orthologs might be taken as direct records of historical organismal lineage splittings. Thus, in an early (1972) edition of the Atlas of Protein Structure, Margaret Dayhoff and R. V. Eck (37) wrote: “One of the grand biological ideals is to be able to work out the complete, detailed, quantitative phylogenetic tree, the history of the origin of all living species, back to the very beginning. Biologists have had this hope for a long time; biochemistry now has the actual capability of accomplishing it.”
Microbial Phylogenetics and the Path Not Taken
For microbiologists, it was this promise of extending hierarchical classification into the prokaryotes, not any notion of testing Darwin's theory, that was the relevant and exciting message from Zuckerkandl and Pauling. There seemed no possibility of assessing the overall congruence of organismal and molecular trees, because microbial systematists had given up on the former and since the mid 1950s have been content with more practical schemes aimed at reliable species-level identification (38, 39). Indeed, in his seminal 1987 review Bacterial Evolution, Carl Woese (34) stressed the incompatibility of ribosomal RNA phylogenies with even those few higher taxa that microbiologists still believed in and noted that “not only did we know very little about eubacterial phylogeny before the advent of the rRNA approach, but what we thought we knew tended to be wrong.”
Woese did express the concern that a natural hierarchy might not extend into the prokaryotes (which embrace perhaps two-thirds of the biota and the first two-thirds of life's history).
In classifying bacteria microbiologists make two implicit assumptions: (i) that bacteria have a phylogeny, and (ii) that the taxonomic system that works well for the metazoa is actually applicable to, i.e., meaningful in, the microbial world. These two points require explications and discussion, for they are far from self evident.
Neither would hold, Woese realized, if LGT were a significant evolutionary force. If transfer were rampant, “a bacterium would not actually have a history in its own right: it would be an evolutionary chimera.” But “fortunately,” he wrote, “the matter is experimentally decidable. Were an organism an evolutionary chimera, then its various chronometers [phylogenetic markers] would yield different, conflicting phylogenies.” Woese then hazarded that such chimerism probably would not prove a significant problem, from the example provided by congruent alpha-proteobacterial trees based on cytochromes c and rRNA, from the general robustness of the rRNA phylogeny, and from its ability to predict certain domain-level phenotypic characters (for instance, the differences in bacterial and archaeal cell envelopes, transcription, and translation).
Woese thus importantly recognized that if a hierarchical pattern of organization is to be taken as a fact of nature and a legitimate basis for Darwin's theory, evidence for its existence must come not from the congruence of traditional organismal with molecular phylogenies, but from congruence among multiple molecular phylogenies. Put another way, if an hierarchical pattern of groups subordinate to groups can claim to be the natural order only on the basis of molecular data, then that claim could be refuted by substantial disagreement among molecular data sets.
LGT and the Disappearance of the Explanandum
Leaving aside the weakness of phylogenetic signal, which plagues most phylogenetic analyses (18, 40), molecular data sets substantially disagree at many levels of analysis, particularly because of LGT. Although there is increasing evidence for the importance of LGT in the evolution of eukaryotes, especially unicells (41), again we focus on prokaryotes; because LGT is almost certainly more frequent among them, much more extensive and relevant comparative genomic data are available for them, and two-thirds of the history of life (and thus, of the TOL) is theirs. For prokaryotes, LGT, mediated by transduction, conjugation, and transformation, can effect exchange over even the greatest evolutionary distances, whereas homologous recombination is a potent force among closer relatives (3, 42). Both LGT and homologous recombination convert trees into networks, albeit on different scales (43).
Homologous recombination, once thought to be rare in prokaryotes, is now known to be the principle cause of sequence divergence in many bacteria [and at least some archaea (44–46)]. Among strains of a designated species, recombination may be so vigorous that housekeeping genes shared between them can have different within-species trees. As with sexually reproducing animals, there is thus no single true phylogeny for individuals within a species (47), although trees based on concatenated sequences are often erroneously taken to represent such a phylogeny.
