Abstract
The universally conserved ribonucleoprotein RNase P is involved in the processing of tRNA precursor transcripts. RNase P consists of one RNA and, depending on its origin, a variable number of protein subunits. Catalytic activity of the RNA moiety so far has been demonstrated only for bacterial and some archaeal RNase P RNAs but not for their eukaryotic counterparts. Here, we show that RNase P RNAs from humans and the lower eukaryote Giardia lamblia mediate cleavage of four tRNA precursors and a model RNA hairpin loop substrate in the absence of protein. Compared with bacterial RNase P RNA, the rate of cleavage (kobs) was five to six orders of magnitude lower, whereas the affinity for the substrate (appKd) was reduced ≈20- to 50-fold. We conclude that the RNA-based catalytic activity of RNase P has been preserved during evolution. This finding opens previously undescribed ways to study the role of the different proteins subunits of eukaryotic RNase P.
Keywords: catalytic RNA, ribozyme, RNA processing
Ribonucleoprotein complexes play fundamental roles in several cellular processes vital for growth. From an RNA-world perspective, the hypothesis is that during evolution, proteins have taken over the role of RNA (1). However, for the ribosome, the spliceosome, and the endoribonuclease P (RNase P), their RNA moieties have been either suggested or demonstrated to be directly involved in catalysis (2) and, as such, can be considered to be remnants from an RNA world. RNase P, which is responsible for generation of the matured 5′ end of tRNAs in all three kingdoms of life, consists of one RNA and a variable number of protein subunits: 1 in Bacteria, 4 in Archaea, 9 in yeast, and 10 in mammalian (human) nuclear RNase P (3, 4). Bacterial RNase P RNA is catalytically active in the absence of protein, which is also the case for some archaeal RNase P RNA that show weak activity without proteins at high salt concentrations (5). However, no catalytic activity for eukaryotic RNase P RNA has been detected so far. It has been discussed that through evolution, the RNase P proteins in Eukarya and Archaea have evolved to play more fundamental roles and their functions might be to (i) ensure an active RNase P RNA conformation and (ii) assist in substrate binding and/or catalysis (3, 4, 6–8). Here, we demonstrate that eukaryotic RNase P RNA indeed is able to cleave its substrate in the absence of protein(s), suggesting that the catalytic activity resides in the RNA subunit of RNase P, irrespective of the domain of life from which it is derived.
Results and Discussion
Previous trials to demonstrate catalytic activity for eukaryotic RNase P RNA in the absence of protein(s) have been performed mainly at pH >7 (refs. 4, 9, and 10 and references therein). At this and higher pH, spontaneous and Me2+ (e.g., Mg2+)-induced hydrolysis of RNA become prominent, in particular during long incubations (data not shown; see below). Hence, the likelihood of detecting weak cleavage activity is reduced. We therefore decided to investigate whether eukaryotic RNase P RNA-mediated cleavage could be detected at pH 6.0, a condition where spontaneous and Mg2+-induced hydrolysis of the RNA substrate and RNase P RNA is likely to be reduced. The chemistry of cleavage of the scissile bond also is suggested to be rate-limiting at this pH (ref. 11 and references therein). As substrates, we used different tRNA precursors and pATSerUG, a well characterized RNA hairpin loop model substrate [Fig. 1 and supporting information (SI) Fig. 6; refs. 11–15].
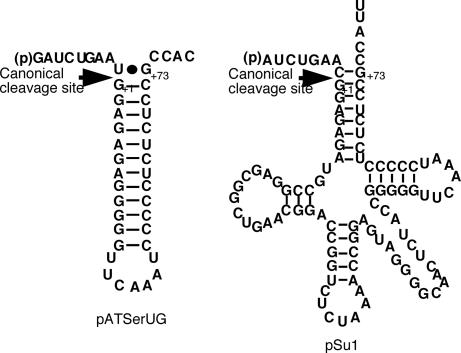
Fig. 1.
Secondary structures of pATSerUG and the precursor to tRNASer, pSu1. The canonical RNase P cleavage sites are indicated with arrows, and numbering is in accordance with standard numbering of tRNA. For the secondary structures of the well characterized tRNA precursors pSu3 and pHis[UAG], see SI Fig. 6 and ref. 14, whereas characterization of pTS-L(-1U) as substrate for E. coli RNase P RNA is unpublished (but see ref. 15, where the secondary structure and construction of the original pTS-L are outlined).
