Abstract
Due to their small size, nanoparticles have distinct properties compared with the bulk form of the same materials. These properties are rapidly revolutionizing many areas of medicine and technology. Despite the remarkable speed of development of nanoscience, relatively little is known about the interaction of nanoscale objects with living systems. In a biological fluid, proteins associate with nanoparticles, and the amount and presentation of the proteins on the surface of the particles leads to an in vivo response. Proteins compete for the nanoparticle “surface,” leading to a protein “corona” that largely defines the biological identity of the particle. Thus, knowledge of rates, affinities, and stoichiometries of protein association with, and dissociation from, nanoparticles is important for understanding the nature of the particle surface seen by the functional machinery of cells. Here we develop approaches to study these parameters and apply them to plasma and simple model systems, albumin and fibrinogen. A series of copolymer nanoparticles are used with variation of size and composition (hydrophobicity). We show that isothermal titration calorimetry is suitable for studying the affinity and stoichiometry of protein binding to nanoparticles. We determine the rates of protein association and dissociation using surface plasmon resonance technology with nanoparticles that are thiol-linked to gold, and through size exclusion chromatography of protein–nanoparticle mixtures. This method is less perturbing than centrifugation, and is developed into a systematic methodology to isolate nanoparticle-associated proteins. The kinetic and equilibrium binding properties depend on protein identity as well as particle surface characteristics and size.
Nanoparticles offer many new interesting developments in biomedicine and technology, for example in diagnostics, treatment and novel functional materials. These particles are small enough to enter almost all areas of the body, including cells and organelles, leading potentially to a new approach to medicine (NanoMedicine) or even a source of biological hazard (1–4). Despite the remarkable speed of development of nanoscience, relatively little is known about the interaction of nanoscale objects with biological systems, and this is now a serious bottleneck in the whole nanomedicine and nanotoxicology enterprise. For example, gene transfection and other forms of intracellular delivery depend on these issues, and rational approaches to that field have been limited by poor understanding of the nature of the surface of the transfection vector and how this affects its efficiency. When nanoparticles enter a biological fluid, they become coated with proteins that may transmit biological effects due to altered protein conformation, exposure of novel epitopes, perturbed function (due to structural effects or local high concentration), and/or avidity effects arising from the close spatial repetition of the same protein.
A deep understanding of the biological effects of nanoparticles requires knowledge of the equilibrium and kinetic binding properties of proteins (and other molecules) that associate with the particles. However, the isolation and identification of particle-associated proteins, a fundamental prerequisite for nanobiology, nanomedicine and nanotoxicology, is not a simple task. Furthermore, in terms of the biological response, the more abundantly associated proteins do not necessarily have the most profound effect. A less abundant protein with high affinity and specificity for a particular receptor may instead be a key player. It is thus essential to develop methods to identify both major and minor particle-associated proteins, and to study the competition between proteins to bind when the system is under kinetic or thermodynamic control. A central methodological problem is to separate free protein from protein bound to nanoparticles, ideally employing nonperturbing methods that do not disrupt the protein-particle complex or induce additional protein binding. The preferred method to-date has been centrifugation, identifying the major serum proteins albumin, IgG and fibrinogen as being associated with a wide range of particles of seemingly disparate molecular composition (5–18). Due to its high abundance, albumin is almost always observed on particles and may be retrieved even if it has relatively low affinity. Other proteins observed with several particle types in these centrifugation assays are immunoglobulins, apolipoproteins and alpha-1-antitrypsin. There is little doubt that many more proteins are associated to much lower degrees.
Our understanding of protein–nanoparticle interactions and their biological consequences may be advanced if we find a means to go beyond mere identification of particle-associated proteins. Of highest relevance would be information on the binding affinities and stoichiometries for different combinations of proteins and nanoparticles, and ranking of the affinities of proteins that coexist in specific bodily fluids or cellular compartments. The rates by which different proteins bind to and dissociate from nanoparticles, i.e., the time scales on which particle-associated proteins exchange with free proteins, are other critical parameters determining their interaction with receptors, and biological effects. A tightly associated protein that exchanges slowly may follow the particle if it endocytoses from the extracellular fluid into an intracellular location, whereas a protein with fast exchange will be replaced by an intracellular protein during or after such transfer. The biological outcome may also differ depending on the relative protein exchange rates between nanoparticles and cellular receptors. It is clear that, in understanding how particles will interact with cells, these issues, currently almost unstudied, are amongst the most fundamental.