Although some prokaryotic species may be as tightly bounded by barriers to recombination as vertebrate species are, there is no reason that they should always be so, and there are instances where they clearly are not (48–51).
Much (up to 30%) within-species genome-to-genome variation in gene content results from LGT and gene loss: in some species, the “pan-genome” seems of unlimited size (52–54).
Genes that are patchily distributed among strains of a species, species of a phylum or indeed phyla of Bacteria and Archaea (55), include many that are essential in determining key phenotypic differences at those levels and are of special interest to systematists and physiologists, such as drug resistance, virulence, catabolism, phototaxis, photosynthesis, nitrogen fixation, and aerobiosis.
Within major bacterial divisions (“phyla”), such as gamma-proteobacteria, widely shared (“core”) genes generally make up at most 20% of any genome (9, 12, 56, 57). Although many such genes might have a common within-division phylogeny, this claim is very difficult to prove: individual gene signals are generally too weak to distinguish between many alternative topologies (10, 12). If one assumes a single within-division tree, then the collective signal (such as might be obtained by concatenating core genes) could reveal its structure by robustly supporting one or a few topologies. But such collective robustness does not in fact prove that there is a shared tree (40), because concatenation by itself will increase robustness (bootstrap values), even for random data.
In addition, many core genes clearly do have histories of within- and between-division (or domain) LGT, even for supposedly conservative groups like the cyanobacteria (57).
Gene sharing by LGT does not respect domain boundaries. The hyperthermophilic bacterium Thermotoga maritima likely obtained a quarter of its genes from archaea, and the archaean Methanosarcina mazei may have enjoyed an equally generous donation from bacteria (58, 59). There is also increasing evidence for transdomain LGT into the nuclear genomes of eukaryotes, especially phagotrophic unicells (41, 60).
At the domain level, the evidence for shared phylogeny among shared genes is even scantier than for phyla. The number of genes that can be shown (by their universal or very frequent presence in many bacterial and archaeal divisions) to comprise the “universal core” make up <5% of the average prokaryotic genome (56). Again, it is hard to prove that these few have a common evolutionary history, and rooting the universal tree remains highly problematic (61, 62).
Evolutionists still disagree about the meaning of such data and especially about their failure to prove phylogenetic congruence. Charles Kurland et al. (5) wrote in these pages three years ago “that HGT [LGT] has been ascribed such an inflated role” because “its frequency has been overestimated by the failure to distinguish it from other phylogenetic anomalies.” Indeed, there have been many instances in which insufficient phylogenetic signal or methodological artifact have been improperly claimed as LGT, most notably perhaps in the case of our own genomes (63). But to make “vertical descent” the null hypothesis against which claims for LGT must be tested is to assume that which is to be proved: that an inclusive hierarchy exists independently of our beliefs. And phylogenetic incongruence is only one part (often the weakest part) of the evidence for LGT. Stronger are the various sorts of direct evidence for the operation of LGT promoting agents (phages, plasmids, integrons, pathogenicity islands, and the like). Strongest is the inescapable conclusion that the majority of genes in most genomes, because they are patchily distributed within their respective species, phyla, or domains, perforce have complex (and non-“vertical”) evolutionary histories.
Because there is substantial disagreement among prokaryotic molecular data sets and little strongly supported congruent signal among data sets that do not clearly disagree, a claim that a hierarchical pattern of groups subordinate to groups is the universal natural order cannot be sustained as an explanandum (6, 8, 18). (That many seemingly different analyses of these data nevertheless do agree in some ways is not surprising and is discussed later.) And from what we know of the nature and frequency of processes of gene exchange and gain and loss through homologous recombination and LGT, which obey a model of inheritance different from Darwin's concept of descent and offer a more modern explanans, there is no strong expectation that a universal hierarchy that embraces all life should be produced with molecular markers.
