Abstract
In this article, the effect of a d(CG) DNA dinucleotide repeat sequence on RNA polymerase II transcription is examined in yeast Saccharomyces cerevisiae. Our previous report shows that a d(CG)n dinucleotide repeat sequence located proximally upstream of the TATA box enhances transcription from a minimal CYC1 promoter in a manner that depends on its surrounding negative supercoiling. Here, we demonstrate that the d(CG)9 repeat sequence stimulates gene activity by forming a Z-DNA secondary structure. Furthermore, the extent of transcriptional enhancement by Z-DNA is promoter-specific and determined by its separation distance relative to the TATA box. The stimulatory effect exerted by promoter proximal Z-DNA is not affected by helical phasing relative to the TATA box, suggesting that Z-DNA effects transcription without interacting with the general transcription machinery by looping-out the intervening DNA. A nucleosome-scanning assay reveals that the d(CG)9 repeat sequence in the Z conformation blocks nucleosome formation, and it is found in the linker DNA with two flanking nucleosomes. This result suggests that Z-DNA formation proximally upstream of a promoter is sufficient to demarcate the boundaries of its neighboring nucleosomes, which produces transcriptionally favorable locations for the TATA box near the nucleosomal DNA-entry site and at dyad positions on the nucleosome. These findings suggest that Z-DNA formation in chromatin is a part of the “genomic code” for nucleosome positioning in vivo.
Keywords: chromatin remodeling, promoter, transcription, nucleosome positioning
Chromatin is a highly dynamic and intricate nucleoprotein structure that plays an active role in gene expression and genome organization (1, 2). Although the DNA molecule generally occurs in the right-handed B form, alternative structures such as left-handed Z-DNA, DNA triplex, cruciform, and G-quadruplex have been implicated in transcription, telomere organization, and recombination (reviewed in refs. 3–5). In vitro evolution and selection of DNA molecules possessing biochemical activities corroborate the notion that DNA molecules are structurally and functionally diverse (6–8). Formation of Z-DNA in vivo requires negative DNA supercoiling (9, 10), which can be supplied by a moving RNA polymerase. Z-DNA formation at a promoter region has been shown to correlate with transcriptional activity (5, 11–14). Although Z-DNA often acts as an enhancer of transcription, its formation in the upstream regulatory region of the rat nucleolin gene decreases the level of expression (15). In addition, the Z-DNA binding domain of vaccinia virus E3L protein has been shown to be necessary for the transactivation of several genes resulting in antiapoptotic activity in HeLa cells (16).
The formation of Z-DNA upstream of a gene may lead to transcriptional activation by mediating the loss of nucleosome(s) that occlude important promoter elements (3). Z-DNA formation might act by absorbing the negative supercoils released by removal of the nucleosome(s) at the promoter region. In addition, it may play a more active role in removing or repositioning overlying nucleosomes because of the fact that formation of nucleosomes on Z-DNA or methylated d(CG) dinucleotide repeats is highly disfavored (17–22). In both scenarios, an “open” chromatin region is stabilized by the DNA conformational change (3). The role of Z-DNA in chromatin remodeling and enhancement of transcription has been demonstrated in the human CSF1 promoter, where activation requires the cooperation among the transcription factor nuclear factor I, the chromatin-remodeling BAF complex, and an upstream promoter-proximal Z-DNA structure (12, 23). More recently, induction of the human heme oxygenase HO-1 gene has been shown to require Z-DNA formation near the promoter region (14).
We previously have demonstrated that DNA with an alternating deoxycytidine-deoxyguanosine dinucleotide repetitive sequence [d(CG)n] can potentiate transcription in yeast when placed approximately three helical turns (28 bp) upstream of the CYC1 TATA box (24) (Fig. 1A). Here, we show that transcriptional activation by the d(CG)9 repeat sequence depends on the Z conformation. Activation is core promoter-specific and most effective when situated 28 bp or fewer upstream of the TATA box. Changing the distance between the d(CG)9 repeat sequence and TATA box modulates the extent of activation. Using a nucleosome-scanning assay, we demonstrate that a linker-DNA region is formed at the Z-DNA structure with two flanking nucleosomes. Our results suggest a mechanism of transcriptional regulation in which Z-DNA creates an open chromatin state at the promoter by displacing nucleosomes from its environs and establishing the boundaries of its neighboring nucleosomes. Therefore, increasing evidence suggests that Z-DNA formation in chromatin is one of the mechanisms used by chromatin-remodeling proteins to mobilize nucleosomes and may be considered part of the in vivo “genomic code” of nucleosome positioning (25).
