Abstract
C57BL/6 (B6) mice subjected to a high-fat diet develop metabolic syndrome with obesity, hyperglycemia, and insulin resistance, whereas 129S6/SvEvTac (129) mice are relatively protected from this disorder because of differences in higher basal energy expenditure in 129 mice, leading to lower weight gain. At a molecular level, this difference correlates with a marked higher expression of uncoupling protein 1 (UCP1) and a higher degree of uncoupling in vitro in mitochondria isolated from muscle of 129 versus B6 mice. Detailed histological examination, however, reveals that this UCP1 is in mitochondria of brown adipocytes interspersed between muscle bundles. Indeed, the number of UCP1-positive brown fat cells in intermuscular fat in 129 mice is >700-fold higher than in B6 mice. These brown fat cells are subject to further up-regulation of UCP1 after stimulation with a β3-adrenergic receptor agonist. Thus, ectopic deposits of brown adipose tissue in intermuscular depots with regulatable expression of UCP1 provide a genetically based mechanism of protection from weight gain and metabolic syndrome between strains of mice.
Keywords: brown fat, UCP1, diabetes, genetics, obesity
Both rodents and humans exhibit a great variability in predisposition to obesity, insulin resistance, and development of diabetes. In humans, this variability is shown by the increased prevalence of obesity and diabetes in certain populations and the segregation of diabetes in families (1–3). In rodents, we and others have shown that different mouse strains exhibit differential susceptibility to diabetes and diet-induced obesity (4–10). For example, C57BL/6 (B6) mice on a high-fat diet develop severe obesity, hyperglycemia, and insulin resistance (8, 11), whereas 129S6/SvEvTac (129) mice, which are often crossed with the B6 mouse to make genetic knockouts, are considered resistant to dietary-induced obesity and glucose intolerance (6). Using intercross and F2 mice, we have recently shown that this diabesity phenotype of B6 mice is inherited in a dominant fashion, with at least four loci on three different chromosomes contributing to the phenotype (6).
In rodents and human infants, a major defense mechanism against obesity is brown adipose tissue (BAT), which serves to increase energy expenditure through dissipation in the form of heat (thermogenesis) (12). In rodents and humans, brown fat tends to be localized to the intrascapular and paraspinal regions, and the amount of BAT appears to decrease as mammals go from the neonatal to adult stage (13). Increases in thermogenesis in BAT occur in response to the cold or to calorie intake, and ablation of BAT by use of a toxigene results in hyperphagia and obesity (14). The protein responsible for thermogenesis in brown fat is uncoupling protein 1 (UCP1) (15–17). UCP1 is located in the inner mitochondrial membrane and serves to uncouple oxidative phosphorylation by promoting a proton leak across the mitochondrial membrane, thereby generating heat and lowering ATP synthesis (18). The major endogenous stimulator of thermogenesis and UCP1 expression is β3-adrenergic stimulation through the sympathetic nervous system (16). β3-Adrenergic receptors are expressed predominantly in BAT, and stimulation of these receptors in mice increases oxygen consumption and UCP1 mRNA expression (19, 20). Knockout of UCP1 in mice causes a decrease in oxygen consumption after β3-adrenergic receptor agonist treatment and increased cold sensitivity (21). Two other uncoupling proteins, UCP2 and UCP3, are more widely expressed than UCP1, but they appear to play important roles in mitochondrial function in only a few tissues. Although it was initially thought that UCP2 and UCP3 might allow tissues such as skeletal muscle also to contribute to thermogenesis and energy expenditure (22), subsequent studies have found that this theory is not the case (23, 24).
In the present work, we have explored differences in basal energy expenditure and its role in development of obesity and the metabolic syndrome in B6 and 129 mice. We found that 129 mice have higher rates of energy expenditure than B6 mice, which appears to be the result of unexpectedly high levels of UCP1 mRNA and protein and uncoupling in mitochondria isolated from hindlimb skeletal muscle of adult 129 mice. Careful morphometric analysis, however, demonstrates that the UCP1 is actually located in brown adipocytes interspersed in the perimuscular and intermuscular adipose tissue of the leg, that this “ectopic” brown fat is markedly more abundant in mice of the 129 strain, and that it is also subject to regulation by β3-adrenergic stimulation. This previously unrecognized depot of brown fat-expressing regulatable UCP1 is sufficient to contribute to differences in energy homeostasis between strains of mice, and thus it represents a potential mechanism contributing to differences in the propensity to gain weight and development of the metabolic syndrome between animals of different genetic background.
Results
Effect of Diet on Fat Percent and Weight Gain.