The rates of association and dissociation are likely to vary quite considerably with protein and particle type. The lifetimes of typical protein–protein complexes range from microseconds to weeks, and protein–ligand complexes typically have lifetimes of microseconds to days. The association rate constants of some complexes approach the diffusion-controlled limit, whereas conformational changes upon binding may slow down the process by orders of magnitude. Although most kinetic studies of adsorbed proteins concern extended surfaces of larger particles, reported time scales of exchange of adsorbed proteins from silica, polymer and TiO2 nanoparticles range from 100 s to many hours (19–24). The affinities and/or exchange rates depend on molecular details and the stability of the protein toward unfolding (19, 25).
In the present work, we implement a range of methods for studying kinetic and equilibrium parameters of protein–nanoparticle interactions that will facilitate a deeper understanding of the molecular basis of cell–nanoparticle interactions and the potential risks associated with nanoparticles. We use a set of tailored copolymer nanoparticles that allows us to systematically investigate how the size and composition (hydrophobicity) of the particles affects their interaction with proteins, relative affinities for different proteins and rates of association and dissociation. In particular, we introduce an approach (based on size-exclusion chromatography gel filtration) that yields both the identity of the proteins on nanoparticles as well as rates of exchange with plasma proteins. This method is potentially less perturbing of protein–particle complexes than centrifugation. We also show that isothermal titration calorimetry (ITC) can be used to assess the stoichiometry and affinity of protein binding and surface plasmon resonance (SPR) studies (in which nanoparticles are linked to gold via a thiol anchor) yield additional data on protein association to and dissociation from nanoparticles. We find a clear dependence of the binding and dissociation parameters on protein identity and on the particle surface characteristics.
Results
ITC Reveals a Dependence on Particle Hydrophobicity and Size (Radius of Curvature).
ITC was investigated for its potential to assess the stoichiometry, affinity and enthalpy of protein-nanoparticle interaction. Protein is injected into a nanoparticle solution in the sample cell and the difference in heat that needs to be added to the sample and reference cells to keep both cells at the same temperature is monitored. If the reaction is exothermic, less heat needs to be added to the sample cell and a negative signal is obtained. If the concentrations of both the nanoparticles and injected protein are known, data from multiple injections provide information on the number of protein molecules bound per particle, the apparent affinity, and the enthalpy change.
Human serum albumin (HSA) was titrated into four kinds of N-isopropylacrylamide (NIPAM): N-tert-butylacrylamide (BAM) copolymer nanoparticles to estimate the stoichiometry, affinity, and enthalpy of protein–nanoparticle interaction, as exemplified in Fig. 1. The negative injection signals imply an exothermic process. The data are well fitted assuming a single process; hence, no interprotein interaction needs to be invoked and protein association appears to occur without any significant degree of (negative or positive) cooperativity. The equilibrium association constant appears to be the same for all particles irrespective of size and composition, ≈2 × 106 M−1. As clearly seen in Fig. 1, a larger number of protein injections are needed to reach saturation of the more hydrophobic particles, implying that the number of protein molecules bound (stoichiometry) increases with the particle hydrophobicity. The stoichiometries obtained are 60 ± 20 and 350 ± 70 for 70 nm 85:15 (more hydrophilic) and 50:50 (more hydrophobic) particles, respectively, and 980 ± 700 and 5,400 ± 1700 for 200 nm 85:15 and 50:50 particles, respectively (mean and standard deviation from three or four repeats). The stoichiometries thus depend on particle hydrophobicity and size.
Fig. 1.
ITC. Titration of HSA into solutions of 70 nm nanoparticles with 50:50 (Left) and 85:15 (Right) NIPAM/BAM in 10 mM Hepes/NaOH, 0.15 M NaCl, 1 mM EDTA, pH 7.5, is shown. (Upper) Raw data. (Lower) Integrated heats in each injection versus molar ratio of protein to nanoparticle together with a fit using a one site binding model (Eqs. 3–5 in SI Text). (Inset) Size comparison of albumin and particles of 70 or 200 nm diameter.
Gel Filtration Shows That Protein Exchange Rates Depend on Nanoparticle Hydrophobicity.