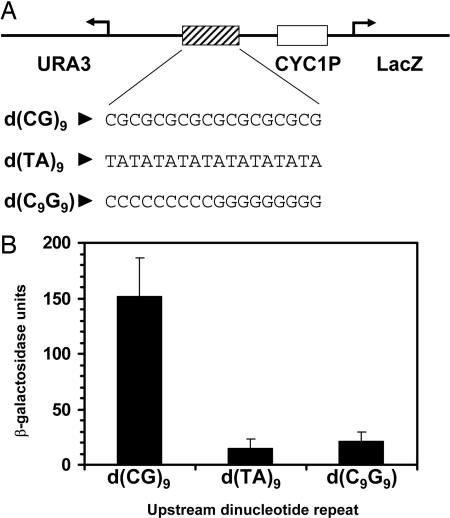
Fig. 1.
The transcriptional enhancing effect of a d(CG) dinucleotide repeat depends on its adopted Z-DNA structure. (A) Diagram of the promoter region of the episomal β-galactosidase reporter showing the position of the dinucleotide repeat/palindromic sequences located 28 bp upstream of the TATA box of yeast CYC1 promoter. (B) β-Galactosidase activities of various reporters in yeast containing DNA repeat sequences d(CG)9, d(TA)9, or d(C9G9). Average β-galactosidase activities and SDs were derived from a single representative experiment performed with each of four independent colonies per yeast reporter strain.
Results
The Ability of d(CG) Dinucleotide Repeat Sequence to Adopt the Z Conformation Is Responsible for Its Transcriptional Activation.
d(CG)n dinucleotide repeat sequences provide the highest propensity to adopt the Z conformation (26). Previous study has demonstrated the transcriptional enhancing activity of the d(CG) repeat and its transcriptional enhancement in the presence of negative supercoiling at the promoter (24) (Fig. 1). However, its palindromic and CG-rich character may account for this transcription stimulation. To establish that d(CG)9 repeats adopt the Z conformation in vivo and that transcriptional enhancement is attributable to this structure, d(TA)9 and d(C9G9) were inserted in place of d(CG)9 proximally upstream of the minimal CYC1 promoter (Fig. 1A). These sequences have a low potential to form Z-DNA (26), but they share the same alternating pyrimidine-purine character and CG richness, respectively. Their tendency to adopt other non-B-DNA structures but not Z-DNA has been demonstrated (27, 28). The level of β-galactosidase activity associated with a d(TA)9 repeat sequence was substantially less than that with a d(CG)9 repeat sequence (Fig. 1B). The fact that we did not observe enhancement in gene expression by the d(TA)9 sequence suggests that the increase in promoter activity is not a result of general alternating pyrimidine-purine character. Likewise, the d(C9G9) sequence did not stimulate promoter activity of the reporter gene (Fig. 1B), further confirming that it is the uniqueness of the alternating d(CG) dinucleotide repeat sequence and not its CG-richness that is required to enhance transcription.
The Extent of Transcriptional Enhancement by d(CG) Repeat Sequence Is Core-Promoter Specific.
Next, we asked whether different RNA Pol II promoters would respond to the upstream d(CG)9 dinucleotide repeat sequence to different extents. Three yeast promoters, CYC1, GAL1, and HIS3, were cloned into the LacZ reporter plasmid with the upstream d(CG)9 repeat sequence located at an equal distance proximally upstream of the TATA box. All three promoters tested were activated by the d(CG)9 repeat sequence; however, the level of gene expression among the three promoters differ by up to 3-fold, with CYC1 and HIS3 being the highest and lowest, respectively (Fig. 2). This variable response is probably a result of the intrinsic affinity of each promoter DNA sequence for the transcription apparatus.
Fig. 2.
The extent of transcriptional stimulation by the d(CG)9 repeat sequence is promoter-specific. The β-galactosidase activities of reporters driven by the yeast GAL1 and HIS3 promoters are compared with the CYC1 promoter. Average β-galactosidase activities and SDs were derived from a single representative experiment performed with each of four independent colonies per yeast reporter strain.
The Extent of Transcriptional Stimulation by Upstream Promoter-Proximal Z-DNA Depends on Its Position Relative to the TATA Box.
The effect of varying the distance separating the upstream promoter-proximal Z-DNA and the TATA box was examined (Fig. 3). As shown in Fig. 3A, the level of expression decreases gradually with increasing distance from 28 to 111 bp; however, where Z-DNA is 73 bp from the TATA box, the expression is increased 2-fold compared with a separation distance of 58 bp (Fig. 3A). The decreasing trend in transcription level proceeds thereafter: when the reporter has a separation distance of 89 bp, it shows a 9-fold drop in activity compared with the activity seen with a separation distance of 73 bp (Fig. 3A).
