Abstract
A factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is a representative of the γ-butyrolactone autoregulators that trigger secondary metabolism and morphogenesis in the Gram-positive, filamentous bacterial genus Streptomyces. Here, we report the A factor biosynthesis pathway in Streptomyces griseus. The monomeric AfsA, containing a tandem repeat domain of ≈80 aa, catalyzed β-ketoacyl transfer from 8-methyl-3-oxononanoyl-acyl carrier protein to the hydroxyl group of dihydroxyacetone phosphate (DHAP), thus producing an 8-methyl-3-oxononanoyl-DHAP ester. The fatty acid ester was nonenzymatically converted to a butenolide phosphate by intramolecular aldol condensation. The butenolide phosphate was then reduced by BprA that was encoded just downstream of afsA. The phosphate group on the resultant butanolide was finally removed by a phosphatase, resulting in formation of A factor. The 8-methyl-3-oxononanoyl-DHAP ester produced by the action of AfsA was also converted to A factor in an alternative way; the phosphate group on the ester was first removed by a phosphatase and the dephosphorylated ester was converted nonenzymatically to a butenolide, which was then reduced by a reductase different from BprA, resulting in A factor. Because introduction of afsA alone into Escherichia coli caused the host to produce a substance having A factor activity, the reductase(s) and phosphatase(s) were not specific to the A factor biosynthesis but commonly present in bacteria. AfsA is thus the key enzyme for the biosynthesis of γ-butyrolactones.
Keywords: A factor, Streptomyces griseus, AfsA, cell-to-cell signaling
The Gram-positive, soil-dwelling bacterial genus Streptomyces is characterized by its complex morphological differentiation resembling that of filamentous fungi and by the ability to produce a wide variety of secondary metabolites, including various biologically active substances. Because of its complex morphogenesis and industrial and medical importance, Streptomyces has been one of the model prokaryotes for the study of multicellular differentiation and secondary metabolite formation. In this genus, A factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone; 1, Fig. 1) in Streptomyces griseus is a representative of the γ-butyrolactone autoregulators, which trigger secondary metabolism or aerial mycelium formation, or both (2, 3). A factor produced in a growth-dependent manner reaches a critical concentration at or near the middle of the exponential growth phase and switches on the transcription of adpA, encoding a transcriptional activator, by binding to ArpA, the A factor receptor, which has bound to the adpA promoter, and dissociating the bound ArpA from the DNA. The effective concentration of A factor is 10−9 M. AdpA then activates a number of genes of various functions required for morphological development and secondary metabolism, forming an AdpA regulon (2, 3). A factor, which is presumed to diffuse freely through the membrane and within an individual multinucleoid hypha, synchronizes morphological development and secondary metabolism not only between neighboring hyphae by means of cell-to-cell signaling but also between different points of a hypha by point-to-point signaling. We therefore call γ-butyrolactone signals in Streptomyces“microbial hormones.” In addition to γ-butyrolactones in Streptomyces, quorum-sensing signals, such as acylhomoserine lactones in Gram-negative bacteria and modified oligopeptides in Gram-positive bacteria, serve as chemical messages for communication with one another (4, 5).
Fig. 1.
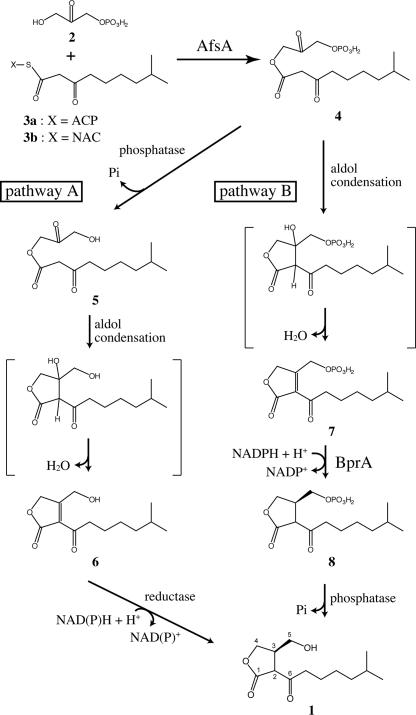
A proposed pathway for A factor biosynthesis. Condensation of DHAP and a β-keto acid derivative is catalyzed by AfsA (this work). After this condensation step, there are two possible biosynthesis routes to A factor. Pathway A was proposed for the biosynthesis of VBs by Sakuda et al. (1) on the basis of feeding experiments. A phosphate group is removed by a phosphatase, followed by nonenzymatic intramolecular aldol condensation. A C–C double bond is reduced by NAD(P)H-dependent reductases, resulting in A factor formation. Pathway B was also predicted by Sakuda et al. (1) and has been elucidated by this study. Nonenzymatic intramolecular condensation are followed by a C–C double-bond reduction step catalyzed by BprA. The last dephosphorylation step yields A factor.