The eukaryotic RNase P RNA variants that we decided to test for catalytic activity were derived from humans (16) and the lower eukaryote Giardia lamblia (9), H1 RNA and G1 RNA, respectively. The P15/P16/P17 region of bacterial RNase P RNA plays an important role in substrate binding and catalysis (11, 17, 18). This region is missing in eukaryotic RNase P RNA (9). We therefore included, together with wild-type Escherichia coli RNase P RNA (M1 RNA), a derivative (M1ΔP15–17 RNA) in which the P15/P16/P17 regions have been deleted. The secondary structures of these RNase P RNAs are depicted in Fig. 2. The experiments were performed as outlined in Materials and Methods.
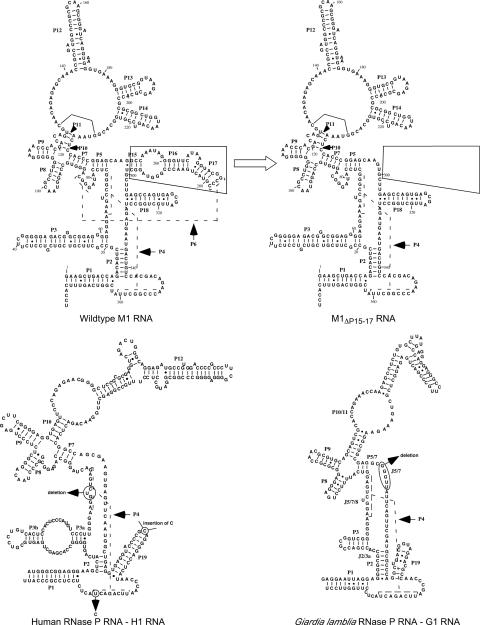
Fig. 2.
Illustration of the predicted secondary structures of the different RNase P RNAs in the present study: E. coli RNase P RNA (M1 RNA) and a variant lacking the P15-17 region (indicated area in M1 RNA; M1ΔP15–17 RNA); human RNase P RNA (H1 RNA) variants with changes at the indicated positions: H1Δ298C325 RNA, H1Δ298U325 RNA, H1C298C325 RNA, H1C298U325 RNA, and H1Δ80–82Δ298 RNA; and the lower eukaryote G. lamblia RNase P RNA, G1 RNA, and G1ΔJ5/6 RNA. For references, see refs. 3, 4, 9, and 16. These RNase P RNAs were generated as described in Materials and Methods.
Eukaryotic RNase P RNA Mediated Cleavage of a Model Hairpin-Loop Substrate.
As shown in Fig. 3A, the eukaryotic H1 RNA and G1 RNA indeed cleaved pATSerUG at pH 6.0. Based on the mobility of the 5′ cleavage products, we inferred that H1 RNA and G1 RNA cleaved the substrate at the same position, the canonical cleavage site, as the wild-type M1 RNA and M1ΔP15–17 RNA. We confirmed that cleavage mediated by H1 RNA, G1 RNA, and M1ΔP15–17 RNA generated the expected pGp, the hallmark of RNase P-mediated cleavage, by thin-layer chromatography with α-32P-GTP internally labeled pATSerUG as a substrate (Fig. 3B; data not shown for G1 RNA).
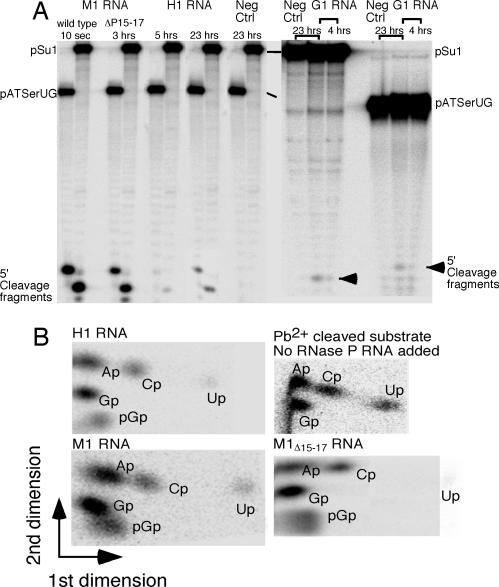
Fig. 3.
Cleavage of pATSerUG and pSu1 with different RNase P RNAs and TLC analysis of cleavage products. (A) Cleavage of 5′ end-labeled pATSerUG and pSu1 with different RNase P RNAs as indicated. Time of incubation is as indicated, and the position of the 5′ cleavage fragments is marked with arrows. Neg ctrl, substrates incubated for 23 h in the reaction buffer without RNase P RNA. The different positions of the 5′ cleavage fragments in G1 RNA vs. M1 RNA- and H1 RNA-mediated cleavage were due to the fact that the G1 RNA and M1 RNA/H1 RNA experiments were run on different polyacrylamide gels. (B) Two-dimensional TLC demonstrating the presence of pGp (the hallmark in RNase P RNA-mediated cleavage) at the 5′ end of the 5′ matured cleavage product after cleavage of [α-32P]GTP internally labeled pATSerUG (specific activity ≥5 Ci/mmol) with wild-type M1 RNA, M1ΔP15–17 RNA, H1 RNA, and no RNase P RNA added as indicated.