The TOL Is Not the Tree of Cells
Microbial phylogeneticists have not in general taken it to be their duty to confirm the existence of a natural inclusive hierarchy or tested the TOL hypothesis that this hierarchy is to be explained by an historical branching process. The ways in which they generally analyze and think about the molecular data presuppose a tree model, and cannot but produce trees. Even when methods that permit reticulated representations of evolution are used, the most common intent has been to discount LGT as noise, in pursuit of the legitimate “phylogenetic signal” assumed to be vertical unless significantly in conflict (11, 64, 40, 66). That is, weak signal is by default taken as vertical. Most importantly, the vast majority of analyses have consisted of the comparative evaluation of one tree with others (in search of the “true” branching topology). Seldom have investigators asked whether non-tree (reticulated) models might not better explain the data at hand. (Exceptions most often involve within-species data sets, where recombination is expected.)
Nevertheless, some phylogeneticists accept that LGT has been so pervasive over the history of life that no hierarchical classification can claim to provide the unique and true accounting of similarities and differences between organisms (and thus, an explanandum for Darwin's explanans). Woese himself asks “What does it mean, then, to speak of an organismal genealogy when nearly all of the genes in the cell, genes that give it its general character, do not share a common history? This question goes beyond the classical Darwinian context” (2).
Many of these same phylogeneticists would claim that their goal all along has actually been the reconstruction of something we might call “the tree of cells,” an “organismal genealogy” (2) retracing all division events back to a single last universal common ancestor. This genealogy might be shown by very few or even no individual genes and yet somehow be recoverable from gene sequence or presence or absence of data. They would base their belief that such a genealogy must exist and be recorded in genes on common-sense observations about processes of genome replication and cell division and the notion, expressed most forcefully by Zuckerkandl and Pauling forty years ago, that an organism's “main memory banks are those polynucleotides that are capable of self-duplication” (36).
In just the last few years, there have been claims to discovery of this tree of cells (nevertheless, still called the “Tree of Life”), using variously concatenated core gene data sets (64–66), genes preselected as likely untransferable (67, 68), distance matrices based on gene presence or absence (69–72), methods using presence/absence as characters (14, 73), shared structural motifs and protein domains (74, 75), and supertrees based on different data at different nodes (6, 31). The two most popular approaches apply maximum likelihood or Bayesian methods to concatenates of the very small number of core genes that are retained by all (or most) genomes, or instead construct distance trees that are based on the much larger numbers of apparent orthologs shared between pairs of genomes. Using the first approach, Ciccarelli et al. (76) have recently presented an updatable “automatic reconstruction of a highly resolved tree of life” that uses only 31 genes, and claim that “the resulting tree of life will be an invaluable tool in many areas of biological research, ranging from classical taxonomy, via studies on the rate of evolution, to environmental genomics where DNA fragments of unknown phylogenetic origin need to be assigned” (76). Of trees based on the second approach, Snel et al. (7) have claimed that, because of their apparent consistency with each other or with rRNA trees, they “have yielded the fundamental insight that genome evolution is largely a matter of vertical transmission.”
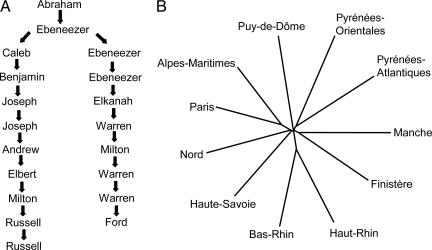
Though such methods may or may not retrace the tree of cells, it is simply misleading to assert they will have such high predictive or retrodictive value (allowing reconstruction of past events) or that they prove that vertical transmission has been the dominant process over all of evolution. So firm is the grip of tree-thinking (77) that it may be easiest to show why these approaches can mislead us through simple analogies or thought experiments such as those presented in Fig. 1. Most importantly in the current context, there is a logical disconnection between the “great tree” of Darwin's simile, which purported to explain the branching patterns of affinities between organisms on which natural classification could be based, and the tree of cells, which can make no such claims, because a majority of phenotype-determining genes have different histories. [Indeed, there is no guarantee that any gene in such a tree, which Dagan and Martin (78) call “the tree of 1%,” has its topology as their phylogeny.] If the tree of cells is taken as a biological fact, it is in any case not the same fact that Darwin accepted as the explanandum of his theory.