The γ-butyrolactone regulatory system is distributed widely in Streptomyces and its related genera, because A factor and A factor homologues with a slight difference in their chemical structures and ArpA homologues are together present in a given actinomycete. Virginiae butanolides (VBs) and BarA that control virginiamycin production in Streptomyces virginiae (6), IM-2 and FarA that control pigment production in Streptomyces lavendulae (7), and SCBs and ScbR that control actinorhodin and undecylprodigiosin production in Streptomyces coelicolor A3(2) (8) are examples. In all of the cases, the ArpA homologues behave as a transcriptional repressor and the cognate γ-butyrolactones release the repression by binding to the respective receptors, as has been shown for the A factor/ArpA system. Although these studies have established the mechanism by which γ-butyrolactones trigger secondary metabolism and/or morphological differentiation, no definitive route or enzymes for the biosynthesis of γ-butyrolactones have been elucidated.
Concerning the A factor biosynthetic enzymes, we proposed that AfsA was a key enzyme of A factor biosynthesis because (i) afsA mutants lost A factor productivity (9), (ii) introduction of afsA into A factor nonproducing Streptomyces strains caused overproduction of A factor with a gene dosage effect (10), and (iii) introduction of afsA into Escherichia coli caused the host to produce a substance having A factor activity (11). However, the AfsA function has been controversial (12, 13). Concerning the biosynthesis pathway, Sakuda et al. (1, 14, 15) proposed a route on the basis of feeding experiments in VB-producing Streptomyces antibioticus (see pathway A in Fig. 1). According to the proposal, the skeleton is formed by coupling between a carbon-3 (C3) unit derived from glycerol and a β-keto acid derivative. However, the C3 substrate and the catalyzing enzymes were still unknown.
We here describe in vivo and in vitro experiments to show that AfsA catalyzes β-ketoacyl transfer from 8-methyl-3-oxononanoyl-acyl carrier protein (ACP) [3a] to the hydroxyl group of dihydroxyacetone phosphate (DHAP) [2] (see Fig. 1). DHAP is derived from glycerol metabolism, and 8-methyl-3-oxononanoyl-ACP is an intermediate of fatty acid metabolism. This condensation is followed by modifications with phosphatases and reductases, resulting in A factor formation. Because these modifications also occur in E. coli, the phosphatase(s) and reductase(s) are not specific to A factor synthesis. As a reductase responsible for this reduction, BprA that is encoded just downstream of afsA was identified. On the basis of these findings, together with the results of transcriptional analysis of afsA, we will discuss the regulation of A factor biosynthesis.
Results
AfsA as a Possible Enzyme for A Factor Biosynthesis.
We observed that afsA restored A factor production in S. griseus HH1 that contained a large deletion including afsA (10). S. griseus mutant ΔafsA containing a sole mutation in afsA showed the same phenotype as HH1. AfsA shows no homology to known proteins or motifs, although in silico analysis predicted a domain (Pfam03756) of ≈80 aa in AfsA (16) [see supporting information (SI) Fig. 5]. The presence of two Pfam03756 domains in AfsA means that a monomeric AfsA molecule takes an intramolecular dimer-like structure. Almost all proteins having a Pfam03756 domain(s) in the current protein databases are confined to AfsA homologues in Streptomyces. One example from organisms other than Streptomyces is a gene product (Gll2829) from Gloeobacter violaceus, having a Pfam03756 domain at its C-terminal region, annotated as a type I polyketide synthase whose C-terminal region is expected to be a thioesterase domain for polyketide chain release. Structure modeling of AfsA by S. Nakamura (personal communication) showed that, like FabZ (17), AfsA predicts a tunnel that would accept an acyl chain of acyl-ACP. The presence of such a tunnel in AfsA supported the idea that AfsA is involved in A factor biosynthesis at a step of condensation of a C3 compound and a C10 fatty acid derivative containing a β-ketoacyl chain.
Cell-Free Synthesis of A Factor.
The expression of afsA alone in E. coli caused the host to produce a substance(s) having A factor activity but with a retention time slightly different from that of A factor on HPLC (11). In the culture broth of E. coli carrying afsA, two new peaks that were absent in the broth of E. coli without afsA were detected on the basis of the [M–H]− ions on liquid chromatography–electrospray ionization mass spectrometry (LC-ESIMS) (data not shown). One peak contained a compound with its m/z 241 and MS/MS fragmentation pattern with peaks of 197, 167, and 143. The m/z value and the fragmentation into three were the same as those for A factor (see below). Because the culture broth showed A factor activity and because E. coli does not produce branched fatty acids, we assumed that this compound was an A factor analogue having a straight side chain of C10. The other peak contained a compound with its m/z 213 and three fragmented peaks of 169, 139, and 115. These values are all smaller by 28, corresponding to C2H4, than those of the compound in the first peak, which suggested that this compound represented an A factor analogue with a C8 straight side chain. We did not further analyze these compounds produced by E. coli carrying afsA.