The rate constants (Table 1) of cleavage (kobs) under saturating single-turnover conditions were several orders of magnitude lower compared with the kobs value for wild-type M1 RNA. But, the kobs values were only 100-fold lower (for H1 RNA) compared with that determined by using M1ΔP15–17 RNA that lacked the P15–17 region. We could not estimate the rate of background cleavage at the RNase P cleavage site. However, based on the data in SI Fig. 7, the estimated rate of degradation at −1 in pATSerUG was 8 × 10−13 pmol/min (see below; SI Fig. 7). Moreover, the rate constant for spontaneous hydrolysis of an unrelated RNA at pH 6.0 without Mg2+ has been estimated to be ≈10−9 to 10−10 min−1 (19). Hence, using this number, the rate constant for H1 RNA corresponded to at least an increase of four to five (for G1 RNA, three to four) orders of magnitude. However, we do not know the fraction of active RNA, and, thus, the given rates (Table 1) could be higher. We also determined the rate of cleavage of pATSerUG as a function of [Mg2+]. These data revealed that the two M1 RNA variants and G1 RNA have similar Mg2+ requirements (plateau at 160 mM), whereas optimal cleavage by H1 RNA required ≈2-fold higher Mg2+ concentration (plateau at 400 mM; Fig. 4). Our data further indicated that both H1 RNA and G1 RNA interact with pATSerUG with reasonable affinities (Table 1). This should be compared with the estimated KD value (2.2 μM) for binding of arylazido-matured tRNA by using a cross-linking assay and RNase P RNA derived from Schizosaccharomyces pombe (20).
Table 1.
The kinetic constants kobs and kobs/Ksto for the different RNase P RNAs
| RNase P RNA variant | kobs, min−1 | kobs/Ksto, min−1·μM | appKd,μM |
|---|---|---|---|
| M1 RNA* | 8.4 ± 0.33 | 13 ± 1.6 | 0.014 ± 0.0014 |
| M1ΔP15–17 RNA | 2.5 × 10−3 ± 6 × 10−4 | 2.7 × 10−4 ± 7.5 × 10−5 | 1.8 ± 0.69 |
| H1 RNA | 2.6 × 10−5 ± 5.2 × 10−6 | 4.9 × 10−6 ± 4.5 × 10−7 | 0.79 ± 0.3 |
| G1 RNA | 3.5 × 10−6 ± 4.8 × 10−7 | 5.2 × 10−6 ± 1.1 × 10−6 | 0.31 ± 0.17 |
kobs was determined under saturating single turnover conditions and is referred to as the rate constant of cleavage at the canonical RNase P cleavage site by using pATSerUG (0.02 μM final concentration) as substrate, whereas kobs/Ksto determined under saturating single-turnover conditions is equal to kcat/Km. Each value is an average of at least three independent experiments and is given as a mean value ± the deviation of this value. ∗, data taken from ref. 13.
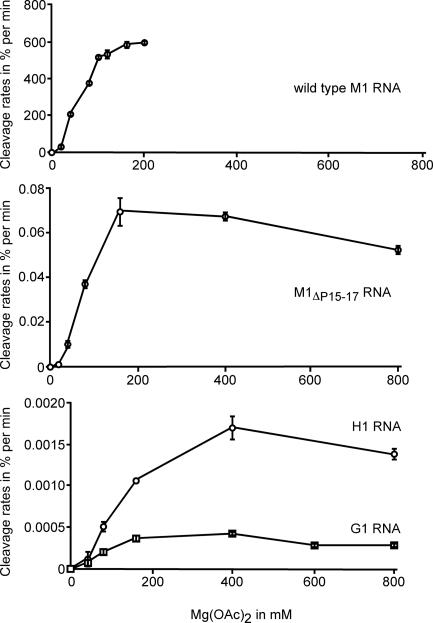
Fig. 4.
RNase P RNA-mediated cleavage of pATSerUG expressed as a percentage of cleavage per minute as a function of [Mg2+]. The different RNase P RNAs used are indicated, and the concentrations were 3.2 μM for M1 RNA, M1ΔP15–17 RNA, and H1 RNA; 6.4 μM for G1 RNA; and 0.02 μM for the substrate. The reactions were performed in buffer C at 37°C (see Materials and Methods). Time of incubation was adjusted to ensure that they were in the linear part of the curve of kinetics. For the calculations, we used the 5′ cleavage fragments. The given data are the average of two independent experiments, and error bars indicate the experimental range. The data for wild-type M1 RNA are based on three independent experiments and taken from ref. 13.