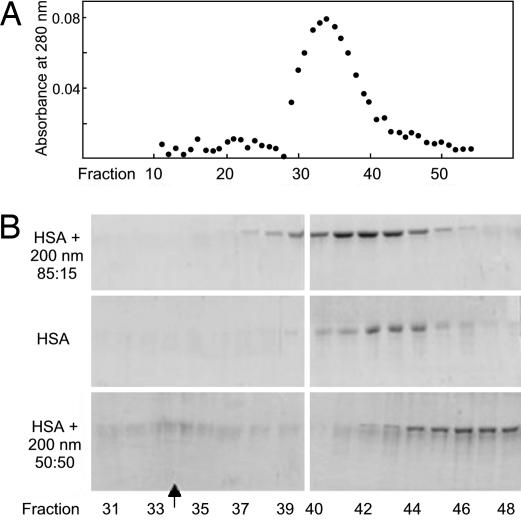
Size exclusion chromatography was the second method investigated for its potential to reveal quantitative information on protein–nanoparticle interactions. Chromatographic elution profiles of protein mixed with nanoparticles were compared with protein alone and particles alone. The chromatographic resin (sephacryl S1000 SF, separation range 5 × 105 to >108 kDa) allows protein and nanoparticles to be resolved, but not different proteins. There is a clear difference in the elution profile of HSA mixed with nanoparticles, compared with free albumin, which implies an interaction between the protein and the particles. HSA mixed with 200 nm 85:15 NIPAM/BAM particles elutes earlier than HSA without particles. The shift is small and with particles, HSA elutes behind the top of the particle peak, indicating that HSA dissociates during passage through the column. Data for HSA and 200 nm particles with 85:15, 65:35 and 50:50 NIPAM/BAM (Fig. 2 and data not shown) reveal that more protein elutes early with the more hydrophilic particles, implying a longer residence time on these nanoparticles. With the most hydrophobic nanoparticles, a large fraction of the protein elutes later than HSA alone. Fibrinogen with 200 nm 65:35 NIPAM/BAM elutes as a double peak (not shown) with elution times equivalent to the free protein and earlier than HSA on the same particles, suggesting that fibrinogen dissociates at a lower rate. Fibrinogen mixed with 50:50 NIPAM/BAM particles also elutes as a double peak with elution times equivalent to the free protein and later than the free protein. Our data imply that single purified proteins associate with the NIPAM/BAM copolymer nanoparticles and that the rates of association/dissociation are very different from protein to protein, depending on the particle composition.
Fig. 2.
Size-exclusion chromatography (gel filtration) on a 1.5 × 95 cm sephacryl S1000 SF column. (A) Chromatogram of 200-nm 50:50 NIPAM/BAM particles alone. (B) SDS/PAGE of precipitated fractions from the elution of HSA with 200 nm 85:15 particles (Top), without (Middle) and with 200 nm 50:50 particles (Bottom). The arrow indicates the elution time of the particles (peak position).
Estimation of Exchange Rates Based on Size-Exclusion Chromatography.
The elution profile of a single protein in a protein–nanoparticle mixture depends on the dissociation rate, the chromatographic run time and flow rate, and other factors. If protein exchange from the particles is very slow, with a residence time several times longer than the separation time, one fraction of the protein would elute with the particles and one at the same position as for protein injected alone. If the exchange is very fast, the protein would elute at the same position as without particles. Intermediate dissociation rates produce divided or broadened peaks and the detailed elution pattern will be determined by the rates of protein–particle exchange. By comparing our experimental data with simulated chromatograms for different dissociation rates (26), we can estimate the dissociation rate to be ≈4 × 10−4 s−1 (half life, 30 min) for the complex of HSA with 200 nm 85:15 particles. The dissociation rate needs to be ≈10-fold higher to yield a protein peak at the same position as for protein alone. For HSA, we observe later elution when applied in a mixture with the 200 nm 50:50 nanoparticles, compared with free HSA. Therefore, the exchange rate for HSA from these more hydrophobic particles can be inferred to be at least one order of magnitude faster than from the more hydrophilic particles. The fact that HSA together with 50:50 particles elutes after HSA alone is puzzling. One possible explanation is that the protein particle complex has a low affinity for the size exclusion resin. The important finding is the clear difference in exchange rates due to particle hydrophobicity (and due to protein identity as shown below).
SPR Measurements of Protein Association to and Dissociation from Nanoparticles.