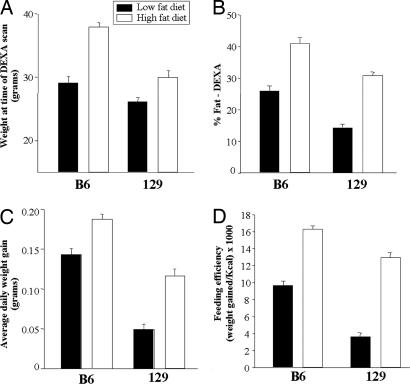
Mice were maintained on either a low-fat (14% of calories) or high-fat diet (55%) from the age of 6 weeks to the age of 24 weeks. As shown in ref. 6, after ≈6 weeks the 129 mice exhibited significantly lower weights on both low-fat (P = 0.043) and high-fat diet (P = 0.021) compared with B6 mice (Fig. 1A), and DEXA scans revealed that the percent body fat was ≈15% lower in 129 mice on both diets (P < 0.03) (Fig. 1B). Thus, the calculated total fat mass of the 129 mice were 3.7 ± 0.3 and 9.3 ± 0.6 g on low- and high-fat diets, respectively, versus 7.5 ± 0.7 and 15.6 ± 0.7 for B6 mice. This pattern continued throughout the 18-week study such that the average weight gain in 129 mice was ≈52% and 45% lower than that of B6 mice on the low-fat (P = 0.006) and high-fat diets (P = 0.001), respectively (Fig. 1C).
Fig. 1.
Weight, fat percentage, daily weight gain, and feeding efficiency of B6 and 129 mice. Mice were maintained on either a low-fat diet (14% of calories from fat) or high-fat diet (55% fat) starting by the end of puberty (around the age of 6 weeks). Dual-energy x-ray absortionmetry (DEXA) scanning was performed after ≈6 weeks on the diets. (A) Body weight at the time of DEXA scanning in B6 and 129 mice on low-fat (open bars) and high-fat diets (filled bars). B6 versus 129 mice on low-fat diet, P = 0.043; on high-fat diet, P = 0.021. (B) Fat percentage determined by DEXA scanning of B6 and 129 mice on low- and high-fat diets. B6 versus 129 on low-fat diet, P = 0.021; on high-fat diet, P = 0.029. (C) Average daily weight gain during the 18 weeks on low- and high-fat diets. B6 versus 129 on low-fat diet, P = 0.006; on high-fat diet, P = 0.001. (D) The mice were placed in individual cages, and food was measured every 3rd day. Feeding efficiency was calculated as weight gain per kcal of food intake in mice on both diets. Feeding efficiency was 2.6-fold lower in 129 versus B6 on the low-fat diet (P = 0.021) and 1.3-fold lower in 129 on the high-fat (P = 0.001). This latter result was previously reported as part of an analysis of the genetic determinants of energy expenditure and insulin resistance (6); together with the new data in A–C in this figure and the data in Fig. 2, it demonstrates the full range of metabolic characteristics of diet-induced obesity in this cohort of these two strains of mice. All results are expressed as mean ± SEM with four mice in each group.
Food Intake, Feeding Efficiency, Activity Level, and Energy Expenditure.
To determine whether the reduced fat percent and weight gain of 129 mice were the results of a lower food intake or higher energy expenditure, we measured the number of calories consumed by each strain for a 9-day period in the middle of the diet study. Surprisingly, despite their lower rate of weight gain, when adjusted for body weight, 129 mice consumed 39.9% more calories than B6 mice (P = 0.021) on the low-fat diet and an equal number of calories on the high-fat diet (data not shown). The average caloric intake per mouse was 18.9 ± 0.4 and 11.2 ± 0.4 for the 129 mice on low- and high-fat diets compared with 16.7 ± 0.2 and 14.1 ± 0.4 for the B6 mice. Thus, feeding efficiency, i.e., weight gain per calorie ingested, was 62% lower in 129 versus B6 mice on the low-fat diet (P = 0.021) and 33% lower in 129 on the high-fat diet (P = 0.001) (Fig. 1D) (6).
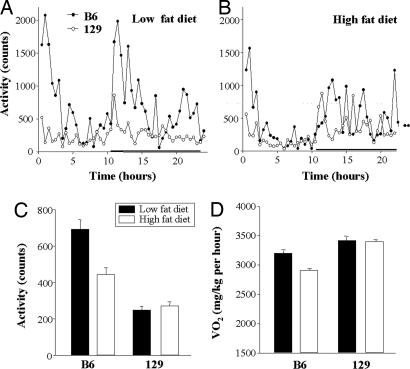
Energy expenditure was assessed by using indirect calorimetry (25), and activity was assessed by housing mice in metabolic cages with movement sensors. As expected, both B6 and 129 mice exhibited their highest level of activity during the dark period (Fig. 2 A and B). When calculated over 24 h, the activity level was ≈50% lower in the 129 than the B6 mice on both low- and high-fat diets (P < 0.001) (Fig. 2C) (6). Metabolic rate, i.e., O2 consumption over 24 h, on the other hand, was 5–6% higher in 129 mice than in B6 mice on both diets during the light period, and it was as much as 14% higher on the high-fat diet during the dark period (6). Furthermore, body temperature, especially on the high-fat diet, was higher in the 129 mice compared with the B6 mice (36.7 ± 0.2°C versus 35.6 ± 0.1°C; P = 0.001).
Fig. 2.
Activity and metabolic rate of B6 and 129 mice on low- and high-fat diets. Mice were placed in indirect calorimetry chambers and allowed to adapt for 48 h. Activity was measured as beam break counts during 24 h on low-fat (A) and high-fat diets (B) in B6 (filled circles) and 129 mice (open circles). The black horizontal bar denotes the dark period. (C) The bars represent the mean activity level over 24 h ± SEM with four mice in each group. Mean activity level of 129 versus B6 mice was significantly lower (P < 0.001) on both diets.