Eukaryotic RNase P RNA Mediated Cleavage of tRNA Precursors.
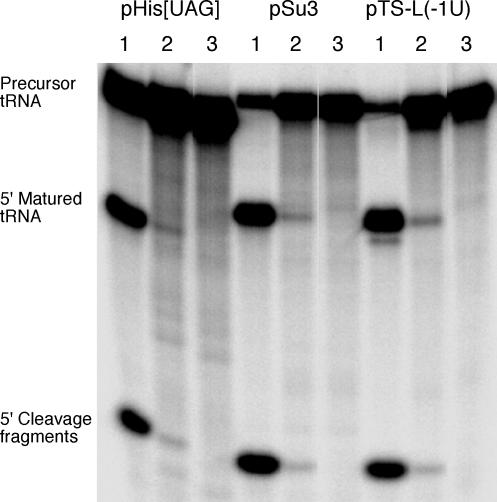
We also tested cleavage of four different tRNA precursors (Figs. 1 and SI Fig. 6). H1 RNA and G1 RNA also cleaved these precursors with low efficiencies (Figs. 3A and 5; cleavage of pSu3, pHis[UAG], and pTS-L(-1U) with G1 RNA not shown). Although we did not observe complete cleavage of any of our substrates (maximum fraction of substrate converted into product was ≤3.2%), the percentage of cleavage of pSu1 with H1 RNA increased over time (SI Fig. 8). Higher frequency of cleavage was also observed when the amount of H1 RNA was increased (data not shown). Based on the mobilities of the 5′ cleavage fragments, we conclude that these precursors were cleaved at the canonical RNase P cleavage site, but at present, we cannot exclude the possibility of cleavage at other positions. However, quantification of the radioactivity in the bands corresponding to the 5′ leaders and matured tRNAs, respectively, resulted in the expected 1:3 ratio (within error; data not shown). We emphasize that to detect cleavage activity, a high specific radioactivity of substrates is required (≥5 Ci/mmol; 1 Ci = 37 GBq; see Materials and Methods).
Fig. 5.
Cleavage of various internally 32P-labeled full-size tRNA precursors [pSu3, pHis[UAG], and pTS-L(-1U)] by wild-type M1 RNA and H1 RNA. For experimental details, see Materials and Methods. Lanes: 1, wild-type M1 RNA (10-sec reaction time); 2, H1 RNA (23-h reaction time); 3, negative control (23-h reaction time). pretRNAHis, precursor to tRNAHis; pSu3, precursor to tRNATyrSu3; pTS-L, chimeric tRNA precursor generated by replacement of the pSu3 acceptor stem with the acceptor stem of tRNASerSu1 (see also ref. 15). Specific activity for all substrates was ≥5 Ci/mmol.
Structural Changes in Conserved Regions of H1 RNA and G1 RNA Resulted in No Detectable Cleavage.
To investigate whether structural changes in conserved regions of H1 RNA and G1 RNA influenced cleavage activity, we deleted three residues in the P4 helix of H1 RNA (H1Δ80–82 RNA, in a Δ298 background; see below) and five in J5/7 in G1 RNA (G1ΔJ5/7 RNA; Fig. 2). These regions were selected based on their importance in cleavage mediated by bacterial RNase P RNA (ref. 4 and references therein). When these variants were incubated with pSu1 or pATSerUG (for G1ΔJ5/7 RNA only pATSerUG), we did not observe any cleavage activity after 24-h incubation (H1Δ80–82 RNA; SI Fig. 8) and 19.5 h (G1ΔJ5/7 RNA; data not shown). These data suggested that these regions in eukaryotic RNase P RNA are important for activity; the observed cleavage is not related to any contaminating activity that copurified with H1 RNA (see also below).
H1 RNA variants have been reported in the literature (16, 21), one with C at position 325 and one with U. When we generated the deletion (Δ80–82) variant of H1 RNA, we noted that the clone encoding H1 RNA used above carried a deletion of C298 (Fig. 2). To study the effect of these variations, we generated the following H1 RNA derivatives: H1Δ298U325 RNA, H1C298U325 RNA, and H1C298C325 RNA (our original variant is referred to as H1Δ298C325 RNA). All three H1 RNA variants were active and cleaved pATSerUG and pSu1 at the same position as the original H1 RNA derivative, H1Δ298C325 RNA (SI Fig. 8). These variants showed lower cleavage activity compared with H1Δ298C325 RNA but in the same range as G1 RNA. Thus, it appears that the residues at these positions influence the activity. When residues 80–82 in P4 were deleted in H1C298U325 RNA, no cleavage activity was detected as expected (data not shown). In conclusion, the original H1 RNA, H1Δ298C325 RNA, is likely to have been instrumental in detecting cleavage mediated by H1 RNA alone. At present we cannot explain this, but it might be related to the folding of H1 RNA.