Gold surfaces with thiol-conjugated nanoparticles were used to study the kinetics of association and dissociation of plasma, HSA and fibrinogen with nanoparticles. Both the association and dissociation rates are clearly dependent on the hydrophobicity of the particles (Fig. 3). The data for plasma (Fig. 3 B and C) could not be fitted assuming a single process with a uniform dissociation rate constant. Neither could the association data be fitted assuming a single process with uniform association and dissociation rate constants. Rate constants were therefore estimated by curve fitting to the data using Eqs. 1 and 2, which account for two processes. Of course, there are more than two kinds of proteins in plasma, but the very large improvement in the fit on going from one to two processes does not motivate the inclusion of additional rate constants. The important results are that the particles bind more than one kind of protein from plasma, and that these proteins differ from one another in terms of their association and dissociation rates. For plasma proteins on the 70 nm 85:15 NIPAM/BAM particles, the two dissociation rate constants are 3.7 × 10−4 s−1 and 6.1 × 10−5 s−1 and for plasma proteins on the 70 nm 50:50 NIPAM/BAM particles, the rate constants are 2.0 × 10−3 s−1 and 3.4 × 10−5 s−1. A similar difference is seen between the 200 nm particles with 85:15 and 50:50 NIPAM/BAM. SPR studies with pure HSA and fibrinogen show dissociation rate constants consistent with the fast dissociation event, suggesting that these proteins account for the faster of the observed kinetic processes. Again for HSA and fibrinogen, we observe faster dissociation from the more hydrophobic compared with the more hydrophilic particles. The rates of association and dissociation are affected by hydrophobicity such that the ratio between them and hence the affinity remains roughly constant, in agreement with the results of ITC. In summary, the dissociation rate depends on both protein and particle identity.
Fig. 3.
SPR studies of plasma–nanoparticle interactions. (A) Cartoon of a gold surface with thiol-tethered particles and associated protein over which buffer is flown. (B and C) SPR data of plasma proteins injected at 60-fold dilution over 70-nm 85:15 NIPAM/BAM (blue) or 50:50 NIPAM/BAM (red) for 30 min (B) followed by buffer flow for 24 h (C, first 6,000 s shown). The black lines are computer fits using Eqs. 1 and 2 (SI Text).
Gel Filtration Reveals Preferential Binding of Specific Plasma Proteins.
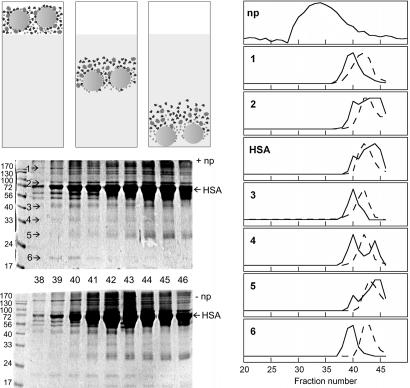
Plasma is a complex fluid that contains >3,700 different proteins, and the relative amounts of the proteins vary over the population (27) and also in the same individual over the day. In addition, there is a large sequence variation among individuals (28). Gel filtration was evaluated for its potential to be a nonperturbing method for studying differential protein binding in such complex fluids and reveals that several plasma proteins are preferentially enriched on the nanoparticles. The protein elution profile is distinctly different with and without particles (Fig. 4). HSA elutes in different fractions when plasma is mixed with 200 nm 50:50 NIPAM/BAM particles compared with chromatographic runs with plasma alone, indicating that HSA in plasma binds to the particles. In addition, there are at least six plasma proteins (marked 1–6 in Fig. 4) that elute earlier with 200 nm 50:50 NIPAM/BAM particles than in plasma without particles. This finding implies that these proteins associate with the particles, and their elution profiles (Fig. 4) indicate slower exchange than for HSA and many other plasma proteins. Interestingly, proteins 2, 4, and 5 show a double peak with elution before and after the position of protein alone. The approximate molecular weights of proteins 1–6 are 180, 80, 50, 35, 28, and 20 kDa, respectively. None of these agree with the molecular weights of HSA or IgG, the most abundant plasma proteins.
Fig. 4.
Size exclusion chromatography of plasma proteins on a 1.5 × 95 cm sephacryl S1000 SF column. (Top Left) Cartoon showing the principle of the method where the darker protein has low affinity but high on and off-rate, and the smallest light gray protein has high affinity and lower dissociation rate. (Middle and Bottom Left) SDS/PAGE of precipitated plasma proteins from a chromatogram with 200 nm 50:50 NIPAM/BAM particles (Middle) and without particles (Bottom). Arrows indicate six protein bands migrating differently when particles are present. (Right) Chromatographic elution profiles of the particles (np), and proteins 1–6 and HSA alone (dashed line) and with nanoparticles (solid line). In 1–6, the y axis represents the density from the SDS/PAGE for each protein and fraction.
Particle-Associated Proteins by Centrifugation.