Fig. 3.
Transcriptional stimulation by promoter-proximal Z-DNA with respect to its distance from TATA box. (A and B) β-galactosidase reporter activities as a function of separation distances between upstream Z-forming d(CG)9 repeat sequence and TATA box. Control indicates the absence of promoter-proximal Z-DNA-forming d(CG)9 sequence, where it is replaced by a random sequence of approximately equal length (shortened by 3 bp). Average β-galactosidase activities and SDs were derived from a single representative experiment performed with each of four independent colonies per yeast reporter strain.
If Z-DNA stimulates transcription by interacting with transacting factors at the TATA box and “looping-out” of any intervening DNA, the stimulatory effect should decrease incrementally as the separation distance increases. Further, a small change in the distance, which alters the helical register, would be predicted to make a large difference, because it would misorient the transacting factor with respect to its target. When a deletion of 4 bp is introduced into the intervening sequence, there is little or no effect on expression level (Fig. 3B), in contradiction to the prediction of the loop-out model. Further, the observed transcriptional profile as a function of separation distance does not support such a model because of the unexpectedly high expression of the reporter construct with a separation distance of 73 bp. These results led us to propose that the Z-DNA structure influences positioning of neighboring nucleosomes relative to the promoter to facilitate an open chromatin organization conducive to a higher level of transcription (see Discussion).
Z-DNA Formation Demarcates the Boundaries of Its Flanking Nucleosomes.
A role for Z-DNA in chromatin remodeling and transcriptional activation in vivo has been demonstrated in the human CSF1 promoter, where transcriptional activation by the human BAF complex depends on the presence of a Z-forming TG/CA dinucleotide repeat sequence near the promoter (12). Similar results were found with the human heme oxygenase HO-1 gene (14). The presence of this Z-forming DNA sequence was shown to increase nuclease accessibility of the promoter DNA region of CSF1, which is associated with a nucleosome in the basal state (23). Thus, we decided to examine the nucleosome positions at the yeast CYC1 promoter in the absence and presence of the upstream d(CG)9 dinucleotide repeat sequence. First, substitution and inactivating mutations were generated at the CYC1 promoter TATA box to change its sequence from TATATAAA to TGTGTGGG so that the effect of transcription on chromatin organization at the promoter region is eliminated. Any change in nucleosome positioning in the presence of an upstream promoter-proximal d(CG)9 repeat sequence likely is to be caused by its adoption of the Z conformation. When the TATA sequence is mutated, the upstream promoter-proximal Z-DNA no longer activates transcription, and reporter gene expression is reduced to the basal level of the wild-type promoter without Z-DNA (Fig. 4A).
Fig. 4.
Z-DNA formation demarcates the boundaries of neighboring nucleosomes. (A) Reporter gene activities are shown for the promoter constructs with either wild-type (+) or mutant (−) TATA box in the presence (+) or absence (−) of the upstream d(CG)9 repeat sequence. (B) Nucleosome-scanning assay. (Upper) Nucleosome occupancy is measured for four overlapping regions (≈100–140 bp) surrounding the d(CG)9 repeat sequence at the mutant TATA box promoter (regions 1 to 4). DNA of the promoter region is depicted as a line with 10-bp intervals (vertical lines). Location of d(CG)9 repeat sequence is denoted as an insertion at 0 bp. In plasmids without this Z-forming sequence, it is replaced by a random sequence of approximately equal length (shortened by 3 bp). Regions 1 through 4 are indicated by horizontal lines below the DNA. (Lower) Nucleosome occupancy of the four overlapping regions is measured by the relative amounts of PCR products generated in the absence (−) or presence (+) of the upstream promoter-proximal d(CG)9 repeat sequence at the mutant TATA box promoter. Relative to the ends of the d(CG)9 repeat sequence, the locations of the regions 1 to 4 determined by PCR primers are as given in parentheses: region 1 (−109 to −5), region 2 (−71 to +26), region 3 (−28 to +72), and region 4 (+3 to +145). Specific PCR products of interest in the electrophoresis gels are indicated by arrowheads on the right. The asterisk in region 4 indicates a nonspecific PCR product also observed in the absence of reporter plasmid.
