Abstract
Trypanosomes are unique eukaryotic cells, in that they virtually lack mechanisms to control gene expression at the transcriptional level. These microorganisms mostly control protein synthesis by posttranscriptional regulation processes, like mRNA stabilization and degradation. Transcription in these cells is polycistronic. Tens to hundreds of protein-coding genes of unrelated function are arrayed in long clusters on the same DNA strand. Polycistrons are cotranscriptionally processed by trans-splicing at the 5′ end and polyadenylation at the 3′ end, generating monocistronic units ready for degradation or translation. In this work, we show that some trans-splicing/polyadenylation sites may be skipped during normal polycistronic processing. As a consequence, dicistronic units or monocistronic transcripts having long 3′ UTRs are produced. Interestingly, these unspliced transcripts can be processed into mature mRNAs by the conventional trans-splicing/polyadenylation events leading to translation. To our knowledge, this is a previously undescribed mRNA maturation by trans-splicing uncoupled from transcription. We identified an RNA-recognition motif-type protein, homologous to the mammalian polypyrimidine tract-binding protein, interacting with one of the partially processed RNAs analyzed here that might be involved in exon skipping. We propose that splice-site skipping might be part of a posttranscriptional mechanism to regulate gene expression in trypanosomes, through the generation of premature nontranslatable RNA molecules.
Keywords: gene expression, posttranscriptional regulation, RNA processing
Regulation of gene expression is central in defining the phenotype of a cell or organism. In the vast majority of cases, this regulation is controlled by transcriptional regulation in which DNA sequence elements, together with DNA-binding proteins, target genes for transcription by RNA polymerase II. In addition to regulating single genes, master regulators of transcription may give rise to a gene expression phenotype, a trait that may be inherited (1). Mature transcripts can also be regulated in their expression at the posttranscriptional level. A number of cis-motifs mainly located in the 5′ and 3′ UTRs determine the fate of the transcript by the use of highly specific and sometimes evolutionarily conserved machineries. This process is essentially achieved through RNA-binding proteins (RBPs) forming, together with the mRNA, ribonucleoprotein (RNP) complexes. It is the composition of the RNP complex that determines whether an mRNA is transported to the cytoplasm for translation, degradation, or other processing events (2, 3).
Posttranscriptional regulation is common among Kinetoplastid parasites, like trypanosomes and Leishmania (4, 5), which are the causative agents of several infections affecting both humans and domestic animals worldwide (6). In these parasites, transcription by RNA polymerase II starts at a few genomic locations within chromosomes. In contrast to operons in bacteria, polycistronic units in trypanosomatids require processing before translation (7). Cotranscriptional processing of polycistronic units occurs through trans-splicing and polyadenylation, resulting in the production of mature mRNAs (8).
Trans-splicing is a process similar to cis-splicing but occurring between two precursor RNAs that are transcribed from different genomic locations. This discontinuous transcription was first discovered in African trypanosomes in mRNAs encoding variant surface glycoproteins (9, 10), which were found to contain the same leader sequence at their 5′ end (reviewed in ref. 11). Trans-splicing involves this leader sequence of ≈40 nt long, the spliced leader (SL), which is transferred from the 5′ end of a small RNA (≈140 nt long), to an AG splice-acceptor site on pre-mRNA molecules. So far, all mature mRNAs in trypanosomes were found to contain the 5′ terminal miniexon, and thus all transcripts in these cells are supposed to be cotranscriptionally processed by trans-splicing. Ultimately, trans-splicing, together with polyadenylation, allows polycistronic transcripts to be processed into monocistronic units ready for translation.
Given the unusual mechanism to produce mature mRNAs, transcription initiation is likely to be irrelevant for the regulation of gene expression in trypanosomes, and thus regulated protein production relies almost entirely on posttranscriptional processes. One of these mechanisms, analyzed in some detail, is the modulation of mRNA stabilization/destabilization (reviewed in refs. 4 and 5). Given that the SL sequence is added 30–100 nucleotides upstream of the translation initiator ATG codon, long UTRs are located only at the 3′ end of trypanosome transcripts. These regions contain cis-elements involved in the control of transcript half-life and in the modulation of translation efficiency. In particular, AU-rich sequences present within the 3′ UTR of mRNAs, collectively termed AU-rich elements, were shown to bind RNA-recognition motif (RRM)-type RBPs, which then modify the half-lives of the RNAs (12). Dozens of RRM-type RBPs have been identified in the genome of Kinetoplastid parasites (13). Two of these RBPs, named Trypanosoma cruzi U-rich binding protein 1 (TcUBP1) and TcUBP2, were found to be in complex with the poly(A)-binding protein 1 (PABP1) on the 3′ UTR of the transcripts encoding the core of the mucin antigens of T. cruzi (14), molecules essential for the protection of the parasite against the host response.