Most studies have used centrifugation as the method of separation of particle-associated proteins, and here it is compared with size exclusion chromatography. Centrifugation was performed with the aim of separating particle-bound from unbound plasma proteins. The copolymer nanoparticles do not readily sediment when they are dispersed in solution. The nanoparticles were dissolved at high concentration at 5°C together with plasma, and incubated at 23°C to form aggregates that can be collected by centrifugation. After three cycles of centrifugation and resuspension in buffer, total centrifugation time ≈20 min, the proteins in the nanoparticle pellets were separated and visualized by SDS/PAGE (Fig. 5A). Five proteins were observed at 175, 75, 50, 35, and 28 kDa, which correspond to proteins 1–5 from gel filtration. The 28-kDa protein is the by far most prevalent protein. This band was cut out from similar gels as in Fig. 5A, digested with trypsin, and identified as apolipoprotein A-1 (28 kDa; P < 10−6) by mass spectrometry. The protein pattern is the same for all particles, but the relative amounts of the bound proteins vary. The 50:50 NIPAM/BAM particles bind ≈50-fold more of apolipoprotein A-I than the 65:35 NIPAM/BAM particles, indicating a strong correlation between the amount of bound apolipoprotein A-I and the hydrophobicity of the particles. In contrast, the amount of the 75-kDa protein is similar for the two particles. Although the results from ITC, SPR, and gel filtration show that pure albumin or fibrinogen associate with the 200 nm 50:50 particles, none of these abundant plasma proteins are preferentially associated to the particles in the centrifugation experiments. This finding most likely reflects the faster dissociation of albumin and fibrinogen compared with proteins 1–5. Indeed, in test experiments with significantly shortened washing periods, we do observe some albumin on the 85:15 particles (data not shown).
Fig. 5.
Centrifugation and size exclusion chromatography on isolated protein and particle complexes. (A) SDS/PAGE of proteins retrieved from nanoparticles in centrifugation experiments. The size and NIPAM/BAM ratio of the copolymer particles are given under the respective gel lanes. (B) Size-exclusion chromatogram of plasma protein associated with 200-nm 50:50 NIPAM/BAM particles. (C) SDS/PAGE on fractions 36–71.
The Dissociation Rate of Apolipoprotein A-I from Size Exclusion Experiments.
To compare the exchange rates of apolipoprotein A-I with albumin and fibrinogen, the 200-nm 50:50 NIPAM/BAM copolymer particles with proteins retrieved from plasma by centrifugation were subjected to gel filtration (Fig. 5B). In contrast to HSA (Fig. 2) and fibrinogen, apolipoprotein A-I coelutes with the particles (separation time, ≈2 h), indicating a much slower exchange rate than for albumin and fibrinogen. Clearly, plasma proteins that interact with the particles span a wide range of affinities and exchange rates.
Discussion
The results presented here indicate that many proteins form transient complexes with nanoparticles, and the outcome is determined by competitive binding. The protein “corona” that results then constitutes a major element of the biological identity of the nanoparticle. In our view, a firm understanding of this particle–corona complex is a prerequisite for future developments; thus, a thorough characterization is essential. Here we show that SPR, ITC, and size exclusion chromatography (gel filtration) are suitable tools for quantifying kinetic and equilibrium binding parameters. These and other methods have the potential to enable mapping of the nature and state of the proteins expressed on the surface of nanoparticles, thereby opening the way toward a rational understanding of their biological interactions.
In the present study, we find a strong dependence of protein adsorption on particle surface characteristics and size but also on protein identity. Albumin and fibrinogen display higher rates of both association and dissociation than apolipoprotein A-I and many other plasma proteins. The molecular composition of the particles also has a strong influence on the exchange rates. It is interesting to note that, for example, for albumin, the residence time is shorter on the more hydrophobic compared with the more hydrophilic particles, whereas the more hydrophobic particles seem to obtain a larger degree of surface coverage at equilibrium.