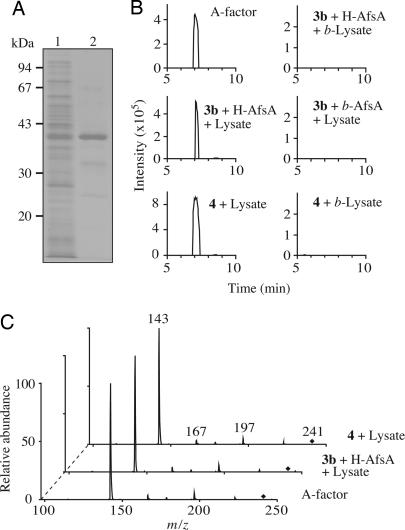
We next tried to produce A factor by using a cell-free extract from S. griseus that should contain branched fatty acids. The fatty acids of Streptomyces consist primarily of branched-chain fatty acids that are synthesized from isobutyryl- and methylbutyryl-CoA (18). For this purpose, we purified histidine-tagged AfsA (H-AfsA) from E. coli harboring pQE80-AfsA (Fig. 2A). This plasmid was constructed so that the revised afsA-coding sequence (SI Fig. 6A) was placed under the control of the T5 promoter. pQE80-AfsA would direct the synthesis of H-AfsA with a sequence of Met-Arg-Gly-Ser-His6-Gly-Ser-Ala-Cys-Glu-Leu-Gly-Thr-(AfsA: 2Pro to 314Gly)-Cys-Ser-Gln-Ala. Incubation of 25.6 μg of H-AfsA with a crude lysate (387 μg of protein) of mutant ΔafsA did not give any significant signals with m/z values between 240 and 242 on LC-ESIMS analysis (data not shown), probably because the intracellular pool of β-keto acid derivatives, which are intermediates of fatty acid synthesis and the predicted substrates for A factor biosynthesis, is small in cells. We therefore synthesized 8-methyl-3-oxononanoyl-N-acetylcysteamine (NAC) [3b] (see SI Materials and Methods) that is a mimic of the corresponding β-ketoacyl-ACP [3a] and added it to the reaction mixture. Incubation of the reaction mixture containing the crude lysate (387 μg of protein), 25.6 μg of H-AfsA, and 28.7 μg of 3b in a total of 100 μl at 16°C for 120 min yielded a compound that eluted at the same retention time as authentic A factor on LC-ESIMS (Fig. 2B). The compound had the same m/z value 241.1 [M–H]− and the same MS/MS fragmentation pattern with peaks of 197, 167, and 143 as authentic A factor (Fig. 2C). These results further supported the idea that AfsA is a biosynthetic enzyme of A factor that uses 8-methyl-3-oxononanoyl-ACP as the C10 substrate.
Fig. 2.
Cell-free synthesis of A factor in the presence of exogenously added H-AfsA. (A) SDS/PAGE analysis of the H-AfsA protein used. Lane 1, the soluble fraction of E. coli harboring pQE80-AfsA; and lane 2, purified H-AfsA. (B) Selected ion chromatograms by LC-ESIMS of authentic A factor and the compounds produced in the lysate of mutant ΔafsA supplemented with (i) H-AfsA and the β-ketoacyl-NAC [3b] or (ii) the 8-methyl-3-oxononanoyl-DHAP ester [4]. NADH and NADPH were also added to the reaction. b-Lysate and b-AfsA mean the lysate and H-AfsA, respectively, that were boiled for 5 min. Peaks were measured on the basis of m/z 241.1 ± 0.3 [M–H]−. (C) MS/MS fragmentation spectra of the peaks obtained in B. Precursor ions are indicated with diamonds.
AfsA as an Enzyme Catalyzing Acyl Transfer Between DHAP and a Fatty Acid Derivative.
We tested pyruvate, phosphoenolpyruvate, 3-phosphoglyceric acid, glyceraldehyde-3-phosphate, dihydroxyacetone phosphate (DHAP), dihydroxyacetone, glycerol-3-phosphate, and glycerol as candidates of the C3 substrate for AfsA. Each of these C3 compounds was incubated in the mixture containing 8-methyl-3-oxononanoyl-NAC and H-AfsA, and the reaction products were analyzed by LC-ESIMS. Although no newly produced peaks were detected in any of the reactions, a decrease of the mass peak of 8-methyl-3-oxononanoyl-NAC was observed only when DHAP was used (data not shown). As described below, DHAP actually served as the C3 substrate. The presence of a phosphate group in the product hampered the detection of a peak representing the coupling product between DHAP and 8-methyl-3-oxononanoyl-NAC.