In this work, we show that, during the primary RNA processing of polycistronic cleavage, some signals are likely to be masked, generating stable long transcripts containing a 5′ SL end. These precursor RNA molecules, referred to as intermediate RNAs, are further trans-spliced and polyadenylated generating functional monocistronic units. Two different classes of molecules that can originate by this mechanism were identified, a dicistronic premRNA and a precursor transcript containing a long unspliced 3′ UTR. We suggest that trans-splicing/polyadenylation site skipping is part of a mechanism to control gene expression through which RNAs in a “translational latency state” can be stored for further processing into a mature transcript when required by the cell.
Results
Identification of a Stable Dicistronic Transcript That Can Be Further Processed into Monocistronic Mature mRNAs.
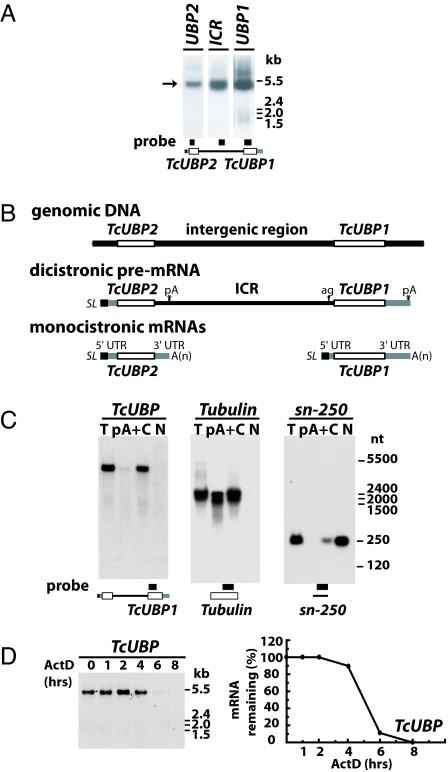
Given that TcUBP1 and TcUBP2 are important regulators of mRNA turnover, we asked how these two proteins are regulated. We have previously reported that both genes are located within a genomic fragment separated by an intergenic region of ≈3 kbp (15). Using Northern blots, we showed that TcUBP1 and TcUBP2 are present in a single dicistronic unit, referred to as TcUBP (Fig. 1A). This transcript was cloned by RT-PCR and shown to contain TcUBP2 upstream of the TcUBP1 ORF together with the expected intercistronic region (ICR) (Fig. 1B). Moreover, 5′ RACE and sequence analysis demonstrated that TcUBP pre-mRNA has the canonical 5′ end 39 nt corresponding to the SL RNA (see Fig. 1B).
Fig. 1.
Identification of a highly stable dicistronic pre-mRNA in the cytoplasm. (A) Northern blot of TcUBP RNA. Total RNA from the insect stage of the parasite was electrophoresed in an agarose gel and hybridized with the probes indicated below the images. (B) Schematic structure of the polycistronic gene cluster that contains TcUBP2 and TcUBP1 genes, the pre-mRNA unit, and the TcUBP1 and TcUBP2 monocistronic RNAs. (C) TcUBP RNA is localized in the cytoplasm. Total RNA (T), poly(A)+ RNA (pA+), and cytoplasmic (C) and nuclear (N) RNA fractions from insect-stage parasites were separated in an agarose gel and hybridized with the probes indicated below the images (schemes are not to scale). (D) TcUBP mRNA half-life was determined by ActD treatment. Insect-stage parasites were incubated with the drug, and total RNA was extracted at 0, 1, 2, 4, 6, and 8 h of treatment. Samples were separated on agarose gels and hybridized with the TcUBP probe. Northern blot signals were quantified by using Kodak (Rochester, NY) Image Software and plotted. The relationships between the percentage of mRNA remaining vs. time (hours in ActD) are shown. Molecular weight markers are shown on the right. The systematic gene name for TcUBP1 is Tc00.1047053507093.220 and for TcUBP2, Tc00.1047053507093.229.
We next studied the dicistronic unit subcellular localization and poly(A)-tail content by using Northern blots performed with total (unfractionated) RNA, a poly(A)+ RNA fraction, and cytoplasmic and nuclear fractions. Two different RNAs were used as nuclear and cytoplasmic markers, a small nuclear RNA of 250 nt (known as sn-250) and β-tubulin, respectively. TcUBP is mainly localized in the cytoplasm (Fig. 1C) and appears to lack or to have a very short poly(A) tail because it was not recovered after an oligo(dT)-affinity column (Fig. 1C) and because we were unable to clone its 3′ end by 3′ RACE experiments by using an oligo(dT)18 primer (results not shown). To further analyze whether the absence or reduced poly(A)-tail content of the TcUBP dicistron affects its mRNA stability, we measured its half-life by using RNA extracted from control and actinomycin D (ActD)-treated parasites (Fig. 1D). The half-life of this transcript is ≈5 h, indicating a normal or slower turnover rate compared with other short-lived mRNAs such as Amastin, EP1, Procyclin, and TcSMUG, the average half-life of which is ≈1 h (16–19).