The degree of nanoparticle surface coverage by albumin can be calculated from the ITC results. Dividing the surface area at half a protein diameter above the particle surface by the smallest cross section of HSA yields 620 protein molecules per 70-nm particle, and 4,650 proteins per 200-nm particle. Whereas hexagonal close packing of ideal spheres on a flat surface would yield 90.6% surface coverage, the stoichiometries we obtain correspond to surface coverages of 60 ± 11 and 116 ± 37% for the more hydrophobic particles of 70 and 200 nm, respectively, and to 10 ± 3 and 21 ± 15% for the more hydrophilic particles of 70 and 200 nm, respectively. This finding suggests that a single layer of albumin is adsorbed to the surface of the largest and most hydrophobic particle, whereas a sparser layer is associated with the more hydrophilic particles. Whereas the stoichiometries reported should be interpreted with care because of the fact that the particles are somewhat expanded at 5°C, the important finding is the clear effect of particle hydrophobicity. This finding suggests that the more bulky hydrophobic tertbutyl groups of BAM are important for providing binding sites on the particle surface. For each level of hydrophobicity, we also find an influence of particle size. The lower degree of surface coverage observed for the smaller particles suggests that the higher curvature interferes with binding. To what degree the particle surface can be considered a smooth sphere is unclear (there are many dangling ends from the polymer chains) but efficient close packing on the surface of a sphere becomes more difficult as the radius of curvature becomes significant compared with the protein size (see Fig. 1), and this could account for the different degrees of surface coverage of the 70- and 200-nm particles. In this context, we note that for the five proteins retrieved by centrifugation on the 50:50 NIPAM/BAM particles, the yield of the largest protein is higher on 200-nm compared with 70-nm particles. When a larger amount of plasma is used to achieve extensive competition among the proteins for the particle surface, the smallest of the five proteins dominates. Competitive binding on (sometimes highly) curved surfaces is clearly a new area for which there are few classical experiments. Thorough characterization of these systems will be important for the development of nanobiology, and will require joint biological and physical science approaches.
Another striking observation is the extraordinary variation in dissociation rates for proteins on nanoparticles, and the technical challenges implied thereby. The implication is that depending on the experimental procedure and times of the different steps in the protocol, different sets of proteins may be identified as part of the particle–protein corona. For example, in centrifugation experiments, one may expect quite different results depending on the incubation and wash times used. The longer these times, the lower the fraction of quickly dissociating proteins like albumin and fibrinogen. Also the concentrations of particles as well as bodily fluid will influence the outcome of identification experiments. The total protein concentration in bodily fluids and intracellular environments can be up to 35% (0.35 g/ml) representing several thousand different proteins spanning a wide range of concentrations. As a result, there will be competition between the proteins for the available nanoparticle surface area in a typical biological environment. HSA and fibrinogen may dominate on the particle surface at short times, but will subsequently be displaced by lower abundance proteins with higher affinity and slower kinetics, for example apolipoprotein A-I. In contrast, when the available nanoparticle surface area is in excess over the total protein concentration, lower affinity proteins like albumin may also be found in isolation experiments. To identify a set of associated proteins that more closely reflects the situation in vivo will require the protein mixture (e.g., plasma) to be in excess over the available particle surface area, and at best that the particle concentration used reflects a true biological situation (such as a typical therapeutic or imaging particle concentration). It is clear then that we will need new physiochemical and biophysical methods to characterize the protein corona fully. In this work we show that gel filtration with careful choice of column dimensions and relative concentrations of protein and particles isolates both major and minor particle associated proteins, and modern methods of proteomics may be applied to identify them. We have shown that a fraction-by-fraction comparison of the proteins eluting with and without particles allows the identification of both slowly and more rapidly dissociating proteins, and their exchange rates may be estimated from their elution profiles. Further development of the technique, using a wide range of column designs, lengths, and other parameters is expected to make the approach increasingly flexible, and to contribute to our understanding of the protein corona.
Materials and Methods
For further details, please see supporting information (SI) Text.
Nanoparticles.
N-isopropylacrylamide-co-N-tert-butylacrylamide (NIPAM/BAM) copolymer particles of 70 and 200 nm diameter and with three different ratios of the comonomers (85:15, 65:35, and 50:50 NIPAM/BAM) were synthesized in SDS micelles as described (29). Monomers in appropriate ratio, and cross-linker were dissolved in H2O with SDS (the concentration of SDS determines the particle size). Polymerization was induced by ammonium persulfate and heating at 70°C for 4 h. Particles were dialyzed against H2O until no traces of monomers, cross-linker, initiator or SDS could be detected by proton NMR (30).
Thiol-Linked Nanoparticles.
NIPAM/BAM/acrylic acid copolymer nanoparticles were synthesized as above with the addition of appropriate amounts of acrylic acid to the monomer solution to obtain particles with on average less than one carboxyl group on the particle surface. The covalent attachment of homocysteine to the acrylic acid groups was achieved by formation of amide bonds between the primary amino group of the amino acid and carboxylic acid (31).
SPR Experiments.