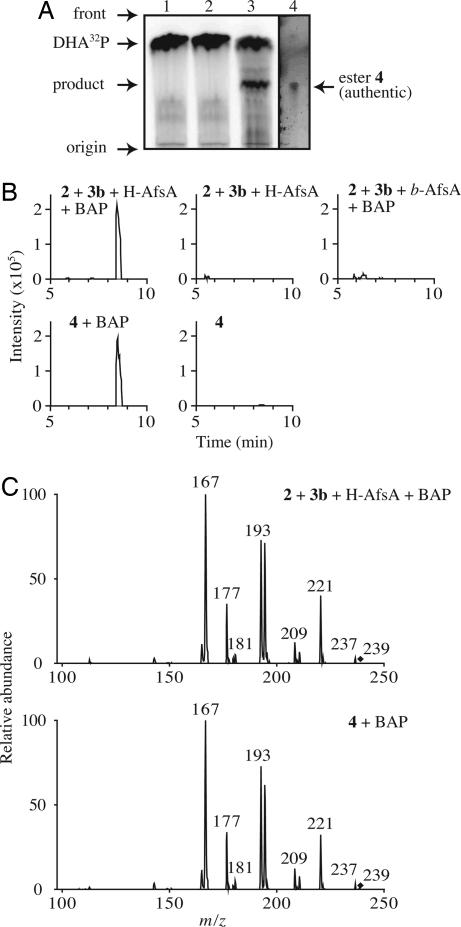
We then prepared 32P-labeled DHAP (DHA32P) and used it in the AfsA reaction. Radio-TLC analysis revealed a product with radioactivity, in addition to DHA32P (Fig. 3A). Assuming that the product was an 8-methyl-3-oxononanoyl-DHAP ester [4], we chemically synthesized this ester to use it as a reference (see SI Materials and Methods). The synthesized ester [4] showed the same Rf value on TLC under the same conditions (Fig. 3A). To identify the structure of the compound by LC-ESIMS, we removed a phosphate group of 4 by phosphatase treatment to facilitate ionization of the compound under the LC-ESIMS conditions. The dephosphorylated ester [5] was expected to result in forming a butenolide [6] nonenzymatically (Fig. 1). The dephosphorylated product from the chemically synthesized ester [4] and from the product of the AfsA reaction between DHAP and 8-methyl-3-oxononanoyl-NAC eluted at the same retention time on LC-ESIMS (Fig. 3B) and had the same m/z value of 239.1 [M–H]−, representing the butenolide [6], and the same MS/MS fragmentation pattern (Fig. 3C). Thus, we concluded that AfsA catalyzed the condensation of DHAP and 8-methyl-3-oxononanoyl-ACP to produce the β-ketoacyl-DHAP ester [4]. In agreement with this conclusion, A factor was produced when the ester [4] was added to the cell lysate of mutant ΔafsA, which showed that the function of AfsA was to produce this ester (Fig. 2 B and C).
Fig. 3.
In vitro synthesis of the 8-methyl-3-oxononanoyl-DHAP ester [4] from DHAP and β-ketoacyl-NAC [3b] by the action of AfsA. (A) Radio-TLC analysis of the AfsA reaction. Lane 1, DHA32P + 3b + boiled H-AfsA; lane 2, DHA32P + H-AfsA (without 3b); lane 3, DHA32P + 3b + H-AfsA; and lane 4, the chemically synthesized ester [4]. The synthesized ester 4 in lane 4 was visualized with 2,4-dinitrophenylhydrazine. (B) The in vitro reaction mixtures were treated with BAP and analyzed by LC-ESIMS. Extracted ion chromatography was prepared at m/z 239.1 ± 0.3 [M–H]−. (C) MS/MS fragmentation spectra of the peaks obtained in B. Precursor ions are indicated with diamonds.
Biochemical Properties of AfsA.
By using radio-TLC analysis, we determined enzyme properties of AfsA. The pH optimum for AfsA was determined to be 6.5–7.5 in Tris·HCl buffer and 7.0–8.0 in sodium phosphate–citrate buffer. Outside this pH range, the activity greatly decreased. The temperature optimum was 10–20°C, with an optimum at 15°C. At 30°C, the activity decreased to 32% of that measured at 15°C, and almost no activity was detected at 35°C. No metals were required for the activity, and EDTA (5 mM) had no effect on the reaction. The Km value for DHAP was 204.6 ± 23.4 μM, and the kcat value was 2.0 ± 0.4 min–1 (SI Fig. 7).
We used gel-filtration chromatography to determine the subunit structure of H-AfsA under nondenaturing conditions. A single peak representing H-AfsA eluted at 45.4 kDa. Because H-AfsA had a calculated molecular mass of 36.3 kDa and migrated at a position of 37.7 kDa on SDS/PAGE, we conclude that this protein is monomeric.
The Downstream Steps in A Factor Biosynthesis After the AfsA Reaction.
The conversion of the ester [4] to A factor [1] requires two further enzymatic steps after the condensation between DHAP and 8-methyl-3-oxononanoyl-ACP. According to pathway A in Fig. 1, predicted by Sakuda et al. (1), the first step is the removal of the phosphate group derived from DHAP by a phosphatase, and the second is the reduction of the C2–C3 double bond of the butenolide [6] formed from 5 by intramolecular aldol condensation. In fact, A factor formation in the crude lysate of mutant ΔafsA after the AfsA reaction was inhibited by addition of sodium orthovanadate, a phosphatase inhibitor (data not shown). Furthermore, no A factor formation occurred unless NADPH was added, suggesting that the reductase required NADPH (data not shown). Because introduction of afsA in E. coli caused the host to produce an A factor analogue, perhaps having a straight side chain, these phosphatases and reductases may not be specific to the biosynthesis of γ-butyrolactones but are present commonly in bacteria. Pathway A appeared to work for the conversion of the ester [4] to A factor [1] via the butenolide [6], because (i) incubation of 4 in the lysate of mutant ΔafsA without NADPH yielded the butenolide [6], and (ii) 6 was reduced to A factor by the lysate of ΔafsA in the presence of NADPH.