No Cleavage at the Canonical Cleavage Site Detected in the Presence of Unrelated RNA.
Given the observed low levels of cleavage activities for the eukaryotic RNase P RNAs, it was important to rule out the possibility of contamination of e.g., M1 RNA and to demonstrate that the RNase P RNA variants were not degraded during these long incubations (with respect to pATSerUG, see also above).
From our data it was apparent that H1 RNA (i.e., H1Δ298C325 RNA), G1 RNA, and M1ΔP15–17 RNA were remarkably stable with <1% degradation during the incubation period (data not shown). Moreover, using [32P]pCp or [γ-32P]ATP and standard 3′ or 5′ end-labeling protocols, we could not detect any contamination of M1 RNA or any other RNA in our solutions that could explain our results. Dot blot analysis and RT-PCR did not reveal any traces of M1 RNA in our H1Δ298C325 RNA and G1 RNA (and tisAB mRNA; see below) solutions that could explain our data (SI Fig. 9). We also tested other unrelated RNAs, the 354-nt-long tisAB mRNA (22) and bulk yeast tRNA, but no activity in the presence of any of these was observed under the assay conditions (for yeast tRNA; data not shown). In the case of tisAB mRNA, no activity was detected even after 6 days of incubation in the presence of pATSerUG (SI Fig. 7).
The pATSerUG substrate was remarkably stable (SI Fig. 7) under the reaction conditions with only ≈5% of the substrate degraded after 6 days of incubation at 37°C. Based on these data (SI Fig. 7), we estimate that the limit of detection of cleavage at a single position (position −1; we could not estimate the spontaneous hydrolysis at the RNase P cleavage site) must be <4 × 10−5 % of cleavage per min (≈8 × 10−13 pmol/min). In keeping with the discussion above, our unpublished data have shown that during long incubation times, the substrate pATSerUG is significantly less stable in 50 mM Tris·HCl (pH 7.2) supplemented with 1.25% PEG 6000/100 mM NH4Cl/160 mM MgCl2 compared with the reaction conditions used here. For this reason, we did not test whether H1 RNA and G1 RNA were active under this or any other conditions. Together, this emphasizes the importance of choosing suitable reaction conditions to differentiate between H1 RNA-mediated cleavage and nonspecific hydrolysis.
Conclusion and Concluding Remarks
In summary, we conclude that the eukaryotic RNase P RNA from humans (H1 RNA) and the lower eukaryote G. lamblia (G1 RNA) can mediate cleavage of a model RNA hairpin substrate and several tRNA precursors at the correct position in the absence of protein. Thus, eukaryotic RNase P RNA is an RNA enzyme, as previously demonstrated for bacterial and some archaeal RNase P RNAs (see above). Consequently, the catalytic activity of RNase P has been preserved in its RNA subunit through evolution.
As reported here, the cleavage activity for some RNase P RNA derived from Archaea is low and requires high ionic conditions (5). Likewise, RNase RNA from Chlamydia trachomatis shows low cleavage activity (23, 24). The secondary structures of some of these RNase P RNAs reveal differences in functionally important regions (25), e.g., P15/P17 and P7-P11, which could at least partly explain the observed low cleavage rates. This explanation is also valid for eukaryotic RNase P RNAs that lack the P15/P17 domain and the internal bulge in P10 (Fig. 2). Moreover, the compositions of human and archaeal RNase P with 10 and 4 proteins, respectively, are more complex compared with bacterial RNase P that consists of only one protein and one RNA. This, together with the observed low cleavage rates for H1 RNA and G1 RNA compared with cleavage mediated by M1 RNA, supports the idea that the proteins play important roles in cleavage mediated by eukaryotic RNase P. For example, the proteins might assist in folding of the RNA, substrate binding, and/or catalysis (see above). From this perspective, our finding opens previously undescribed possibilities to study the roles of the different protein subunits in eukaryotic RNase P and to identify specific residues/regions in the RNA that are important for function.
Materials and Methods
Preparation of Substrates and RNase P RNA.
pATSerUG was either purchased from Dharmacon USA (Lafayette, CO) or prepared as run-off transcripts by using T7 DNA-dependent RNA polymerase. This model substrate is cleaved as efficiently as precursor tRNAs (12). The different tRNA precursors were generated as run-off transcripts (26–28). Following standard procedures, the different substrates were labeled with 32P either at the 5′ end with γ-32P [100 pmol of pATSerUG or pSu1; 10 ml of [γ-32P]ATP (specific activity 3,000 Ci/mmol) in 30 ml of final reaction volume and no cold ATP] or internally by using [α-32P]GTP (pATSerUG; final specific activity ≥5 Ci/mmol) or [α-32P]UTP [pATSerUG, pSu3, pTS-L(-1U) and pHis[UAG]; final specific activity ≥5 Ci/mmol]. The various RNA substrates were gel-purified according to standard procedures.