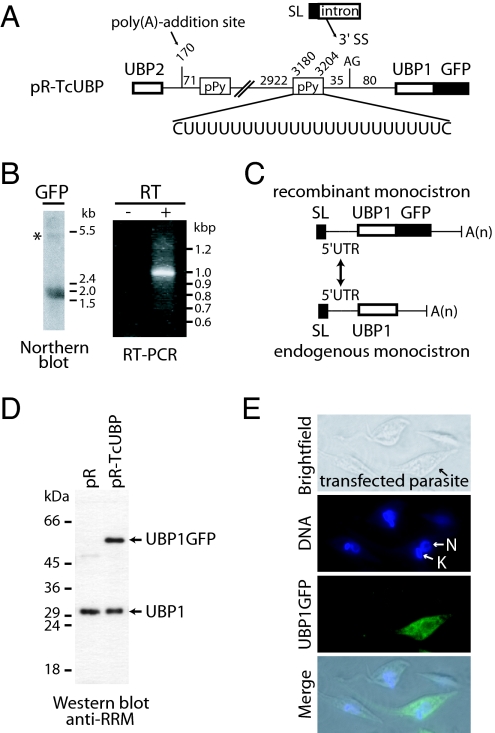
The TcUBP dicistron is expressed in all life-cycle stages of T. cruzi (15) and is likely to give rise to mature TcUBP1 and TcUBP2 mRNAs. Even though monocistrons are barely detectable in Northern blot assays (see Fig. 1A), they were cloned by RT-PCR and RACE experiments and were shown to have a SL sequence at its 5′ end, a small 5′ UTR preceding the coding region, and a 3′ UTR ending in a poly(A) tail (12, 14). The presence of both dicistronic and monocistronic species in the cell prompted us to evaluate whether the TcUBP dicistron can be further processed by trans-splicing and polyadenylation to give rise to mature mRNAs. To test this possibility, we fused the TcUBP dicistronic unit with a GFP gene downstream of the TcUBP1 coding sequences, a tag that allowed us to follow its transcription and translation. This construct was named pR-Tc UBP and was used in transient transfections. pR, an empty vector, was used as control (Fig. 2A). After transfection, we detected a band corresponding to the TcUBP1-GFP monocistronic RNA (Fig. 2B), thus showing that a mature transcript can be produced from the dicistronic construct (Fig. 2A). Next, we asked whether trans-splicing/polyadenylation processing occurs at the same sites as those used to generate monocistronic RNAs from natural dicistrons. A PCR product of ≈1 kbp was identified and cloned. By sequencing, we confirmed that it contained the expected TcUBP1 and GFP coding sequences (Fig. 2B Right). More importantly, trans-splicing at the 5′ UTR occurred at the same site as the endogenous transcript (Fig. 2C). In these experiments, the dicistronic RNA was hardly detectable (Fig. 2B), thus suggesting that under the conditions used it might be rapidly processed.
Fig. 2.
TcUBP pre-mRNA is processed by trans-splicing. (A) Scheme of the pR-TcUBP DNA construct (dicistron with TcUBP1 CDS fused to GFP) made in pRIBOTEX vector. (B) Northern blot using RNA obtained from transfected parasites and hybridized with a GFP probe (Left); ∗ denotes the position of a band of ≈5 kb detectable when the film was overexposed. RT-PCR was performed by using total RNA extracted from pR-TcUBP transfected parasites. cDNA was synthesized by using a COOH-GFP primer, and the PCR was performed with TcSL and GFP as primers (see SI Table 3). The PCR products were separated in an agarose gel and visualized by ethidium bromide staining (Right). (C) Schematics of endogenous and recombinant TcUBP1 monocistronic mRNAs, showing that both share the same 5′ UTR. (D) Western blot was performed by using total protein extracts from transfected parasites (pR and pR-TcUBP) and probed with polyclonal rabbit anti-RNA-recognition motif sera (1/1,000 dilution). (E) Fluorescence microscopy of a T. cruzi insect-stage parasite transfected with the pR-TcUBP construct shown in A. 3′ SS, acceptor splice site; pPy, polypyrimidine tract; N, nuclear DNA; K, kinetoplast DNA. The molecular weight markers are shown on the right.