Microarray Analysis of Gene Expression in Skeletal Muscle.
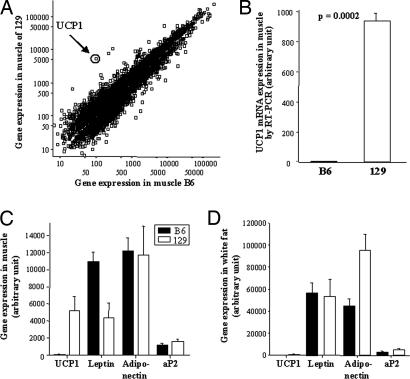
In an attempt to define the mechanism of this difference in basal energy expenditure, gene expression was assessed by using Affymetrix microarrays on RNA from skeletal muscle of 6-month-old B6 and 129 mice that had been maintained on a regular chow (21.6% fat) diet. Interestingly, of the 12,488 genes and ESTs on the chip, the gene with the highest difference in expression between the strains was the gene encoding UCP1, which exhibited 52.4-fold higher expression in muscle of 129 versus B6 mice (P = 0.056) (Fig. 3A). Quantitative RT-PCR demonstrated an even greater difference, with muscle UCP1 gene expression being 110-fold higher in 129 than in B6 (P = 0.0002) (Fig. 3B). By comparison, UCP2 and UCP3 expression in muscle was similar between the strains.
Fig. 3.
Gene expression in skeletal muscle and expression of fat-specific genes in skeletal muscle and epididymal fat. (A) Gene array analysis of skeletal muscle by using Affychip U72Av.2. The scatter plot depicts the genes and ESTs with differential expression between B6 and 129 of >1.15-fold. The UCP1 gene expression (encircled) exhibited the highest fold difference between the two strains (52-fold higher in 129 versus B6). (B) The UCP1 gene expression of skeletal muscle was confirmed by quantitative RT-PCR showing a 110-fold higher expression of UCP1 in 129 than in B6 (P = 0.0002). Gene expression of UCP1 and of white fat-specific genes (leptin, adiponectin, and aP2) in muscle (C) and epididymal white fat (D) of B6 (filled bars) and 129 mice (open bars) is shown. Only leptin in muscle was significantly higher in B6 mice compared with 129 mice (P = 0.023). Data represent the mean of four chips with cRNA from two or three mice on each chip. The mice used for the study had been maintained on a regular chow (21% of calories from fat) for 6 months.
Expression of Fat-Specific Genes.
The finding of UCP1 in muscle of 129 mice was surprising because every effort had been made to remove all adjacent adipose tissue. To determine whether the finding might be caused by fat contamination, we analyzed the expression of three fat-specific genes, leptin, adiponectin, and aP2 (26–28), in the same muscle samples, as well as in epididymal fat (Fig. 3 C and D). This examination revealed similar levels of expression of adiponectin and aP2 in the B6 and 129 muscle samples and a significantly higher level of leptin in B6 compared with 129 (P = 0.023) (Fig. 3C). In white fat from the same animals, although the expression of UCP1 was slightly higher in 129 mice, there were no significant differences in expression of leptin, aP2, or adiponectin (Fig. 3D) (note the difference in scales). Together, these data suggest that to the extent that there may be fat contamination of muscle samples, analysis of adipose marker genes indicated that it was very similar in B6 and 129 mice.
Proton Uncoupling.
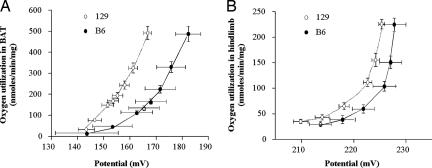
To determine whether increased UCP1 in muscle was functionally active, mitochondria were isolated from hindlimb muscle and BAT of B6 and 129 mice, and proton flux was determined. Interestingly, O2 use and membrane potential in mitochondria from BAT of 129 mice respiring on succinate revealed an upward-left shift, indicating increased H+ flux at any given membrane potential, compared with B6 (Fig. 4A). A similar, but smaller, increase in respiration of 129 versus B6 mice was seen in mitochondria isolated from hindlimb skeletal muscle (Fig. 4B).
Fig. 4.
Kinetics of the proton leak in mitochondria isolated from 129 and B6 strain mice. Respiration and mitochondrial inner membrane potential were determined simultaneously in BAT (n = 10 for each strain) (A) and hindlimb muscle (n = 8 for each strain) (B) of B6 (filled squares) and 129 mice (open circles). Data points and error bars represent mean ± SEM for both oxygen use and potential, and they are thus directed along both the x and y axes. Please note that different scales have been used for A and B. The mice used for the study had been maintained on a regular chow.