SPR studies of protein associating to and dissociating from nanoparticles were performed using a BIAcore 3000 instrument (BIAcore, Uppsala, Sweden). Thiol-linked nanoparticles were dissolved in 20 mM sodium phosphate buffer, 100 mM NaCl (pH 7.5), on ice and applied to gold surface. Alkylated (to avoid coupling to gold via free thiols groups) single proteins or plasma diluted in flow buffer (10 mM Tris·HCl, pH 7.4/3 mM EDTA/150 mM NaCl/0.005% Tween-20) were injected for 30 min to study the association kinetics. After 30 min, buffer was flown over the sensorchip surface for 10–24 h to study dissociation kinetics.
ITC.
HSA was titrated into nanoparticles in buffer (10 mM Hepes·NaOH, pH 7.5/150 mM NaCl/1 mM EDTA) at 5°C. The reaction cell contained particles composed of 85:15 or 50:50 NIPAM/BAM. The first protein injection was 1 μl followed by a series of 5-μl injections. Data were fitted using a simple 1:1 binding isotherm with affinity, stoichiometry, and ΔH as variable parameters.
Gel Filtration of Copolymer Nanoparticles and Plasma Proteins.
Nanoparticles were mixed with human plasma or the single proteins on ice in 10 mM Tris·HCl (pH 7.5), 0.15 M NaCl, 1 mM EDTA. The mixture was loaded onto sephacryl S-1000 SF column operated at 5°C in the same buffer. Eluted fractions were analyzed by absorbance at 280 nm. The proteins were analyzed in detail by SDS/PAGE after TCA precipitation. For gel filtration of isolated proteins on the particles (mainly apolipoprotein AI), particles were mixed with plasma on ice and pelleted by centrifugation after heating to 23°C. The pellet was washed three times, dissolved in buffer on ice, and loaded onto a 95 cm sephacryl S-1000 SF column. Fractions were analyzed as above.
Protein Identification by Mass Spectrometry.
After the separation of proteins by SDS/PAGE (12%), bands were excised from the gel, reduced, alkylated, and digested with trypsin, and the resulting peptide mixtures were separated and analyzed by nanoscale liquid chromatography quadrupole time-of-flight MS/MS (32). Spectra were analyzed by MASCOT software to identify tryptic peptide sequences matched to the international protein index (IPI) database (www.ebi.ac.uk/IPI/IPIhelp.html)
Supplementary Material
Acknowledgments
This work was supported by the Marie Curie Research Training Network Colloidal and Interfacial Properties of Synthetic Nucleic Acid Complexes (K.D. and T.C.), European Union Sixth Framework Programme Consortium NanoInteract (K.D., I.L., and S.L.), Science Foundation Ireland (S.L., Walton Fellow; K.D. and I.L., RFP-Spatiotemporal aspects of nanoparticle interaction with cells), and the Swedish Research Council (S.L.).
Abbreviations
- ITC
isothermal titration calorimetry
- SPR
surface plasmon resonance
- HSA
human serum albumin
- NIPAM
N-isopropylacrylamide
- BAM
N-tert-butylacrylamide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 2029.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608582104/DC1.
References
- 1.Colvin VL. Nat Biotechnol. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- 2.Lynch I, Dawson KA, Linse S. Science STKE. 2006;327:pe14. doi: 10.1126/stke.3272006pe14. [DOI] [PubMed] [Google Scholar]
- 3.Lynch I. Physica A. 2007;373:511–520. [Google Scholar]
- 4.Radomski A, Jurasz P, Alonso-Escolano D, Drews M, Morandi M, Malinski T, Radomski MW. Br J Pharmacol. 2005;146:882–893. doi: 10.1038/sj.bjp.0706386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allémann E., Gravel P, Leroux Jean-Christophe J-C, Balant L, Gurny R. J Biomed Mater Res. 1997;37:229–234. doi: 10.1002/(sici)1097-4636(199711)37:2<229::aid-jbm12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Blunk T, Hochstrasser DF, Sanchez JF, Müller BW, Müller RH. Electrophoresis. 1993;14:1382–1387. doi: 10.1002/elps.11501401214. [DOI] [PubMed] [Google Scholar]