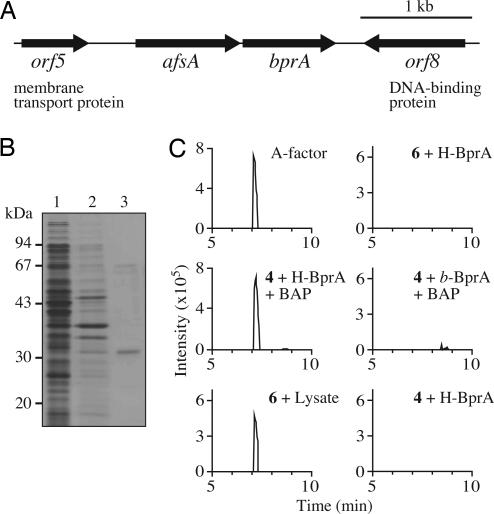
In bacteria, functionally related genes are usually encoded as neighbors. The nucleotide sequence in the neighborhood of afsA revealed the presence of an ORF encoding an oxidoreductase-like protein (Fig. 4A). We named it bprA (b utenolide p hosphate reductase) because of its function described below. A similar oxidoreductase gene (SCO6267, 76% identity in amino acid sequence to BprA) is also located downstream of scbA, an afsA orthologue in S. coelicolor A3(2). We expected that BprA would be responsible for the reduction of the butenolide [6]. We prepared histidine-tagged BprA (H-BprA) (Fig. 4B), with a sequence of Met-Gly-Thr-(BprA)-Arg-Ser-His6. The butenolide [6] was prepared by a phosphatase treatment of the ester [4]. Incubation of 6 with H-BprA in the presence of NADH and NADPH did not reduce 6, however. Therefore we hypothesized pathway B in which a reductase first reduces a butenolide phosphate [7] to produce 8 and subsequently a phosphatase dephosphorylates 8 to produce A factor [1]. Because the butenolide [7] was formed nonenzymatically from the ester [4] in the reaction buffer, H-BprA was incubated with 4 in the presence of NADPH at 25°C for 60 min, followed by the addition of bacterial alkaline phosphatase (BAP) or the lysate of ΔafsA. This reaction yielded a distinct amount of A factor (Fig. 4C) that confirmed the A factor activity in mutant ΔafsA (data not shown). The BprA reaction required NADPH but not NADH. Although we did not determine the stereochemistry at position 3 of the A factor produced in this way, its biological activity suggested that almost the entire population of the molecules are the R-form; the 3R-form is essential for A factor activity (19). Thus, A factor is synthesized through not only pathway A but also pathway B. Because of the operon structure of afsA and bprA, we speculate that pathway B is the major route for A factor biosynthesis in S. griseus.
Fig. 4.
BprA as a reductase involved in the reduction step for A factor biosynthesis. (A) Gene organization in the neighborhood of afsA. afsA and bprA appear to form an operon. (B) SDS/PAGE analysis of the H-BprA protein used. Lane 1, the soluble fraction of E. coli harboring pQE60-BprA; lane 2, insoluble fraction; and lane 3, purified H-BprA. (C) Selected ion chromatograms by LC-ESIMS of authentic A factor and the compounds produced from precursors, 4 or 6, by H-BprA or the lysate of mutant ΔafsA. b-BprA means H-BprA boiled for 5 min. Peaks were measured on the basis of m/z 241.1 ± 0.3 [M–H]−.
Discussion
AfsA as the Key Enzyme for A Factor Biosynthesis.
AfsA catalyzes a crucial step for A factor biosynthesis whose reaction is a new type of acyl transfer. DHAP acyltransferase (DHAPAT) (20) that is involved in lipid formation in mammalian and yeast cells also catalyzes acyl transfer between DHAP and long-chain fatty acyl-CoAs but does not use β-ketoacyl-ACP or -CoA. Because the acyl-DHAP formed by DHAPAT contains no β-keto groups that cause the α-carbon to have much reactivity to facilitate intramolecular cyclization, no γ-butyrolactone ring is formed by the DHAPAT reaction. Although DHAPAT has not been found in bacteria, sn-glycerol-3-phosphate acyltransferase (G3PAT) that is also involved in membrane lipid formation in bacteria is similar to DHAPAT in catalysis (21). G3PAT catalyzes acyl transfer between G3P and acyl-ACP or -CoA without formation of a γ-butyrolactone ring. AfsA probably has no evolutional relation with these enzymes, because DHAPATs and G3PATs show no homology in amino acid sequence with AfsA.
As mentioned above, an AfsA monomer appears to form an intramolecular dimer-like structure involving the repeated Pfam03756 domains. Because no conserved serine or cysteine residues to which the β-ketoacyl moiety attaches are present in Pfam03756 domains, we speculate that AfsA catalyzes the direct β-ketoacyl transfer, like the acyl transfer by G3PAT, without forming a β-ketoacyl-AfsA complex (22). Serine proteases, thioesterases, and lipases employ an Asp(Glu)-His-Ser catalytic triad for their catalysis; the Asp-His charge relay system increases the nucleophilicity of the Ser, which attacks the substrate to form an acyl-Ser intermediate that is subsequently hydrolyzed. In G3PAT, the Asp and His in the HX4D motif may play a similar role in catalysis, with the hydroxyl group of the sn-G3PAT acyl acceptor being analogous to the hydroxyl group of the active site Ser in the catalytic triad and the acyl-G3PAT product being analogous to the acyl-Ser intermediate. An Asp (Glu)-His charge relay in AfsA might increase the nucleophilicity of the hydroxyl group of DHAP as the β-ketoacyl acceptor. His-222, Asp-231, His-232, and Glu-240 are conserved in the C-terminal Pfam03756 domains of AfsA homologues (Fig. 5). Alternatively, the conserved glutamate residues (Glu-78 and Glu-240; Fig. 5) in both of the C- and N-terminal Pfam03756 domains may serve as bases for deprotonation from the hydroxyl group of DHAP, which causes a nucleophilic attack on the β-ketoacyl chain. Determination of the structure of AfsA by x-ray crystallography will clarify the mechanism of the AfsA reaction.