This increase correlated with differences in UCP1 expression by quantitative Western blotting, which revealed 1.4 ± 0.2 μg of UCP1 per mg of mitochondrial protein in hindlimb of 129 mice versus UCP1 protein at or below the level of detection. (<0.3 μg/mg) in mitochondria isolated from muscle of B6 mice. Although low compared with the amount of UCP1 in BAT (73.4 ± 5.5 and 41.6 ± 5.5 μg/mg of mitochondrial protein for 129 and B6, respectively; n = 9–10), when multiplied by the relative mass of hindlimb muscle versus BAT, muscle-associated UCP1 adds at least an additional 10% over intracapsular BAT in 129 mice but represents <2.7% addition in B6 mice.
Morphometric Analysis of Fat in Muscle Samples and UCP1 Protein Expression.
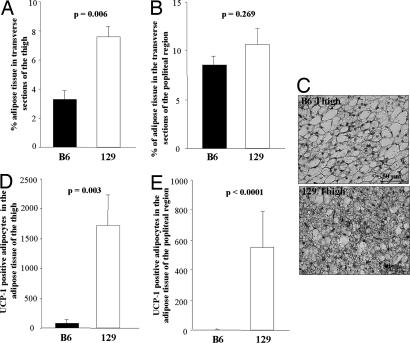
Although all efforts were made to isolate skeletal muscle for gene expression and respiration studies free of all visible adjacent fat and connective tissue, some fat is always associated with muscle, as shown above by fat-specific gene expression analysis. To determine exactly how much fat is interspersed in skeletal muscle, a detailed morphometric analysis [see supporting information (SI) Methods] was done. In agreement with the gene expression for white adipose markers, the total amount of adipose tissue when quantified morphologically was slightly, but nonsignificantly, greater in the thigh muscle (Fig. 5A) and in the popliteal region (Fig. 5B) of 129 versus B6 mice. On the other hand and surprisingly, UCP1 immunostaining (Fig. 5C) revealed small rests of multilocular brown adipocytes intermixed with white adipose tissue adjacent to the thigh muscle. Quantitation demonstrated that the number of UCP1-positive multilocular adipocytes in muscle of 129 mice was 6.8-fold higher than that within the thigh of B6 mice (P = 0.008) (Fig. 5D). More striking, fat in the popliteal region showed a >700-fold increase in UCP1-positive cells in 129 mice compared with B6 (P = 0.008) (Fig. 5E). When all representative sections were analyzed, the total number of UCP1-positive adipocytes interspersed in the posterior limb muscle of 129 mice was 9.1-fold higher than that in B6 mice (P = 0.008). Within the limits of sensitivity, no UCP1 expression could be detected in either the white adipose tissue or the skeletal muscle itself.
Fig. 5.
Morphology of adipose tissue from skeletal muscle of 129 and B6 mice. The bones of the posterior leg were removed from 13-week-old mice, and four transverse serial sections of the thigh (A) and three sections of the popliteal region (B) were obtained from B6 (filled bars) and 129 mice (open bars). The percentage of the area of each transverse section occupied by adipose tissue was calculated by using a morphological imaging system (see Experimental Procedures). The adipose tissue mass was significantly higher in 129 compared with B6 mice of the thigh only, as indicated on the figure. (C) A representative staining of UCP1 immunoreactive multilocular adipocytes is shown. (Scale bar, 50 μm.) Quantification of the UCP1-positive adipocytes of the thigh (D) and popliteal region (E) revealed a statistically significantly higher number of UCP1-positive cells as indicated by the P values on the figures. The bars represent the mean ± SEM of five mice of each strain maintained on regular chow at 22°C.
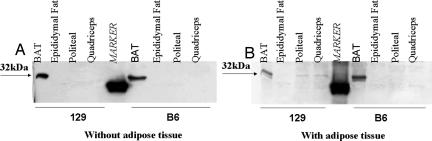
This difference in intramuscular brown fat was further substantiated by immunoblotting. Thus, quadriceps and popliteal muscle samples dissected in the usual fashion from 129 mice revealed immunoreactive UCP1, whereas there was no detectable UCP1 in muscle samples in which the interspersed fat had been removed (Fig. 6). By contrast, muscle samples prepared from B6 mice and epididymal white fat from both strains had no detectable UCP1 on immunoblot analysis.
Fig. 6.
Western blot analysis of UCP1. Adipose-free extract (A) or adipose-containing extract (B) of popliteal and quadriceps was used for Western blot analysis, and it was compared with BAT and epididymal fat. Twenty micrograms of protein was loaded in each lane and blotted with UCP1 antibody. The arrows indicate location of bands at 32 kDa, where UCP1 is located.
Effect of β3-Adrenergic Receptor Agonist on UCP1 Expression.