Fig. 1.
Two analogies to illustrate misapplication of tree-thinking. (A) Shown is a genealogy relating the senior author to another molecular evolutionist, Russell F. Doolittle. This pattern reflects the inheritance of surnames and Y chromosomes, but of <0.5% of our respective genomes. Only an extreme patriarchalist would privilege this particular pattern of relationships or the common ancestor we share over the hundreds that we do not so far back in history. And yet, such privileging is not dissimilar to basing the phylogeny of prokaryotes on only a tiny subset of the genes that any of them contain, knowing that other genes will give different results. (B) Shown is a second thought experiment, a distance tree of the départements of France based on the number of shared surnames (among the top 500) in pairwise comparisons of lists of the most frequent names in each. The pattern is interesting, tells us much about French history and demographics, and is ultimately based on the genealogies of individuals. Some of its nodes would likely also be recovered by independent measures, such as geographical separation or dietary preferences. But this pattern is by no means a phylogeny of French départements, because départements do not arise by a branching process of descent with modification and the nodes on this tree in no way correspond to ancestral departments. Yet similar trees based on shared gene content of genomes (even when extensive LGT is admitted) have often been presented as the phylogenies of those genomes, and the recovery of some nodes by independent measures (rRNA phylogenies) has been claimed in their support.
Process Pluralism and Pattern Pluralism
Evolutionists have long acknowledged a diversity of population-level diversification mechanisms (selection, drift, convergence, and parallelism) and (with reservations) clade-level mechanisms that extend beyond the selectionist and gradualist framework mapped out by Darwin. At the genome level, vertical descent and LGT, gene creation, duplication and loss, in all combinations with population-level processes, expand the evolutionary repertoire. A multifaceted process pluralism is now the common view (79). The belief that nature must nevertheless exhibit a single pattern of true relationships among taxa remains vigorous, and fuels the continued enthusiasm for universal tree building and its broad application on the basis of very few and often contradictory data. We call this belief “pattern monism.” “Pattern pluralism” (the recognition that different evolutionary models and representations of relationships will be appropriate and true for different taxa or for different purposes) is an appealing alternative, and can defuse the crisis within the discipline.
To be sure, much of evolution has been tree-like and is captured in hierarchical classifications. Although plant speciation is often effected by reticulation (80) and radical primary and secondary symbioses lie at the base of the eukaryotes and several groups within them (81, 82), it would be perverse to claim that Darwin's TOL hypothesis has been falsified for animals (the taxon to which he primarily addressed himself) or that it is not an appropriate model for many taxa at many levels of analysis. Birds are not bees, and animals are not plants. But in other taxa or at other levels, reticulation may be the relevant historical process, and nets or webs the appropriate way to represent what is a real but more complex fact of nature. Available software [such as Splitstree (43), NeighborNet (83), Lumbermill (84), or T-Rex (85)] allow biologists to explore phylogenetic patterns that are not necessarily tree-like. Other approaches, such as the analysis of the plurality signal within a data set or the elimination of impossible relationships, also explore phylogenetic signal without seeking to produce, a priori, a common tree as output. In the near future, even more sophisticated methods should be available, because mathematical research into phylogenetic network reconstruction is presently very active (43, 86–88).