- 7.Diederichs JE. Electrophoresis. 1996;17:607–611. doi: 10.1002/elps.1150170332. [DOI] [PubMed] [Google Scholar]
- 8.Gessner A, Lieske A, Paulke B-R, Müller RH. Eur J Pharm Biopharm. 2002;4:165–170. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]
- 9.Gessner A, Lieske A, Paulke B-R, Müller RH. J Biomed Mater Res. 2003;65:319–326. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 10.Gessner A, Waicz R, Lieske A, Paulke B-R, Mäder K, Müller RH. Int J Pharm. 2000;196:245–249. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 11.Gref R, Lück M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Müller RH. Colloids Surf B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 12.Göppert TM, Müller RH. J Drug Target. 2005;13:179–187. doi: 10.1080/10611860500071292. [DOI] [PubMed] [Google Scholar]
- 13.Kasche V, de Boer M, Lazo C, Gad M. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;790:115–129. doi: 10.1016/s0021-9673(02)02001-0. [DOI] [PubMed] [Google Scholar]
- 14.Labarre D, Vauthier C, Chauvierre D, Boris Petri B, Müller R, Chehimi MM. Biomaterials. 2005;26:5075–5084. doi: 10.1016/j.biomaterials.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 15.Lück M, Paulke B-R, Schröder W, Blunk T, Müller RH. J Biomed Mater Res. 1998;39:478–485. doi: 10.1002/(sici)1097-4636(19980305)39:3<478::aid-jbm19>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Müller RH, Rühl D, Lück M, Paulke B-R. Pharm Res. 1997;14:18–24. doi: 10.1023/a:1012043131081. [DOI] [PubMed] [Google Scholar]
- 17.Salvador-Morales C, Flahaut E, Sim E, Sloan J, Green MLH, Sim RH. Mol Immunol. 2006;43:193–201. doi: 10.1016/j.molimm.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Sun D.-H., Trindade MCD, Nakashima Y, Maloney WJ, Goodman SB, Schurman DJ, Smith RL. J Biomed Mater Res A. 2003;65:290–298. doi: 10.1002/jbm.a.10477. [DOI] [PubMed] [Google Scholar]
- 19.Lundqvist M, Sethson I, Jonsson BH. Langmuir. 2004;20:10639–10647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 20.Lundqvist M, Sethson I, Jonsson BH. Langmuir. 2005;21:5974–5979. doi: 10.1021/la050569j. [DOI] [PubMed] [Google Scholar]
- 21.Renner L, Jorgensen B, Markowski M, Salchert K, Werner C, Pompe T. J Mater Sci Mater Med. 2004;15:387–390. doi: 10.1023/b:jmsm.0000021107.12809.8d. [DOI] [PubMed] [Google Scholar]
- 22.Renner L, Pompe T, Salchert K, Werner C. Langmuir. 2004;20:2928–2933. doi: 10.1021/la0362627. [DOI] [PubMed] [Google Scholar]
- 23.Renner L, Pompe T, Salchert K, Werner C. Langmuir. 2005;21:4571–4577. doi: 10.1021/la046801n. [DOI] [PubMed] [Google Scholar]
- 24.Sousa SR, Moradas-Ferreira P, Saramago B, Melo LV, Barbosa MA. Langmuir. 2004;20:9745–9754. doi: 10.1021/la049158d. [DOI] [PubMed] [Google Scholar]
- 25.Lee WK, McGuire J, Bothwell MK. J Colloid Interface Sci. 2004;269:251–254. doi: 10.1016/j.jcis.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Stevens FJ. Biophys J. 1989:1155–1167. doi: 10.1016/S0006-3495(89)82912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthusamy B, Hanumanthu G, Suresh S, Rekha B, Srinivas D, Karthick L, Vrushabendra BM, Sharma S, Mishra G, Chatterje P, et al. Proteomics. 2005;5:3531–3536. doi: 10.1002/pmic.200401335. [DOI] [PubMed] [Google Scholar]
- 28.Nedelkov D, Kienan UA, Niederkofler EE, Tubbs KA, Nelson RW. Proc Natl Acad Sci USA. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Pelton RH, Hamielec AE, Woods DR, McPhee W. Colloid Polym Sci. 1994;272:467–472. [Google Scholar]
- 30.Lynch I, Miller I, Gallagher WM, Dawson KA. J Phys Chem B. 2006;110:14581–14589. doi: 10.1021/jp061166a. [DOI] [PubMed] [Google Scholar]
- 31.Bernkop-Schnürch A, Leitner V, Moser V. Ind Pharm. 2004;30:1–8. doi: 10.1081/ddc-120027505. [DOI] [PubMed] [Google Scholar]
- 32.Berggård T, Arrigoni G, Olsson O, Fex M, Linse S, James P. J Proteome Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.