Sekinger et al. (29) recently have developed a nucleosome-scanning method to study nucleosome positions in yeast in vivo. The method involves extensive micrococcal nuclease (Mnase) digestion of cross-linked chromosomal DNA to create mononucleosome-sized DNA fragments (≈150 bp). After cross-link removal, PCR amplification of DNA regions of interest (≈100 bp) is used to determine their nucleosome occupancy in vivo. DNA sequences with high nucleosome occupancy are expected to produce high levels of PCR products, whereas sequences in the linker DNA are cleaved by micrococcal nuclease and do not yield PCR products. We examined the nucleosome occupancy of the region surrounding the transcription-incompetent CYC1 promoter in the presence or absence of an upstream promoter-proximal d(CG)9 repeat sequence (Fig. 4B). The nucleosome-scanning assay shows that the four overlapping regions examined all are incorporated into mononucleosomes in the absence of a d(CG)9 repeat sequence, suggesting that nucleosomes are positioned randomly at the mutant TATA box promoter over the region examined [≈110 bp upstream and ≈140 bp downstream of the d(CG)9 sequence insertion site]. This result is in contrast with the observation made at the human CSF1 promoter, where nucleosomes are well positioned even in the absence of Z-DNA formation (23). Strikingly, the presence of a d(CG)9 repeat sequence leads to a dramatic decrease in nucleosome occupancy in two of the four examined regions (regions 2 and 3), which include the Z-DNA-forming sequence (Fig. 4B). Regions 1 and 4 flanking the Z-DNA sequence remain equally occupied by nucleosomes in the absence and presence of a d(CG)9 repeat sequence (Fig. 4B). We conclude that the dramatic decline in nucleosome occupancy in regions 2 and 3 is attributed to the Z-DNA structure adopted by the d(CG)9 repeat sequence. The nucleosome-scanning assay result suggests that the Z-forming d(CG)9 dinucleotide repeat sequence forms a micrococcal nuclease-sensitive linker DNA region free of nucleosomes. It is known from in vitro studies that Z-DNA strongly disfavors nucleosome assembly (20). Region 4, located immediately downstream of the d(CG)9 repeat sequence, remains incorporated into a nucleosome in the presence of Z-DNA. The position of this nucleosome is in agreement with the observed effect on transcription activities of separation distance between Z-DNA and the TATA box in which optimal transcriptional stimulation by Z-DNA occurs at separation distances of ≤28 bp and at 73 bp (Fig. 3). At ≤28 bp, the TATA box is located near the nucleosomal DNA-entry site. At 73 bp, it is located near the dyad of this positioned nucleosome (Fig. 5A). Thus, Z-DNA formation in vivo appears to have a regulatory function in nucleosome positioning with a direct influence on gene expression.
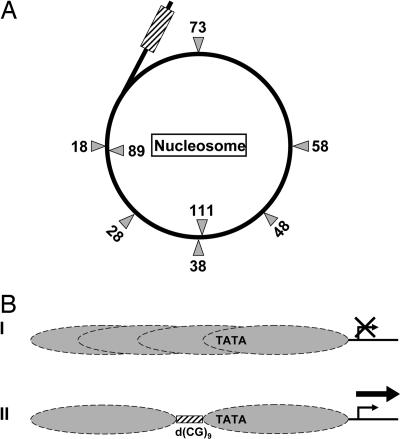
Fig. 5.
A model of transcriptional regulation by upstream promoter-proximal Z-DNA. (A) A proposed model whereby Z-DNA (hatched box) demarcates the boundary of its downstream TATA box-associated nucleosome. The gray triangles indicate the locations of the TATA box (see Fig. 3) at varying separation distances (in base pairs) from the upstream Z-DNA in the transcription reporters used in this study. The TATA box locations in the first turn of nucleosomal DNA relative to Z-DNA are denoted on the outside, and the ones in the second turn are indicated on the inside of the circle. (B) Nucleosomes are positioned randomly over the TATA box in our yeast model reporter system, as found in the absence of the d(CG)9 dinucleotide repeat sequence, repressing transcription (I). Presence of the d(CG)9 dinucleotide repeat sequence and its formation into a Z-DNA conformation upstream of the TATA box displaces its overlying nucleosome by exclusion, creating a linker DNA region over the d(CG)9 dinucleotide repeat sequence and positioning the TATA box near the DNA-entry site of the adjacent nucleosome, facilitating transcription (II).