To a pluralist, it is unsurprising that universal trees based on concatenated core gene data sets, pairwise similarities in gene content, or 16S rRNA can be similar, insofar as Bacteria, Archaea, and many of the phyla within them (although not their branching orders) are often re-created (e.g., see refs. 4, 6–8, and 76). Preferential patterns of gene exchange and similarities in cellular adaptations will favor such coherence (3). (To return to the analogy in Fig. 1B, we would not be surprised if a tree with similar structure could be obtained for French départements with other analyses and other data sets, such as dietary preferences or speech patterns: still, we would neither consider this tree to be phylogenetic nor take its internal nodes to be ancestral départements). But the three-domain rRNA-based scheme and its many subsidiary branchings is not the only true representation of the phylogenetic relationships between their member species, and these domains (and the various kingdoms or phyla within the first two, at least) are not what Darwin (or for that matter, Linnaeus or Hennig) understood higher taxa to be.
Darwin's TOL hypothesis, like most biological theories, is a claim about the process that underlies a pattern. It is important for modern phylogeneticists to remember that reconstructing the TOL was not the goal of Darwin's theory, but rather it was an integral element of his developing model of the evolutionary process. Importantly, this simile prompted generations of scientists to take Darwin's claim that evolution had occurred seriously, for all his lack of a coherent theory of inheritance. The TOL was thus the ladder that helped the community to climb the wall of acceptance and understanding of evolutionary process. But now that we have climbed it, we do not need this ladder anymore. In 2006, our understanding of evolution at the molecular, population genetic, and ecological levels is rich and pluralistic in character and does not require (or justify) a monistic view of the phylogenetic pattern.
Holding onto this ladder of pattern is an unnecessary hindrance in the understanding of process (which is prior to pattern) both ontologically and in our more down-to-earth conceptualization of how evolution has occurred. And it should not be an essential element in our struggle against those who doubt the validity of evolutionary theory, who can take comfort from this challenge to the TOL only by a willful misunderstanding of its import. The patterns of similarity and difference seen among living things are historical in origin, the product of evolutionary mechanisms that, although various and complex, are not beyond comprehension and can sometimes be reconstructed.
In this regard, our task is not different from that of contemporary cultural or social historians. We know much about what can happen and have a variety of tools by which we might unravel what has happened. We should use them all, but without seeking some elusive unifying “metanarrative,” either tree or web. Phylogenetics could become again the rich and realistic science of the genesis of phyla and address within a multifaceted pluralistic framework not only new questions about the past [identification of networks (89), hubs and highways (90) of gene exchange and vertical descent] but also the present (in particular, through integration of metagenomic data with evolutionary and ecological theory).
Acknowledgments
We thank Andrew Roger for critical discussion. This work was supported by the Canada Research Chair Program, the Canadian Institute for Advanced Research, and the Canadian Institutes for Heath Research.
Abbreviations
- TOL
Tree of Life
- LGT
lateral gene transfer.
Footnotes
The authors declare no conflict of interest.
References
- 1.Doolittle WF. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR. Proc Natl Acad Sci USA. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gogarten JP, Doolittle WF, Lawrence JG. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 4.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 5.Kurland CG, Canback B, Berg OG. Proc Natl Acad Sci USA. 2003;19:9658–9692. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daubin V, Gouy M, Perriere G. Genome Res. 2002;112:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snel B, Huynen MA, Dultith BE. Annu Rev Microbiol. 2005;58:191–209. doi: 10.1146/annurev.micro.59.030804.121233. [DOI] [PubMed] [Google Scholar]