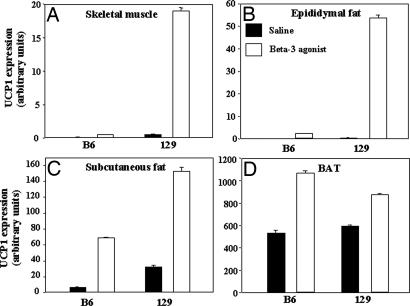
UCP1 in intrascapular brown fat is highly regulated by β3-adrenergic receptor agonists. To determine whether this regulation was also true for UCP1 in intermuscular brown fat, 10-week-old B6 and 129 mice were treated with a β3-adrenergic agonist (disodium(R,R)-5-[2-[2-(3-chlorophenyl)-2-hydroxyethyl]-amino]propyl]-1,3-benzodioxole-2,2-dicarboxylate) (CL 316,243) or saline for 7 days, and UCP1 expression was determined by real-time PCR. Basal levels of UCP1 mRNA expression in the treated control mice was similar in BAT of 129 and B6, and both responded with a 50–100% increase of UCP1 after treatment with the β3-adrenergic agonist (Fig. 7D). By contrast, basal levels of UCP1 were higher in hindlimb skeletal muscle and white fat samples from 129 mice compared with B6. Furthermore, after treatment with β3-adrenergic agonist UCP1, gene expression rose in skeletal muscle, white fat, and brown fat of 129 mice versus B6 mice with the most significant increase in muscle (P = 0.029) (Fig. 7 A–C). Similar results were obtained in mice of 6–7 months of age.
Fig. 7.
β3-Adrenergic receptor agonist-induced UCP1 expression. B6 and 129 mice on a regular chow were treated with saline (filled bars) or β3-adrenergic receptor agonist (open bars) for 7 days. Mice were killed, and UCP1 gene expression was determined by quantitative RT-PCR (see Experimental Procedures) in skeletal muscle (A), epididymal fat (B), s.c. fat (C), and BAT (D). After the 7 days, there was a significantly higher expression of UCP1 in total skeletal muscle of the 129 mice (P = 0.029) compared with B6. Each bar represents the mean ± SEM of eight mice.
Discussion
It has long been recognized that individual humans and different strains of rodents have significant differences in the propensity to gain weight and to manifest the metabolic syndrome. In the present work, we attempted to define the cause for this difference in mice by comparing the diabesity-prone B6 mouse with the diabesity-resistant 129 mouse. We find that the most important difference between these two genetic strains is a higher basal metabolic rate in 129 mice, which appears to be caused, at least in part, by previously unrecognized differences in the presence of brown fat in unexpected intermuscular locations.
The difference in response to dietary fat between these two strains of mice is striking. The 129 mouse gains on average 32% less weight on a high-fat diet than the B6 mouse on either a low- or high-fat diet. DEXA scanning reveals that this difference can be accounted for entirely by lower fat mass in 129 versus B6 mice, which, surprisingly, is not the result of a lower caloric consumption or higher activity of the 129 mouse strain. Indeed, 129 mice actually have a significantly higher caloric intake on the low-fat diet, but the weight gain per gram of food eaten was 62% lower in 129 mice. In addition, oxygen consumption of the 129 strain is significantly higher than that in the B6 on both low- and high-fat diets. Together with other studies (29, 30), this finding suggests that differences in metabolic rate could account for the resistance of the 129 mouse to gain weight. A clue to the potential mechanism of this difference came from the unexpected finding of markedly higher expression of UCP1 in skeletal muscle of 129 versus B6 mice. This finding led us to uncover a potential mechanism for differences in diabesity risk, namely genetically programmed differences in brown fat development, such that in the 129 mouse, there are significant collections of brown fat in regions not usually thought to have brown fat, i.e., within hindlimb skeletal muscle, allowing for significantly higher basal energy consumption.
Skeletal muscle has long been envisioned as an attractive site for unexpected energy expenditure because skeletal muscle represents a large tissue mass. Skeletal muscle is the major site for insulin-mediated glucose disposal and a major determinant of the resting metabolic rate. Although UCP2 and UCP3 are normally expressed in skeletal muscle, most studies have indicated that UCP2 or UCP3 expression per se has no impact on energy expenditure (23, 24), and in our studies and those of Surwit et al. (31), UCP2 and UCP3 expression in muscle was not different between strains (see SI Discussion). It is well recognized that transgenic and knockout mice expressing ectopic UCP1 may exhibit altered energy metabolism (32). For instance, Li et al. (33) have created mice overexpressing UCP1 in skeletal muscle, and they have shown that these mice are resistant to obesity, have lower glucose and insulin levels, and better glucose tolerance than control mice. Indeed, mice with UCP1 expression in muscle at only 1% of the level in BAT had a 98% higher oxygen consumption than wild-type mice (33), demonstrating how efficient UCP1 can be in its antidiabetic and antiobesity effect.
Two recent studies of knockout mice have suggested ectopic expression of UCP1 in muscle. Thus, mice lacking the transcriptional corepressor RIP140 are lean and resistant to high-fat diet-induced obesity and hepatic steatosis, and they have increased oxygen consumption. Interestingly, these mice have a >10-fold up-regulation of UCP1 gene expression in muscle as well as a 100-fold up-regulation in white adipose tissue (34). Similarly, mice with knockout of the liver X receptor are defective in hepatic lipid metabolism, resistant to obesity when challenged with a high-fat/high-cholesterol diet, and exhibit ectopic expression of uncoupling proteins in muscle and white adipose (35). In view of the findings of this work, however, these observations need to be revisited to determine the exact cell type expressing UCP1, which could well be interstitial brown fat in the muscle samples. Indeed, by both immunoblotting and immunohistochemistry, we did not detect any UCP1 expression in myocytes themselves. Furthermore, our data show that assessment of gene expression for markers typical of white fat, e.g., leptin, adiponectin, and aP2, is not sufficient to determine whether there are differences in intermuscular brown fat.