Discussion
Our current report extends a previous observation made in yeast that the d(CG) dinucleotide repeat sequence can function as a cis-regulatory promoter-proximal element (24), and it further characterizes the properties of its transcriptional regulation. The d(CG)9 dinucleotide repeat sequence activates transcription in a manner that depends on its adoption of the Z form of DNA. The extent of stimulation by Z-DNA is promoter-specific and is not affected by the helical phasing between Z-DNA and the TATA box, but it is influenced by their separation distance. Our findings show that the separation distance between Z-DNA and the TATA box conferring maximal transcription stimulation is 18–28 bp (Figs. 3 and 5A; data not shown). The profile of transcriptional activities with respect to the separation distance follows a descending trend with increasing distance, except that an increase of expression occurs at a distance of 73 bp (Fig. 3A). Furthermore, we show that the Z-DNA structure in vivo serves to demarcate its neighboring nucleosomes on DNA because its presence leads to the virtual absence of nucleosome occupancy over the DNA region containing the d(CG)9 dinucleotide repeat sequence. Thus, it creates a linker DNA region (Figs. 4B and Fig. 5B).
This model of Z-DNA-mediated chromatin remodeling suggests that a separation distance of 18–28 bp between the TATA box and Z-DNA, which yields high transcription activity, places the TATA box near the DNA-entry site on a nucleosome with the Z-DNA structure abutting the upstream boundary of this nucleosome (Fig. 5). At an 18-bp separation distance, the transcription activity is the same as that of 28 bp (data not shown). The interaction between histones and nucleosomal DNA is highly dynamic, and recent evidence shows that DNA unwraps from the surface of the octamer by “peeling off” at the DNA-entry sites on the nucleosome in a reversible manner (30–32), providing windows of opportunity for access to DNA sites by DNA binding proteins at these regions. A surprising finding is that when the TATA box is located at 73 bp from the upstream Z-DNA, the descending trend of transcriptional activation is reversed with a modest increase in gene activity compared with adjacent TATA box locations (Fig. 3A). Based on our proposed model, this distance corresponds to a TATA box location at the nucleosomal dyad (33), suggesting that at this position the DNA is more accessible than at other adjacent positions on the nucleosome (Fig. 5A). This observation lends support to the interpretation of a positioned nucleosome located immediately downstream of the Z-DNA element (Fig. 5). Further corroboration comes from the nucleosome-scanning assay that reveals the presence of a positioned nucleosome at this position (Fig. 4B). The high accessibility of the TATA box located at the DNA-entry site on the nucleosome can readily be explained by reversible DNA–histone interactions. However, the observed modest gain in TATA box binding by the TATA box binding protein when located at nucleosomal dyad suggests accessibility, where the DNA “crosses over” from the first to the second nucleosomal turn.
Chromatin generally acts as a barrier to impede the access of transcription factors to DNA. A d(CG)3 sequence has been shown to displace nucleosome positioning upon 5-cytosine methylation in vitro (22), and cytosine methylation of d(CG) dinucleotide repeat sequences is known to promote the formation of Z-DNA (34). Together, a situation can be envisioned in which nucleosomes are positioned at the promoter region, repressing transcription, and the presence of a d(CG) dinucleotide sequence in the Z conformation of DNA functions to displace associated nucleosomes and expose the TATA box to access by the transcription apparatus (Fig. 5B). The role of Z-DNA in enhancing gene expression by means of chromatin remodeling has been demonstrated in the human CSF1 gene (12, 23). A Z-DNA-forming TG/CA dinucleotide repeat sequence is located 26 bp upstream of the human CSF1 CATA box, a distance closely corresponding to the most transcriptionally optimal separation distance observed in this study. This sequence acts in concert with the human BAF chromatin-remodeling complex to promote an open state of chromatin. It is worth noting that in the yeast system used in this study, a d(TG/CA)23 dinucleotide repeat sequence, even in the presence of negative supercoiling induced by the opposite transcribing URA3 gene (Fig. 1), was unable to act as a promoter-proximal enhancing element (data not shown). This result suggests that the d(TG/CA) repeat sequence, which is known to form Z-DNA less readily than d(CG) repeats do, may require additional factors, such as the ATP-hydrolyzing chromatin-remodeling complex, to generate higher levels of negative supercoiling to form Z-DNA. Interestingly, a d(TG/CA) dinucleotide repeat sequence is located 170 bp upstream of the CATA box of the human heme oxygenase HO-1 gene (14). This separation distance corresponds to the length of one nucleosomal DNA repeat, including linker DNA, which places the CATA box at the DNA-entry site of the second downstream nucleosome from the Z-DNA-forming sequence. Thus, it is highly probable that Z-DNA functions as a nucleosome-boundary element at both human CSF1 and HO-1 promoters to allow TATA box accessibility and consequent transcriptional activation, as shown in our yeast model reporter gene system.