- 8.Creevey CJ, Fitzpatrick DA, Philip GK, Kinsella RJ, O'Connell MJ, Pentony MM, Wilkinson M, McInerney JO. Proc Biol Sci. 2004;271:2551–2558. doi: 10.1098/rspb.2004.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerat E, Daubin V, Moran NA. PLoS Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bapteste E, Boucher Y, Leigh J, Doolittle WF. Trends Microbiol. 2004;12:406–411. doi: 10.1016/j.tim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Lerat E, Daubin V, Ochman H, Moran NA. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Susko E, Leigh J, Doolittle WF, Bapteste E. Mol Biol Evol. 2006;23:1019–1030. doi: 10.1093/molbev/msj113. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T. Biol Direct. 2006;1:e19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera MC, Lake JA. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 15.Delsuc F, Brinkmann H, Philippe H. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 16.Darwin C. On the Origin of Species. London: Murray; 1859. [Google Scholar]
- 17.Panchen AL. Classification, Evolution and the Nature of Biology. Cambridge, UK: Cambridge Univ Press; 1992. [Google Scholar]
- 18.Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. BMC Evol Biol. 2005;5:e33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Hara JO. Biol Philos. 1991;6:255–274. [Google Scholar]
- 20.McOuat GR. Stud Hist Phil Sci. 1996;27:473–519. doi: 10.1016/0039-3681(95)00060-7. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 22.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 23.Hempel C. Aspects of Scientific Explanation and Other Essays in the Philosophy of Science. New York: Free Press; 1965. [Google Scholar]
- 24.Haeckel E. Generelle Morphologie der Organismen: Allgemeine Grundzüge der organischen Formen-Wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte Descendenz-Theorie. Berlin: Georg Reimer; 1866. [Google Scholar]
- 25.Taylor FJR. BioSystems. 1978;10:67–89. [Google Scholar]
- 26.Copeland HF. The Classification of Lower Organisms. Palo Alto, CA: Pacific; 1956. [Google Scholar]
- 27.Schwartz RM, Dayhoff MO. Science. 1978;199:395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 28.Sogin ML. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 29.Clarke GD, Beiko RG, Ragan MA, Charlebois RL. J Bacteriol. 2002;184:2072–2080. doi: 10.1128/JB.184.8.2072-2080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- 31.Bininda-Emonds OR. Trends Ecol Evol. 2004;19:315–322. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Baldauf SL. Trends Ecol Evol. 2002;17:450–451. [Google Scholar]
- 33.Schwartz RM, Dayhoff MO. Science. 1978;199:395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 34.Woese CR. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace NR. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 36.Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel HJ, editors. New York: Academic; 1965. pp. 97–166. [DOI] [PubMed] [Google Scholar]
- 37.Dayhoff MO, Eck RV. In: Atlas of Protein Sequence and Structure 1972. Dayhoff MO, editor. Vol 5. Silver Spring, MD: Natl Biomed Res Found; 1972. [Google Scholar]
- 38.Stanier RY, Doudoroff M, Adelberg EA. The Microbial World. 1st Ed. Engelwood Cliffs, NJ: Prentice–Hall; 1957. [Google Scholar]
- 39.Woese CR. Microbiol Mol Biol Rev. 2004;68:173–186. doi: 10.1128/MMBR.68.2.173-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Andersson JO. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gogarten JO, Townsend JP. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 43.Huson DH, Bryant D. Mol Biol Evol. 2006;2:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 44.Feil EJ, Spratt BG. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 45.Hanage WP, Fraser C, Spratt BG. BMC Biol. 2005;3:e6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papke RT, Koenig JE, Rodriguez-Valera F, Doolittle WF. Science. 2004;306:1928–1929. doi: 10.1126/science.1103289. [DOI] [PubMed] [Google Scholar]
- 47.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feil E, Xhou J, Maynard Smith J, Spratt BG. J Mol Evol. 1997;43:631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- 49.Maynard Smith J, Feil EJ, Smith NH. BioEssays. 2000;23:54–61. [Google Scholar]