Although the actual level of UCP1 expression in the mRNA of muscle mixed with ectopic brown fat is low compared with isolated intrascapular brown fat pads, we believe that it is quite significant in terms of metabolism for two reasons. First, the high level of differential gene expression in muscle extracts of 129 mice compared with B6 of similar age in similar environments is indeed striking (110-fold). Second, quantitative analysis of UCP1 protein expression in mitochondria isolated from hindlimb muscle (including the intermuscular brown fat) revealed that in 129 mice the intermuscular depot in the hindlimb would represent an amount of UCP1 equivalent to ≈10% of that in pure intrascapular BAT. Clearly, a 10% change in UCP1-mediated uncoupling over long periods of time would represent a considerable difference in energy expenditure. Furthermore, this increase in UCP1 protein is also paralleled by changes in proton conductance in isolated mitochondria of 129 mice versus B6 and a higher dissipation of energy as heat as measured by higher body temperature in 129 compared with B6 mice.
BAT is usually thought to be located to distinct depots, such as the intrascapular and paraspinal regions (36, 37), although some brown adipocytes have previously been observed interspersed in areas of white fat, and they have been suggested to play a role in the regulation of body weight (38). It has also been proposed that the regulation of UCP1 expression during induction of white to brown fat is genetically determined (38, 39). Indeed, two studies have suggested that this genetic determination may occur at a higher rate in the obesity-resistant A/J mouse compared with B6 (32, 38). The present study indicates that this phenomenon is more widely spread than previously recognized, and it is associated with increased basal energy expenditure, increased body temperature, and resistance to the whole metabolic syndrome. In our case, it also appears that these ectopic collections of brown fat are more sensitive to β3-adrenergic agonists with induction of UCP1 than intrascapular brown fat, especially in the 129 strain.
BAT and UCP1 are attractive targets in the defense against obesity, but thus far they have been unsuccessful in humans (13). As noted above, several studies in rodents suggest that elevated expression of UCPs can decrease the relative weight, although no studies have shown that depletion of UCPs causes increased body weight or adiposity (33). In adult humans, BAT-expressing UCP1 mRNA has been found in patients with pheochromocytoma (40) and in fat tissue around the carotid arteries, in the subscapular region, and around the thoracic aorta in human adults who were heavy alcohol consumers (41). The data of the current study would indicate that a thorough analysis of BAT and UCP1 in all depots, including intermuscular depots, in humans is warranted to determine whether small depots of BAT might be present in these areas and whether they might account for differences in susceptibility to weight gain in different individuals. Furthermore, this hypothesis needs to be explored in populations and individuals with different genetic risk for obesity and the metabolic syndrome. The genetically programmed differences in ectopic expression of BAT and UCP1 adjacent to the muscle in 129 mice may help these mice to have an increased thermic response to a high-fat diet and thereby avoid becoming obese and developing the extreme metabolic syndrome observed in other strains like the B6.
Experimental Procedures
Animal Protocols.
Wild-type B6 and 129 male mice (Taconic, Germantown, NY) were maintained on a low-fat diet containing 14% of calories provided from fat, 25% from protein, and 61% from carbohydrates (Taconic); a high-fat diet containing 55% fat, 21% protein, and 24% carbohydrates (Harlan Teklad, Madison, WI; ref. 6); or a standard chow containing 21% calories from fat, 22% protein, and 57% carbohydrates (PharmaServ, Framingham, MA). Food intake was measured by placing the mice in individual cages and weighing the food every third day during a 9-day period. Body temperature was measured by using a Traceable Expanded Range thermometer with bead type-K probe (Fisher Scientific, Pittsburgh, PA). The mice were maintained on a 12-h light/dark cycle. All protocols for animal use were reviewed and approved by the Animal Care Committee of the Joslin Diabetes Center, and they were in accordance with the National Institutes of Health guidelines.
Indirect Calorimetry, Activity, and DEXA Scanning.
DEXA scanning was performed on a Lunar PIXImus mouse densitometer (GE Healthcare, Piscataway, NJ) on anesthetized 3-month-old B6 and 129 male mice (four per group) that had been on a low-fat or high-fat diet for 6 weeks. The mice were then placed individually in indirect calorimetry chambers (Oxymax OPTO-M3 system; Columbus Instruments, Columbus, OH). After 48 h to allow for adaptation, O2 consumption was measured every 30 min for 24 h, and activity was measured as beam break counts.
RNA Isolation, cRNA Preparation, and Array Hybridization and Analysis.
Skeletal muscle, intrascapular brown fat, and epididymal white fat pads were removed from 6-month-old B6 and 129 mice maintained on a standard chow. Total RNA was extracted and purified with RNeasy (Qiagen, Chatsworth, CA), and a total of 25 μg pooled from two or three mice was used for cRNA synthesis (42). Fifteen micrograms of cRNA was hybridized on murine U74Av.2 chips (Affymetrix, Santa Clara, CA). The data were normalized to 1500 by using the GeneChip software MAS 5.0 (Affymetrix). Three or four chips were used for each strain.