Z-DNA-forming sequences in humans have been reported to be associated with systemic lupus erythematosus (35). A polymorphic sequence with a high potential to form Z-DNA is located upstream of the human natural resistance-associated macrophage protein gene 1 (NRAMP1) promoter, and one particular allele associated with high gene expression correlates with the autoimmune disease rheumatoid arthritis (36). Several human proteins that bind Z-DNA with high affinity are induced by cellular stress, and the induction of these proteins may play a role in the disease process. These proteins, which contain the Z-DNA binding domain (Zα), include the dsRNA adenosine deaminase (ADAR-1) (37) and tumor stromal protein DLM-1 (ZBP1) (38). Further, overexpression of a vaccinia virus protein E3L, which contains a Zα domain, has been shown to induce an antiapoptotic state in HeLa cells (16). This protection from apoptosis correlates with the transactivation of a number of genes. Antiapoptosis and gene activation require a functional Z-DNA binding domain of E3L, and the Zα domains of ADAR-1 and ZBP1 also function in the same manner. Zα stabilizes Z-DNA, and this Z-DNA formation is likely to be involved in regulating gene expression, as shown in this work and others. The induction of Z-DNA binding proteins could be important in the etiology of cancer, where becoming antiapoptotic is a critical step in initiating metastasis.
The location of Z-DNA structures in chromatin may be part of a mechanistic step used by chromatin-remodeling proteins to reposition nucleosomes relative to DNA regulatory elements in the promoter regions. Thus, Z-DNA-forming sequences may function as a part of the genomic code of nucleosome positioning in vivo (25), helping to direct the mobilization of nucleosomes in chromatin. Continued efforts to elucidate the molecular mechanism of transcriptional regulation and chromatin remodeling by Z-DNA may lead to a fuller understanding of the etiology of certain diseases and increase the efficacy of their clinical treatments.
Materials and Methods
Yeast Strains and Media.
Yeast Saccharomyces cerevisiae YM4271 (Mat a ura3–52 his3–200 ade2–101 lys2–801 leu2–3 112 trp1–901 tyr1–501 gal4-Δ512 gal80-Δ538 ade5::hisG; Clontech, Mountain View, CA) was used for all transformations and reporter assays. Synthetic complete media (SD) contains 0.67% yeast nitrogen base (Difco, Franklin Lakes, NJ), 2% glucose, 20 μg/ml adenine, 20 μg/ml uracil, and amino acids (20 μg/ml arginine, 20 μg/ml histidine, 20 μg/ml methionine, 20 μg/ml tryptophan, 30 μg/ml isoleucine, 30 μg/ml lysine, 30 μg/ml tyrosine, 50 μg/ml phenylalanine, 60 μg/ml leucine, and 150 μg/ml valine). Yeast transformants hosting the LacZ reporter plasmid (URA3) were made by using the Yeast EasyComp transformation kit (Invitrogen, Carlsbad, CA) and selected on plates containing 2% agar (Difco) and SD with 2% glucose lacking uracil at 30°C.
Plasmid Construction.
All reporter plasmids used in this study were derived from the pLacZcOp series described in ref. 24. The pLacZcOp-CYC1P reporter is an episomal derivative of pLacZi (Clontech), with the URA3 gene transcribing in the opposite orientation of the LacZ reporter, and it contains the yeast minimal CYC1 promoter (−139 to +3) driving the expression of LacZ. Its UAS DNA sequence between EcoRI and XhoI sites is a random sequence of 25 bp, and it is used as the “control” reporter without d(CG)9 dinucleotide repeat sequence. Plasmids pLacZcOp-d(CG)9-GAL1P and pLacZcOp-d(CG)9-HIS3P contain the yeast GAL1 (−161 to +3) and HIS3 promoters (−86 to +3), respectively, cloned into the XhoI and BamHI sites replacing the CYC1 promoter. DNA oligonucleotides (46-mer) with the d(TA)9 and d(C9G9) sequences were annealed and cloned into the EcoRI and XhoI sites of pLacZcOp-d(CG)9-CYC1P, replacing the d(CG)9 sequence to generate pLacZcOp-d(TA)9-CYC1P and pLacZcOp-d(C9G9)-CYC1P plasmids, respectively. To construct reporter plasmids with varying distances between the upstream d(CG)9 repeat sequence and the CYC1 TATA box, the designed length of oligonucleotides with random sequences was annealed and cloned into the XhoI site of pLacZcOp-d(CG)9-CYC1P plasmid, which has a separation distance of 28 bp between the d(CG)9 sequence and the TATA box. The reporter construct with a separation distance of 24 bp was made by using a 5′ primer with a deletion of the 4 most upstream nucleotides of the CYC1 promoter to yield a shortened CYC1 promoter fragment produced by PCR, which was cloned into XhoI and BamHI sites to replace the existing CYC1 promoter on the plasmid. pLacZcOp-TATA mutant and pLacZcOp-d(CG)9-TATA mutant are reporter plasmids with a mutated CYC1 TATA box sequence (TATATAAA to TGTGTGGG). The mutant CYC1 promoter DNA fragment was produced by PCR with an upstream primer containing the nucleotide substitutions and then cloned into the XhoI and BamHI sites to replace the wild-type CYC1 promoter. All DNA sequences of the oligonucleotide primers used here are available upon request. DNA primers were ordered from Invitrogen. PCR was performed with Platinum pfx DNA polymerase (Invitrogen), and DNA ligation was carried out with the Quick Ligation Kit (New England Biolabs, Inc., Ipswich, MA). All plasmid constructs were confirmed by DNA sequencing (Massachusetts Institute of Technology Biopolymer Facility).