The tRNA precursor pTS-L(-1U) was generated by using QuikChange Directed Mutagenesis (Stratagene) with the plasmid harboring the gene-encoding pTS-L(-2A) as template (27).
The RNase P RNA variants (gene encoding H1 RNA kindly provided by the S.A. laboratory) were generated as T7 DNA-dependent RNA polymerase run-off transcripts and gel-purified as described in refs. 26–30. For construction and generation of M1ΔP15–17 RNA and G1 RNA, see below.
The different H1 RNA derivatives were generated by using QuikChange Directed Mutagenesis (Stratagene) with the plasmid harboring the gene-encoding H1 RNA (see above) as template.
Construction of M1ΔP15–17 RNA.
Two constructs were made by PCR with the E. coli rnpB gene behind the T7 promoter as template. To generate construct A, we used the forward T7 primer (5′-GAATTCGAAATTAATACGACTCACTATA) and the reverse M1wt ΔP15–17 249–220 primer (5′-TTTGAGTCTTGGCCTTGCTCCGGGTGGAGTTTACCGTGCCACGG) that generates an Mly1 restriction site. To generate construct B, we used the forward M1wt ΔP15–17 300–329 primer (5′-TTTGAGTCTAGGCTGCTTGAGCCAGTGAGCGATTGCTGGCCTAG) that also generates an Mly1 restriction site and the reverse M1 RNA 3′ end primer (5′-AGGTGAAACTGACCGATAAG; complementary to residues 358–377 in M1 RNA, see Fig. 2). Both PCR constructs A and B were cleaved with Mly1. The resulting fragments were ligated by using Ready to Go Ligase Kit (GE Healthcare Biosciences). The ligated product was PCR-amplified by using the forward T7 primer and the reverse M1 RNA 3′ end primer and cloned into Topo 2.1 Vector (Invitrogen). The gene construct thus obtained was confirmed by DNA sequencing.
M1ΔP15–17 RNA was generated as run-off T7 RNA polymerase transcriptions by using a PCR-generated template that had been produced with the primer pair FP0520 (5′-GATGTGCTGCAAGGCGATTAAG) and M1 RNA 3′ end and Topo 2.1 Vector carrying the M1ΔP15–17 RNA gene as template. The RNA was gel-purified by using 8% denaturing PAGE and extracted as described by Kufel and Kirsebom (28).
Construction of the Gene Encoding G1 RNA Behind the T7 Promoter.
The coding sequence of Giardia RNase P RNA (G1 RNA) was amplified from G. lamblia strain WB, clone 6 genomic DNA by using the primers T7-GiRNasP5 (5′-CCGAATTCGAAATTAATACGACTCACTATAGAGGAATTAGGAGGGGCGCCACCG-3′) and GiRNasP3 (5′-CCCTGCAGAGGAACCAAGGAGTAGTCTGAATCG-3′). The PCR product was ligated into the pCR4-TOPO vector (Invitrogen), and the amplified gene was verified by sequencing the PCR product. T7 RNA polymerase transcription of G1 RNA was performed on purified PCR products generated by using the T7-G1 RNA gene construct in pCR4-TOPO as template and primers T7-GiRNasP5 and GiRNasdP3. The deletion derivative G1ΔJ5/6 RNA appeared as a side product during the cloning of the G1 RNA gene (S.G.S., unpublished data).
Analysis of the 5′ End of the 5′ Matured Cleavage Product.
The cleavage site was inferred by comparing the mobility of the 5′ cleavage fragments generated by using the different RNase P RNA variants. The presence of pGp at the 5′ end of the large cleavage product was verified by two-dimensional thin layer chromatography as described in ref. 31, using [α-32P]GTP internally labeled pATSerUG (specific activity ≥5 Ci/mmol) as substrate.
To confirm that the appearance of pGp depended on the presence of RNase P RNA, we cleaved [α-32P]GTP internally labeled pATSerUG with Pb2+ (no RNase P RNA added), which generates a 5′OH as one of the cleavage products (for further details, see ref. 32).
Assay Conditions and Determination of the Kinetic Constants Under Single-Turnover Conditions and the Apparent Binding (appKd) Constant.