Quantitative RT-PCR Using the SYBR Green Procedure.
Mice were killed at ≈4 months of age, and tissues were removed and frozen immediately in liquid nitrogen. cDNA was prepared from 1 μg of RNA by using an Advantage RT-PCR kit (BD Biosciences, San Diego, CA). Five microliters of cDNA was used in a 25-μl PCR (SYBR Green; PE Biosystems, Foster City, CA), run in duplicate, and quantitated in the ABI Prism 7700 sequence system (Applied Biosystems, Foster City, CA) with standards on every plate to allow normalization. For the experiment shown in Fig. 3B, individual RNA samples were used (n = 11).
Proton Leak Kinetics.
Mitochondria were isolated as described in refs. 24 and 43 from muscle of 10 mice. Respiration and mitochondrial inner membrane potential were determined simultaneously (24). Mitochondrial inner membrane potential was calculated by using the Nernst equation based on the distribution of the lipophilic cation tetraphenyl phosphonium (24). Mitochondrial matrix volumes were 1.83 ± 0.17 μl for hindlimb of B6 mice, 2.00 ± 0.09 for hindlimb of 129 mice, 1.62 ± 0.22 for BAT of B6 mice, and 1.97 ± 0.32 for BAT of 129 mice (mean ± SEM, n = 5). Corresponding binding corrections were 0.29 ± 0.07, 0.31 ± 0.09, 0.39 ± 0.02, and 0.44 ± 0.03 (mean ± SEM, n = 5). The proton leak was assessed as described in SI Methods.
Morphology and Western Blotting.
Thirteen-week-old male mice (five B6 and five 129) maintained on a regular chow (Charles River Laboratories, Calco, Italy). Mice were anesthetized with ketamine in combination with xylazine, killed, and perfused intraaortically with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). Posterior limbs were removed, and the muscles were prepared for histology and Western blotting as described in SI Methods.
Administration of β3-Adrenergic Receptor Agonist.
B6 and 129 mice were anesthetized as described above. β3-Adrenergic receptor agonist CL 316,243 (Sigma–Aldrich, St. Louis, MO) is a potent β-adrenergic agonist. Mice (eight per group) were administered either saline or the β3-adrenergic agonist (1 mg/kg each day) for 7 days by an Alzet osmotic minipump (Alza, Palo Alto, CA) implanted s.c. along the back. Mice were killed on the seventh day after minipump implantation.
Statistical Analysis.
The Statistical Package of Social Science for Windows, version 11.5 (SPSS, Chicago, IL) was used for the statistical analyses. Significance was analyzed by nonparametric tests (Mann–Whitney U test or Kruskal–Wallis), and a P value <0.05 (two-tailed) was considered significant.
Supplementary Material
Acknowledgments
We thank Scott Lannon, Laureen Mazzola, Pei Lin, Mike Hirshman, and Lorena Capparuccia for technical help and Drs. Efi Kokkotou, Atul Butte, and Isaac Kohane for useful discussions. This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grants DK31036 and DK45935 (to C.R.K.), Diabetes Genome Anatomy Project Core Grant DK60837, and by the Italian Ministry of University Cofin 2002 (to S.C.).
Abbreviations
- 129 mice
129S6/SvEvTac mice
- B6 mice
C57BL/6 mice
- BAT
brown adipose tissue
- DEXA
dual-energy x-ray absortionmetry
- UCP
uncoupling protein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610416104/DC1.
References
- 1.Bogardus C. Diabetes Care. 1993;16:228–231. doi: 10.2337/diacare.16.1.228. [DOI] [PubMed] [Google Scholar]
- 2.Lee ET, Howard BV, Savage PJ, Cowan LD, Fabsitz RR, Oopik AJ, Yeh J, Go O, Robbins DC, Welty TK. Diabetes Care. 1995;18:599–610. doi: 10.2337/diacare.18.5.599. [DOI] [PubMed] [Google Scholar]
- 3.Meigs JB, Cupples LA, Wilson PW. Diabetes. 2000;49:2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 4.Almind K, Kulkarni RN, Lannon S, Kahn C. Diabetes. 2003;52:1535–1543. doi: 10.2337/diabetes.52.6.1535. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni RN, Almind K, Goren HJ, Winnay JN, Ueki K, Okada T, Kahn CR. Diabetes. 2003;52:1528–1534. doi: 10.2337/diabetes.52.6.1528. [DOI] [PubMed] [Google Scholar]
- 6.Almind K, Kahn CR. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 7.Colombo C, Haluzik M, Cutson JJ, Dietz KR, Marcus-Samuels B, Vinson C, Gavrilova O, Reitman ML. J Biol Chem. 2003;278:3992–3999. doi: 10.1074/jbc.M207665200. [DOI] [PubMed] [Google Scholar]
- 8.Bachmanov AA, Reed DR, Tordoff MG, Price RA, Beauchamp GK. Physiol Behav. 2001;72:603–613. doi: 10.1016/s0031-9384(01)00412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak LP, Rossmeisl M. Ann NY Acad Sci. 2002;967:80–87. doi: 10.1111/j.1749-6632.2002.tb04266.x. [DOI] [PubMed] [Google Scholar]