β-Galactosidase Assay with Yeast Liquid Cultures.
The β-galactosidase assay was carried out by using an assay kit supplied from Pierce (Rockford, IL) with minor modifications. Four individual colonies (≈1 mm; 2–3 days old) from each tested strain were picked and separately inoculated into 1 ml of SD with 2% glucose lacking uracil. Cultures were allowed to grow at 30°C for 2 days and then diluted 3-fold with fresh media (SD with 2% glucose lacking uracil) for an additional 6-h incubation with shaking at 30°C. A 100-μl aliquot of each culture subsequently was taken, and its OD660 was measured. Fifty microliters each of extraction buffer and 2× reaction buffer (Pierce) were added to 100-μl cell cultures with brief vortex mixing. Reactions were incubated at room temperature for 90 min and quenched by the addition of 80 μl of stop buffer (1 M sodium carbonate). The total β-galactosidase reporter activity was measured at OD420. The normalized β-galactosidase activity was expressed in units calculated from the equation: (OD420 × 1,000)/(OD660 × time), where the time is in minutes. For each yeast strain, four independent colonies were examined per experiment.
Nucleosome-Scanning Assay.
The procedure is described in ref. 29 and used here with some modifications. Colonies from strains YM4271 containing either pLacZcOp-TATA mutant or pLacZcOp-d(CG)9-TATA mutant plasmids were separately inoculated into 100 ml of SD media (SD with 2% glucose lacking uracil). Cultures were grown for 2 days at 30°C until OD600 reached ≈1–2. Yeast cells then were centrifuged and switched to grow in 100 ml of yeast peptone dextrose for an additional 4 h at 30°C. Formaldehyde was added to cell cultures at final 2% concentration, and cultures were incubated at 30°C for 30 min with shaking. Cross-link reactions were quenched by adding to final concentration of 125 mM glycine and incubating further at 30°C for 5 min. Cells from each strain then were spheroplasted in 1 ml of spheroplast buffer (1 M sorbitol and 2 mM 2-mercaptoethanol) with 200 units (40 μl) of Zymolase (Zymo Research, Orange, CA) for 90 min at room temperature. After two washes with 1 M sorbitol, each cell pellet was resuspended in 1 ml of digestion buffer [1 M sorbitol, 50 mM NaCl, 10 mM Tris (pH 7.5), 5 mM MgCl2, 1 mM CaCl2, 1 mM 2-mercaptoethanol, 0.5 mM spermidine, and 0.075% Nonidet P-40]. A 250-μl aliquot was digested with 450 units of micrococcal nuclease (Worthington, Lakewood, NJ) for 60 min at 37°C to achieve maximal digestion, and digestion was stopped by adding 1/10 vol of solution containing 250 mM EDTA and 5% SDS. Samples were incubated at 65°C overnight to reverse the cross-linking. Phenol:chloroform extraction was carried out before ethanol precipitation. DNA was resuspended in 100 μl of P1 buffer containing RNase A (DNA Miniprep Kit; Qiagen, Valencia, CA) and incubated at 30°C overnight. It then was electrophoresed in 1.5% agarose gel at 100 V for 90 min. Mononucleosome-sized DNA fragments (≈150 bp) were excised from the gel and isolated by using the QIAEX II Gel Extraction Kit (Qiagen). Final DNA volume was 100 μl in water, and DNA concentration was determined by measuring OD260. Equal amounts of mononucleosome-sized DNA fragments (150 ng) were used as a template for PCR (Platinum Pfx DNA polymerase; Invitrogen), and reactions were carried out for 22 cycles. PCR products were electrophoresed in 10% polyacrylamide precast 1× TBE gel and imaged with ImageMaster VDS-CL (Amersham Pharmacia, Uppsala, Sweden) after ethidium bromide staining.