- 50.Silva C, Vinuesa P, Eguiarte LE, Souza V, Martinez-Romero E. Mol Ecol. 2005;14:4033–4050. doi: 10.1111/j.1365-294X.2005.02721.x. [DOI] [PubMed] [Google Scholar]
- 51.Doolittle WF, Papke RT. Genome Biol. 2006;7:e116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser-Liggett CM. Genome Res. 2005;15:1603–1610. doi: 10.1101/gr.3724205. [DOI] [PubMed] [Google Scholar]
- 53.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Konstantinidis KY, Tiedje JM. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 56.Charlebois RL, Doolitttle WF. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, et al. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 59.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Baumer S, Jacobi C, et al. J Mol Microbiol Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- 60.Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ. Proc Natl Acad Sci USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bapteste E, Brochier C. Trends Microbiol. 2004;12:9–13. doi: 10.1016/j.tim.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Zhaxybayeva O, Lapierre P, Gogarten JP. Protoplasma. 2005;227:53–64. doi: 10.1007/s00709-005-0135-1. [DOI] [PubMed] [Google Scholar]
- 63.Andersson JO, Doolittle WF, Nesbø CL. Science. 2001;292:1848–1850. doi: 10.1126/science.1062241. [DOI] [PubMed] [Google Scholar]
- 64.Brochier C, Bapteste E, Moreira D, Philippe H. Trends Genet. 2002;18:1–5. doi: 10.1016/s0168-9525(01)02522-7. [DOI] [PubMed] [Google Scholar]
- 65.Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Nat Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- 66.Brochier C, Forterre P, Gribaldo S. BMC Evol Biol. 2005;5:36. doi: 10.1186/1471-2148-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dutilh BE, Huynen MA, Bruno WJ, Snel B. J Mol Evol. 2004;58:527–539. doi: 10.1007/s00239-003-2575-6. [DOI] [PubMed] [Google Scholar]
- 68.Daubin V, Moran NA, Ochman H. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- 69.Snel B, Bork P, Huynen MA. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 70.Fitz-Gibbon ST, House CH. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.House CH, Fitz-Gibbon ST. J Mol Evol. 2002;54:539–547. doi: 10.1007/s00239-001-0054-5. [DOI] [PubMed] [Google Scholar]
- 72.Lienau EK, DeSalle R, Rosenfeld JA, Planet PJ. Syst Biol. 2006;55:441–453. doi: 10.1080/10635150600697416. [DOI] [PubMed] [Google Scholar]
- 73.Gu X, Zhang H. Mol Biol Evol. 2004;21:1401–1408. doi: 10.1093/molbev/msh138. [DOI] [PubMed] [Google Scholar]
- 74.Lin J, Gerstein M. Genome Res. 2000;10:808–818. doi: 10.1101/gr.10.6.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang S, Doolittle RF, Bourne PE. Proc Natl Acad Sci USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 77.O'Hara RJ. Zool Scripta. 1997;26:323–329. [Google Scholar]
- 78.Dagan T, Martin W. Genome Biol. 2006;7:e118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gould SJ. Proc Natl Acad Sci USA. 1994;91:6764–6771. doi: 10.1073/pnas.91.15.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McBreen K, Lockhart PJ. Trends Plants Sci. 2006;11:G398–G404. doi: 10.1016/j.tplants.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Archibald JM, Keeling PJ. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- 82.Martin W, Müller M. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 83.Nakleh L, Warrow T, Linder CR, St John K. J Comput Biol. 2005;12:796–811. doi: 10.1089/cmb.2005.12.796. [DOI] [PubMed] [Google Scholar]
- 84.MacLeod D, Charlebois RL, Doolittle WF, Bapteste E. BMC Evol Biol. 2005;5:27. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Makarenkov V, Legendre P. J Comput Biol. 2004;11:195–212. doi: 10.1089/106652704773416966. [DOI] [PubMed] [Google Scholar]
- 86.Huber KT, Moulton V. J Math Biol. 2006;52:613–632. doi: 10.1007/s00285-005-0365-z. [DOI] [PubMed] [Google Scholar]
- 87.Chan HL, Lam TW, Yiu SM, Jansson J. J Bioinform Comput Biol. 2006;4:807–832. doi: 10.1142/s0219720006002211. [DOI] [PubMed] [Google Scholar]
- 88.Jin G, Nakhleh L, Snir S, Tuller T. Bioinformatics. 2006;22:2604–2611. doi: 10.1093/bioinformatics/btl452. [DOI] [PubMed] [Google Scholar]
- 89.Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beiko RG, Harlow TJ, Ragan MA. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]