The cleavage reactions were conducted in buffer C (50 mM Mes, pH 6.0 at 37°C/0.8 M NH4Cl) in the presence of different concentrations of Mg(OAc)2 as indicated. For the binding assays, buffer C was supplemented with 0.05% (wt/vol) Nonidet P-40/0.1% (wt/vol) SDS/160 mM CaCl2. In all reactions, before mixing substrate with RNase P RNA, RNase P RNA was preincubated at 37°C in buffer C and 160 mM Mg(OAc)2 (CaCl2 when appKd was determined, see below) to allow proper folding. The high divalent metal ion concentration is rationalized by our previous finding that this is the optimal concentration for binding and cleavage of pATSerUG by using wild-type M1 RNA (for further details, see refs. 12 and 13 and references therein). In Fig. 3, the concentrations of pATSerUG and pSu1 were 0.02 μM; for M1 RNA, M1ΔP15–17 RNA, and H1 RNA, the concentration was 3.2 μM; and for G1 RNA, the concentration was 6.4 μM. In Fig. 5, the concentrations of tRNA precursors and RNase P RNA were 0.02 μM and 6.4 μM, respectively.
The kinetic constants kobs and kobs/Ksto (= kcat/Km) were determined under saturating single-turnover conditions at pH 6.0 and 160 mM Mg(OAc)2 (13). At this pH, the chemistry of cleavage of pATSerUG is suggested to be rate-limiting (ref. 11), and kobs is referred to as the rate constant of cleavage. The final concentration of the substrate was ≤20 nM, and the concentration of RNase P RNA was varied between the 0.040 μM and 53.2 μM range, depending on the RNase P RNA variant used. For the calculations, we used the 5′ cleavage fragment, and the time of cleavage was adjusted to be in the linear part of the curve of kinetics. The kobs and kobs/Ksto values were obtained by linear regression from Eadie–Hofstee plots.
Spin columns were used to determine apparent equilibrium dissociation constants (appKd) in buffer C in the presence of 160 mM CaCl2 (see above and as described in refs. 33 and 34). After the substrate (pATSerUG) and RNase P RNA had been preincubated for 20 min, they were mixed. After an additional 20 min, nonbound substrate was separated from substrate in complex with RNase P RNA on spin columns as described in refs. 33 and 34. The substrate (pATSerUG) concentration was ≤10 nM, and the RNase P RNA concentration (dependent on variant) was varied from 0.01 μM to 30 μM. appKd values were determined by nonlinear regression analysis by using Origin 7.0 software (Originlab) and the equation fc = ft × [RNase P RNA]free/(Kd + [RNase P RNA]free), where fc is the fraction of precursor substrate in complex with RNase P RNA and ft is the maximum fraction of substrate able to bind RNase P RNA, i.e., the reaction endpoint.
Dot Blot Analysis.
Different concentrations of H1 RNA, G1 RNA, tisAB mRNA, and M1 RNA were applied on a nylon membrane filter. The RNA concentrations were as follows: 0.75–10 μg for H1 RNA and G1 RNA, 0.125–2 μg for tisAB mRNA, and 0.1–100 ng for M1 RNA. After application of the RNA, the RNA was UV cross-linked to the filter, and this was followed by hybridization overnight at 42°C by using an oligodeoxynucleotide (5′-AGGTGAAACTGACCGATAAG) complementary to the 3′ end of M1 RNA (residues 358–377; Fig. 2) that had been labeled with γ-32P at the 5′ end (see above). Hybridization (overnight at 42°C) and washing (three 5-min washes at 42°C and one wash for 5 min at room temperature) of the membranes were performed according to standard procedures in ref. 35. The signal was detected on a PhosphorImager (400S; Molecular Dynamics).
Reverse Transcription Followed by PCR: RT-PCR.
One hundred nanograms each of H1 RNA, G1 RNA, tisAB mRNA, and M1 RNA were mixed with 10 mM dNTP mix (a mixture of dATP, dGTP, dTTP, and dCTP) and primer (10 mM final concentration). As primer, we used an oligodeoxynucleotide complementary to the 3′ end of M1 RNA (5′-AGGTGAAACTGACCGATAAG) (see Fig. 2). The mixture was incubated at 65°C for 5 min and then put on ice for 10 min. A mixture of 5× FS buffer (Invitrogen; according to manufacturer's instructions)/0.1 M DTT/RNA guard (GE Healthcare Biosciences) was added according to the protocol (SuperScript II Reverse Transcription protocol given by Invitrogen). After incubation for 2 min at 42°C, 1 μl of SuperScript II Reverse Transcriptase Enzyme (Invitrogen) was added to each reaction and incubated at 42°C for 50 min. The reverse transcriptase was inactivated at 70°C for 15 min, and the samples were put on ice. One microliter of RNase H1 was added, and incubation was prolonged for 20 min at 37°C.