Acknowledgments
We thank Doo-Byoung Oh (ISU Chemical Company, Seoul, Korea) and Yang-Gyun Kim (Sungkyunkwan University, Suwon, Korea) for their generous gift of the parental pLacZcOp-CYC1P reporter plasmid. We thank Ky Lowenhaupt and Alekos Athanasiadis for helpful discussion. This research was supported by grants from the National Institutes of Health and the Ellison Foundation.
Abbreviation
- SD
synthetic complete media.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kornberg RD, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Oliver B, Misteli T. Genome Biol. 2005;6:214. doi: 10.1186/gb-2005-6-4-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Holde K, Zlatanova J. BioEssays. 1994;16:59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]
- 4.Simonsson T. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 5.Rich A, Zhang S. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 6.Joyce GF. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 7.Sreedhara A, Li Y, Breaker RR. J Am Chem Soc. 2004;126:3454–3460. doi: 10.1021/ja039713i. [DOI] [PubMed] [Google Scholar]
- 8.Coppins RL, Silverman SK. Nat Struct Mol Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- 9.Peck LJ, Nordheim A, Rich A, Wang JC. Proc Natl Acad Sci USA. 1982;79:4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordheim A, Lafer EM, Peck LJ, Wang JC, Stollar BD, Rich A. Cell. 1982;31:309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 11.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 13.Muller V, Takeya M, Brendel S, Wittig B, Rich A. Proc Natl Acad Sci USA. 1996;93:780–784. doi: 10.1073/pnas.93.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. Mol Cell Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothenburg S, Koch-Nolte F, Rich A, Haag F. Proc Natl Acad Sci USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon JA, Rich A. Proc Natl Acad Sci USA. 2005;102:12759–12764. doi: 10.1073/pnas.0506011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nickol J, Behe M, Felsenfeld G. Proc Natl Acad Sci USA. 1982;79:1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausio J, Zhou G, van Holde K. Biochemistry. 1987;26:5595–5599. doi: 10.1021/bi00392a003. [DOI] [PubMed] [Google Scholar]
- 19.Casasnovas JM, Azorin F. Nucleic Acids Res. 1987;15:8899–8918. doi: 10.1093/nar/15.21.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner MM, Felsenfeld G. J Mol Biol. 1987;196:581–590. doi: 10.1016/0022-2836(87)90034-9. [DOI] [PubMed] [Google Scholar]
- 21.Davey C, Pennings S, Allan J. J Mol Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 22.Davey CS, Pennings S, Reilly C, Meehan RR, Allan J. Nucleic Acids Res. 2004;32:4322–4331. doi: 10.1093/nar/gkh749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Mulholland N, Fu H, Zhao K. Mol Cell Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh DB, Kim YG, Rich A. Proc Natl Acad Sci USA. 2002;99:16666–16671. doi: 10.1073/pnas.262672699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho PS, Ellison MJ, Quigley GJ, Rich A. EMBO J. 1986;5:2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brukner I, Dlakic M, Savic A, Susic S, Pongor S, Suck D. Nucleic Acids Res. 1993;21:1025–1029. doi: 10.1093/nar/21.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aranda A, Perez-Ortin JE, Benham CJ, Del Olmo ML. Yeast. 1997;13:313–326. doi: 10.1002/(SICI)1097-0061(19970330)13:4<313::AID-YEA93>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Sekinger EA, Moqtaderi Z, Struhl K. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Widom J. Nat Struct Mol Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 31.Li G, Levitus M, Bustamante C, Widom J. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 32.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Proc Natl Acad Sci USA. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 34.Behe M, Felsenfeld G. Proc Natl Acad Sci USA. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adhami E. Med Hypotheses. 2004;62:237–246. doi: 10.1016/S0306-9877(03)00340-2. [DOI] [PubMed] [Google Scholar]
- 36.Searle S, Blackwell JM. J Med Genet. 1999;36:295–299. [PMC free article] [PubMed] [Google Scholar]
- 37.Herbert A, Lowenhaupt K, Spitzner J, Rich A. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Nat Struct Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]