The Variety of γ-Butyrolactone Signal Molecules in Streptomyces.
There are two structural differences in the γ-butyrolactone signal molecules in Streptomyces; one is the length and branching of the fatty acid side chain, and the other is the reduction state of the 6-oxo group (2, 13). The difference of the side chain can be ascribed to the variety of the β-ketoacyl-ACPs used by AfsA and its homologues. In Streptomyces, branched-chain fatty acids, which constitute the majority of the fatty acid pool, were biosynthesized by using amino acid degradation products methylbutyryl- and isobutyryl-CoA as starter units by type II fatty acid synthase (18, 23, 24). SCB1 in S. coelicolor A3(2), having the same C10 branched side chain as A factor, should be formed by ScbA from the same β-ketoacyl-ACP as for the A factor biosynthesis. The difference at position 6, either a keto or a hydroxyl group, can be ascribed to the existence and stereoselectivity of the reductase that reduces this position. Because of the absence of such reductases in S. griseus, position 6 of A factor remains as a keto group. On the other hand, VBs in S. virginiae and SCB1 in S. coelicolor A3(2) have a hydroxyl group at position 6. BarS1 in S. virginiae was isolated as an NADPH-dependent reductase that reduces the 6-oxo group of the penultimate intermediate in the VB biosynthesis (25).
All of the γ-butyrolactone regulators so far isolated have the 3R-form. The stereochemistry at this position depends on the reductases that reduce the C2–C3 double bond of the butenolide intermediate. We assume that the reductases, including BprA, responsible for the reduction of the butenolide have the proper stereospecificity to generate the 3R configuration.
Regulation of A Factor Biosynthesis.
A factor is accumulated in a growth-dependent manner and reaches a maximum, 25 to 30 ng/ml (≈100 nM), at or near the middle of the exponential growth (Fig. 6C). The extremely low concentration of A factor is still enough for triggering secondary metabolism and morphological development, because exogenous supplementation of A factor at 1 to a few nM is enough to restore streptomycin production of A factor-deficient mutants (19). How is the A factor biosynthesis in such small quantities controlled during the growth? The transcription of afsA, starting at two points, was constant during early growth and appeared to increase during the stationary phase (Fig. 6D). Therefore, it is most likely that the time course of A factor production at the extremely low concentration is due to the availability of the substrates for AfsA. The concentration of DHAP [2], derived from glycolysis, depends on the physiological state. The 8-methyl-3-oxononanoyl-ACP [3a], which is synthesized by condensation of three acetate units with the starter substrate isobutyryl-CoA, should be an intermediate in primary fatty acid biosynthesis and is leaked from the pathway. Therefore, the intracellular pool of the β-keto acid derivatives including 3a must be extremely small. Consistent with this, supplementation of 3b, a mimic of 3a, was required for the synthesis of A factor in detectable amounts by using the cell lysate of mutant ΔafsA. Both glycolysis and fatty acid synthesis are active during exponential growth, which is reflected in the time course of A factor production. Therefore, the amount of A factor accumulated, which is synthesized from DHAP and the acyl-ACP by the action of AfsA, reflects the growth period and thus determines the timing of secondary metabolite formation and morphological differentiation. The biosynthesis of A factor reflective of the amount of the intermediates of the primary metabolism or the growth of Streptomyces is analogous to that of N-acylhomoserine lactones in Gram-negative bacteria; N-acylhomoserine lactones are biosynthesized from S-adenosylmethionine derived from the amino acid biosynthesis pathway and the diverse intermediates in fatty acid biosynthesis (26).
Materials and Methods
Bacterial Strains, Plasmids, and Media.
S. griseus IFO13350, media, and pAFB1 were described in refs. 10, 11, 27, and 28. Mutant ΔafsA was generated as described in SI Materials and Methods. pDAF2 containing the 4.8-kb EcoRI fragment downstream from afsA on pUC19 were constructed and used for the gene manipulation described below. E. coli strains and plasmid pUC19 were purchased from TaKaRa Biomedicals, Shiga, Japan. A factor was chemically synthesized essentially according to Mori (29).
Production and Purification of H-AfsA.
The afsA sequence was amplified by PCR with pAFB1 as a template and the following primers: afsA-F, 5′-GGAATTCGGTACCCCCGAAGCAGCAGTCTTGATC-3′ (with an EcoRI site shown by underlining and a KpnI site shown by italic letters) and afsA-R, 5′-AAAACTGCAGCCGTGGGCCCGGGGGCCGGAC-3′ (with a PstI site shown by italic letters). The EcoRI-PstI fragment excised from the amplified afsA sequence was cloned between the EcoRI and PstI sites of pUC19, resulting in pUC19-afsA. The absence of PCR errors was checked by nucleotide sequencing. The KpnI-PstI fragment excised from pUC19-afsA was cloned between the KpnI and PstI sites of pQE80-L (Qiagen, Valencia, CA), resulting in pQE80-AfsA.