This was followed by standard PCRs where 10% of the reverse transcription reaction was used as a template. As primers, we used 5′-TTCGGGGGAGACGGGCGGA (forward; complementary to residues 39–77 in M1 RNA; Fig. 2) and 5′-AGGTGAAACTGACCGATAAG (reverse; complementary to residues 358–377 in M1 RNA; Fig. 2). The resulting PCR products were separated on a 2% agarose gel in TEB buffer (45 mM Tris·borate, pH 8.3/1.25 mM EDTA).
Supplementary Material
Acknowledgments
We thank our colleagues and, in particular, Dr. S. Altman for discussions and suggestions. Drs. S. Altman, S. Dasgupta, and D. Hughes are acknowledged for critical reading of the manuscript, and Dr. F. Darfeuille is acknowledged for tisAB mRNA and discussion. We also thank Mr. B. M. F. Pettersson and Ms. U. Lustig for assistance with the RNase P RNA constructs. This work was supported by a grant from the Swedish Research Council (to L.A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 2031.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607326104/DC1.
References
- 1.Joyce GF. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Cech TR. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 3.Altman S, Kirsebom LA. In: The RNA World. 2nd Ed. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 351–380. [Google Scholar]
- 4.Walker SC, Engelke DR. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. Proc Natl Acad Sci USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y, Altman S. EMBO J. 1995;14:159–168. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrara G, Calandra P, Fuscoloni P, Tocchini-Valentini GP. Proc Natl Acad Sci USA. 1995;92:2627–2631. doi: 10.1073/pnas.92.7.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank DN, Adamidi C, Ehringer MA, Pitulle C, Pace NR. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Mol Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 11.Brännvall M, Kikovska E, Kirsebom LA. Nucleic Acids Res. 2004;32:5418–5429. doi: 10.1093/nar/gkh883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kikovska E, Brännvall M, Kufel J, Kirsebom LA. Nucleic Acids Res. 2005;33:2012–2021. doi: 10.1093/nar/gki344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikovska E., Brännvall M, Kirsebom LA. J Mol Biol. 2006;359:572–584. doi: 10.1016/j.jmb.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 14.Kirsebom LA, Svärd SG. Nucleic Acids Res. 1992;20:425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirsebom LA, Svärd SG. J Mol Biol. 1993;231:594–604. doi: 10.1006/jmbi.1993.1312. [DOI] [PubMed] [Google Scholar]
- 16.Bartkiewicz M, Gold H, Altman S. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 17.Kirsebom LA, Svärd SG. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busch S, Kirsebom LA, Notbohm H, Hartmann RK. J Mol Biol. 2000;299:941–951. doi: 10.1006/jmbi.2000.3789. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Breaker RR. J Am Chem Soc. 1999;121:5364–5372. [Google Scholar]
- 20.Marquez SM, Chen JL, Evans D, Pace NR. Mol Cell. 2006;24:445–456. doi: 10.1016/j.molcel.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baer M, Nilsen TW, Costigan C, Altman S. Nucleic Acids Res. 1989;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel J, Argaman L, Wagner EGH, Altuvia S. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Haas ES, Brown JW. Nucleic Acids Res. 1998;26:4093–4099. doi: 10.1093/nar/26.18.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann B, Pettersson B, Everett KDE, Mikkelsen N-E, Kirsebom LA. Int J Syst Bacteriol. 2000;50:149–158. doi: 10.1099/00207713-50-1-149. [DOI] [PubMed] [Google Scholar]
- 25.Harris ME, Christian EL. Curr Opin Struct Biol. 2003;13:325–333. doi: 10.1016/s0959-440x(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 26.Milligan JF, Groebe DR, Whiterell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brännvall M, Mattsson JG, Svärd SG, Kirsebom LA. J Mol Biol. 1998;283:771–783. doi: 10.1006/jmbi.1998.2135. [DOI] [PubMed] [Google Scholar]
- 28.Kufel J, Kirsebom LA. RNA. 1998;4:777–788. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brännvall M, Pettersson BMF, Kirsebom LA. J Mol Biol. 2003;325:697–709. doi: 10.1016/s0022-2836(02)01195-6. [DOI] [PubMed] [Google Scholar]
- 30.Kufel J, Kirsebom LA. Proc Natl Acad Sci USA. 1996;93:6085–6090. doi: 10.1073/pnas.93.12.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrier-Takada C, van Belkum A, Pleij CWA, Altman S. Cell. 1988;53:267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- 32.Kikovska E, Mikkelsen N-E, Kirsebom LA. Nucleic Acids Res. 2005;33:6920–6930. doi: 10.1093/nar/gki993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beebe JA, Fierke CA. Biochemistry. 1994;33:10294–10304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- 34.Warnecke JM, Held R, Busch S, Hartmann RK. J Mol Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.