- 10.West DB, York B. Am J Clin Nutr. 1998;67:505S–512S. doi: 10.1093/ajcn/67.3.505S. [DOI] [PubMed] [Google Scholar]
- 11.Rossmeisl M, Rim JS, Koza RA, Kozak LP. Diabetes. 2003;52:1958–1966. doi: 10.2337/diabetes.52.8.1958. [DOI] [PubMed] [Google Scholar]
- 12.Spiegelman BM, Flier JS. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 13.Himms-Hagen J. Rev Endocr Metab Disord. 2001;2:395–401. doi: 10.1023/a:1011856617047. [DOI] [PubMed] [Google Scholar]
- 14.Lowell BB, Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 15.Lowell BB. Curr Biol. 1998;8:R517–R520. doi: 10.1016/s0960-9822(07)00336-3. [DOI] [PubMed] [Google Scholar]
- 16.Lowell BB, Spiegelman BM. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 17.Nicholls DG, Locker J. Physiol Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Rousset S, Alves-Guerra MC, Mozo J, Miroux B, Cassard-Doulcier AM, Bouillaud F, Ricquier D. Diabetes. 2004;53(Suppl 1):S130–S135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- 19.Cinti S, Cancello R, Zingaretti MC, Ceresi E, De Matteis R, Giordano A, Himms-Hagen J, Ricquier D. J Histochem Cytochem. 2002;50:21–31. doi: 10.1177/002215540205000103. [DOI] [PubMed] [Google Scholar]
- 20.Susulic VS, Frederich RC, Lawitts J, Tozzo E, Kahn BB, Harper ME, Himms-Hagen J, Flier JS, Lowell BB. J Biol Chem. 1995;270:29483–29492. doi: 10.1074/jbc.270.49.29483. [DOI] [PubMed] [Google Scholar]
- 21.Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 22.Wolf G. Nutr Rev. 2001;59:56–57. doi: 10.1111/j.1753-4887.2001.tb06977.x. [DOI] [PubMed] [Google Scholar]
- 23.Erlanson-Albertsson C. Acta Physiol Scand. 2003;178:405–412. doi: 10.1046/j.1365-201X.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- 24.Fink BD, Hong YS, Mathahs MM, Scholz TD, Dillon JS, Sivitz WI. J Biol Chem. 2002;277:3918–3925. doi: 10.1074/jbc.M107955200. [DOI] [PubMed] [Google Scholar]
- 25.Reitman ML. Annu Rev Nutr. 2002;22:459–482. doi: 10.1146/annurev.nutr.22.010402.102849. [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, et al. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 27.Friedman JM, Halaas JL. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 28.Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 29.Brownlow BS, Petro A, Feinglos MN, Surwit RS. Physiol Behav. 1996;60:37–41. doi: 10.1016/0031-9384(95)02210-4. [DOI] [PubMed] [Google Scholar]
- 30.Ravussin E, Lillioja S, Knowler WC, Christin L, Freymond D, Abbott WG, Boyce V, Howard BV, Bogardus C. N Engl J Med. 1988;318:467–472. doi: 10.1056/NEJM198802253180802. [DOI] [PubMed] [Google Scholar]
- 31.Surwit RS, Wang S, Petro AE, Sanchis D, Raimbault S, Ricquier D, Collins S. Proc Natl Acad Sci USA. 1998;95:4061–4065. doi: 10.1073/pnas.95.7.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. J Clin Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Nolte LA, Ju JS, Han DH, Coleman T, Holloszy JO, Semenkovich CF. Nat Med. 2000;6:1115–1120. doi: 10.1038/80450. [DOI] [PubMed] [Google Scholar]
- 34.Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, et al. Proc Natl Acad Sci USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ. Cell Metab. 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Sell H, Deshaies Y, Richard D. Int J Biochem Cell Biol. 2004;36:2098–2104. doi: 10.1016/j.biocel.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Dalgaard LT, Pedersen O. Diabetologia. 2001;44:946–965. doi: 10.1007/s001250100596. [DOI] [PubMed] [Google Scholar]
- 38.Coulter AA, Bearden CM, Liu X, Koza RA, Kozak LP. Physiol Genomics. 2003;14:139–147. doi: 10.1152/physiolgenomics.00057.2003. [DOI] [PubMed] [Google Scholar]
- 39.Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. Mol Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garruti G, Ricquier D. Int J Obes Relat Metab Disord. 1992;16:383–390. [PubMed] [Google Scholar]
- 41.Kortelainen ML, Pelletier G, Ricquier D, Bukowiecki LJ. J Histochem Cytochem. 1993;41:759–764. doi: 10.1177/41.5.8468458. [DOI] [PubMed] [Google Scholar]
- 42.Yechoor VJ, Patti ME, Saccone R, Kahn CR. Proc Natl Acad Sci USA. 2002;99:10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong Y, Fink BD, Dillon JS, Sivitz WI. Endocrinology. 2001;142:249–256. doi: 10.1210/endo.142.1.7889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.