For production of H-AfsA, E. coli BL21(DE3) cells harboring pQE80-AfsA were grown, collected after induction of the T5 promoter with isopropyl-β-d-thiogalactopyranoside, and sonicated. The sonicate was centrifuged to remove cell debris. H-AfsA was purified by using Ni-nitrilotriacetic acid (NTA) agarose (Qiagen), essentially according to the manual from the manufacturer. The H-AfsA sample was dialyzed against 10 mM Tris·HCl (pH 7.4) containing 330 mM NaCl, 10% glycerol, and 1 mM DTT. H-AfsA was further purified by chromatography on a HiTrap Q column (Amersham Biosciences, Piscataway, NJ) equilibrated with the dialysis buffer. H-AfsA recovered in the flow-through fraction was collected. Proteins were quantified by using a protein assay kit (Bio-Rad, Hercules, CA) with BSA as the standard. Gel filtration analysis was performed by using a Superdex 200 10/30 column (Amersham Biosciences) by fast protein liquid chromatography with isocratic elution with the dialysis buffer containing no DTT.
Production and Purification of H-BprA.
The bprA sequence downstream of afsA was amplified with PCR. The primers were bprA-F, 5′-CGGAATTCCCATGGGTACCATTCTCGTGACCGGTGCGACCGGAGCG-3′ (with an EcoRI site shown by bold letters, an NcoI site shown by italic letters, and a KpnI site shown by underlining) and bprA-R, 5′-CCCAAGCTTAGATCTTTCCGGCGCGAACGCCTCCGCGTG-3′ (with a HindIII site shown by italic letters and a BglII site shown by underlining). The EcoRI-HindIII fragment excised from the amplified bprA sequence was cloned in pUC19. After confirmation of no PCR errors, the NcoI-BglII fragment was excised and cloned between the NcoI and BglII sites of pQE-60 (Qiagen), resulting in pQE60-BprA. H-BprA was purified from E. coli BL21(DE3) harboring pQE60-BprA by using Ni-NTA agarose.
Enzyme Assays.
The dialysis buffer used for preparation of H-AfsA was used for the AfsA reactions and cell lysate preparation. A cell lysate (4.3 mg of protein per ml) was obtained by sonication of mycelium of mutant ΔafsA grown at 30°C for 36 h in yeast–malt–peptone–dextrose (YMPD) liquid medium. To 90 μl of the cell lysate, H-AfsA (25.6 μg) was added and incubated at 16°C for 120 min in the buffer containing 1 mM each of NADH, NADPH, and the thioester-NAC [3b] in a total volume of 100 μl. In another assay, 11.8 μg of the ester [4] was added as a substitute for AfsA and 3b. For in vitro assay of H-AfsA, the reaction mixture containing 32 μg of H-AfsA, 100 μM of DHAP [2], and 100 μM of 3b in a total volume of 100 μl was incubated at 16°C for 90 min. After the incubation, 5 units of bacterial alkaline phosphatase (BAP) (TaKaRa Biomedicals) was added, and the reaction was further continued at 25°C for 30 min. For the assay of BprA, the reaction mixture containing 0.4 μg of H-BprA, 100 μM of NADPH, 1 mM of MgCl2, and substrates in a total volume of 100 μl was incubated at 25°C for 60 min. The substrates were 5.9 μg of the ester [4] or the butenolide [6] obtained by BAP treatment of the ester [4]. When 4 was used as the substrate, the reaction was followed by the BAP treatment described above.
All reactions were extracted with 300 μl of ethylacetate, and the organic layer was evaporated. The residual material was dissolved in 20 μl of methanol for LC-ESIMS analysis. LC-ESIMS analysis was carried out by using the esquire high-capacity trap plus system (Bruker Daltonics, Billerica, MA) equipped with a Pegasil-B ODS reversed-phase HPLC column (2 × 200 mm; Senshu Scientific, Tokyo, Japan) and acetonitrile/water/acetic acid (500:500:1, vol/vol/vol) as an eluant at a flow rate of 0.3 ml/min.
Radio-TLC Analysis of the AfsA Reaction.
DHA32P was prepared by enzymatic phosphorylation of dihydroxyacetone according to Jones and Hajra (30), except for using a glycerokinase from a Cellulomonas sp. (Sigma, St. Louis, MO). For the AfsA reactions, the reaction mixture containing 50 mM Tris·HCl or 50 mM sodium phosphate-citrate, 300 mM NaCl, 10% glycerol, 3.2 μg of AfsA, 50 μM DHA32P, and 100 μM 3b was incubated at various temperatures for 60 min. Portions (10 μl) of the reaction mixtures were directly spotted on a silica gel RP-18 F254S TLC plate (Merck, Darmstadt, Germany), and the plate was developed in water/methanol/acetic acid (30:70:1, vol/vol/vol). The 32P-labeled compounds were detected by using a BAS-MS imaging plate (Fuji, Tokyo, Japan).
Determination of Kinetic Parameters of AfsA.
The reactions, containing 50 mM sodium phosphate-citrate (pH 7.5), 300 mM NaCl, 10% glycerol, 1 mM 3b, and 3.2 μg of H-AfsA, were performed in a total volume of 100 μl. The concentrations of DHAP containing DHA32P varied between 50 and 400 μM. After the reaction mixture had been preincubated at 15°C for 5 min, the reactions were initiated by adding H-AfsA and continued for 10 min. The reactions were stopped by adding 100 μl of ethylacetate. Portions (10 μl) of aqueous phases were analyzed by TLC, and the radioactivities of substrates and products were measured by using the Bio-Rad Quantity One software. The concentrations of products were calculated based on the substrate–product conversion ratios and the concentrations of the substrates used. Steady-state parameters were determined by Hanes–Woolf plot.
Supplementary Material
Acknowledgments
We thank Shugo Nakamura, University of Tokyo, for molecular modeling studies of AfsA. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from Monkasho.
Abbreviations
- ACP
acyl carrier protein
- BAP
bacterial alkaline phosphatase
- CoA
coenzyme A
- DHAP
dihydroxyacetone phosphate
- DHAPAT
DHAP acyltransferase
- DHA32P
32P-labeled DHAP
- G3PAT
glycerol-3-phosphate acyltransferase
- H-AfsA
histidine-tagged AfsA
- LC-ESIMS
liquid chromatography-electrospray ionization mass spectrometry
- NAC
N-acetylcysteamine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607472104/DC1.
References
- 1.Sakuda S, Tanaka S, Mizuno K, Sukcharoen O, Nihira T, Yamada S. J Chem Soc Perkin Trans 1. 1993:2309–2315. [Google Scholar]
- 2.Horinouchi S. Front Biosci. 2002;7:d2045–d2057. doi: 10.2741/A897. [DOI] [PubMed] [Google Scholar]
- 3.Ohnishi Y, Yamazaki H, Kato J, Tomono A, Horinouchi S. Biosci Biotechnol Biochem. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 4.Bassler BL, Losick R. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Camilli A, Bassler BL. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano H, Takehara E, Nihira T, Yamada Y. J Bacteriol. 1998;180:3317–3322. doi: 10.1128/jb.180.13.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitani S, Yamada Y, Nihira T. J Bacteriol. 2001;183:4357–4363. doi: 10.1128/JB.183.14.4357-4363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. Mol Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 9.Hara O, Horinouchi S, Uozumi T, Beppu T. J Gen Microbiol. 1983;129:2939–2944. doi: 10.1099/00221287-129-9-2939. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi S, Kumada Y, Beppu T. J Bacteriol. 1984;158:481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ando N, Matsumori N, Sakuda S, Beppu T, Horinouchi S. J Antibiot. 1997;50:847–852. doi: 10.7164/antibiotics.50.847. [DOI] [PubMed] [Google Scholar]
- 12.Kawachi R, Akashi T, Kamitani Y, Sy A, Wangchaisoonthorn U, Nihira T, Yamada Y. Mol Microbiol. 2000;36:302–313. doi: 10.1046/j.1365-2958.2000.01819.x. [DOI] [PubMed] [Google Scholar]
- 13.Takano E. Curr Opin Microbiol. 2006;9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Sakuda S, Higashi A, Nihira T, Yamada Y. J Am Chem Soc. 1990;112:898–899. [Google Scholar]
- 15.Sakuda S, Higashi A, Tanaka S, Nihira T, Yamada Y. J Am Chem Soc. 1992;114:663–668. [Google Scholar]
- 16.Yeats C, Bentley S, Bateman A. BMC Microbiol. 2003;3:3. doi: 10.1186/1471-2180-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimber MS, Martin F, Lu F, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 18.Wallace KK, Zhao B, McArthur HAI, Reynolds KA. FEMS Microbiol Lett. 1995;131:227–234. doi: 10.1111/j.1574-6968.1995.tb07781.x. [DOI] [PubMed] [Google Scholar]
- 19.Hara O, Beppu T. J Antibiot. 1982;35:349–358. doi: 10.7164/antibiotics.35.349. [DOI] [PubMed] [Google Scholar]
- 20.Hajra AK. Biochim Biophys Acta. 1997;1348:27–34. doi: 10.1016/s0005-2760(97)00120-3. [DOI] [PubMed] [Google Scholar]
- 21.Green PR, Merrill AH, Jr, Bell RM. J Biol Chem. 1981;256:11151–11159. [PubMed] [Google Scholar]
- 22.Turnbull AP, Rafferty JB, Sedelnikova SE, Slabas AR, Schierer TP, Kroon JTM, Simon JW, Fawcett T, Nishida I, Murata N, et al. Structure (London) 2001;9:347–353. doi: 10.1016/s0969-2126(01)00595-0. [DOI] [PubMed] [Google Scholar]
- 23.Rock CO, Cronan JE. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 24.Han L, Lobo S, Reynolds KA. J Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikura N, Yamamura J, Nihira T. J Bacteriol. 2002;184:5151–5157. doi: 10.1128/JB.184.18.5151-5157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 27.Kieser H, Bibb MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. In: Molecular Cloning: A Laboratory Manual. Nolan C, editor. Woodbury, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 29.Mori K. Tetrahedron. 1983;39:3107–3109. [Google Scholar]
- 30.Jones KM, Hajra AK. Clin Chem. 1994;40:946–947. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.