Protein expression of TcUBP1-GFP was examined by immunoblotting and fluorescence microscopy. By Western blotting, a band of ≈50 kDa that corresponds to the TcUBP1-GFP fusion protein was detected (Fig. 2D), together with the endogenous signal corresponding to TcUBP1. To further confirm the expression of the TcUBP1-GFP fusion protein, parasites transfected with the recombinant construct were fixed and analyzed by fluorescence microscopy. Transfected parasites were clearly detected over the background (Fig. 2E). Because these parasites almost exclusively have TcUBP1-GFP monocistronic units and not dicistrons, it is reasonable to conclude that monocistronic RNAs are the ones from which GFP is translated. The maturation of the TcUBP dicistron by trans-splicing/polyadenylation prompted us to analyze whether this phenomenon is a particular case of RNA processing or can be extended to other transcripts.
Trans-Sialidase (TS) Transcripts Can Also Be Processed by Trans-Splicing and Polyadenylation.
TS is an enzyme expressed on the surface of T. cruzi; it is essential for the survival of the parasite in the mammalian host (20). There are two TS gene families, one coding for the TS in the mammalian stage of the parasite [mammalian-stage TS (mTS)] and one for the TS present in the insect stage of the parasite [insect-stage TS (iTS)]. Genes encoding the mTS have an extra domain at the 3′ end encoding an immunodominant repeat named SAPA (from shed acute-phase antigen) (21) absent in the iTS family. Although the coding sequences of mTS and iTS genes are almost identical, except for the SAPA domain, their 3′ UTRs are completely different in sequence and allowed the identification of the two groups of transcripts (A.V.J., R. Muiá, O. Campetella, and A.C.F., unpublished work).
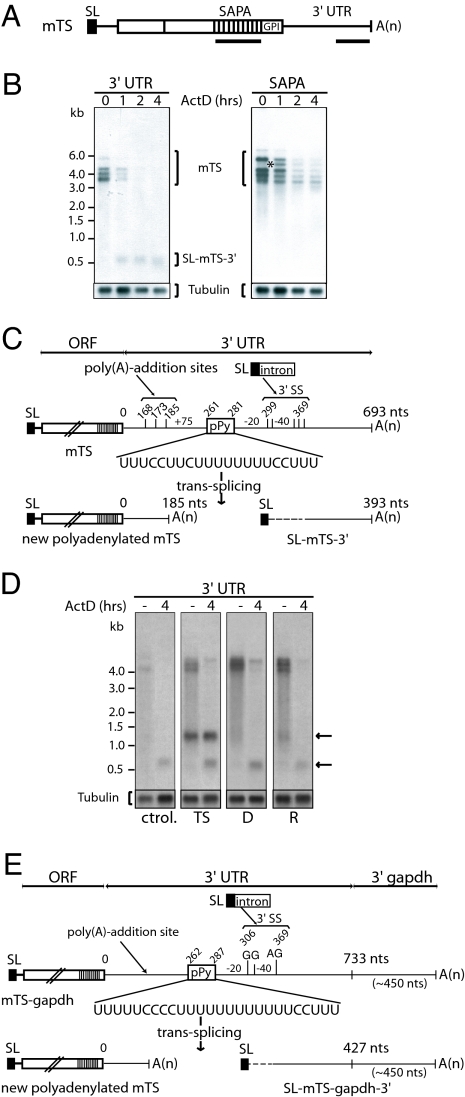
The developmentally regulated expression of the different TSs groups along with their different 3′ UTR sequences encouraged us to evaluate their mRNA stability. To analyze mRNA half-life, we treated cells from the insect-stage parasite with the transcription inhibitor ActD. After different time points, RNA samples were isolated and used to perform a Northern blot with different probes. First, a 3′ UTR probe (Fig. 3A) was used to detect the mTS group of transcripts, showing several bands of high molecular weight (3.3–6 kb). In addition, a small band of ≈600 nt was observed (Fig. 3B). A probe corresponding to the SAPA repeats did not detect this small RNA species (Fig. 3 A and B), thus suggesting that it does not contain sequences from the mTS coding region. A strategy to clone the small RNA product by RACE was devised. Sequencing of the cloned DNA product showed that this small RNA contained the last 324–392 nt of the mTS-3′ UTR, a complete SL sequence on its 5′ end and a poly(A) tail at the 3′ end. Interestingly, probing with the 3′ UTR probe revealed that the small RNA increased in amount after incubation of cells with ActD, whereas the large TS transcript decreased in amount under the same conditions (Figs. 3B and 4B). Taken together, these results suggest that the small transcript is a processed product from the 3′ UTR of the large TS transcript (see below).
Fig. 3.
Trans-splicing/polyadenylation events in the 3′ UTR of TS RNAs. (A) Scheme showing the location of the probes used in these experiments. (B) Northern blots of poly(A)+ RNA from parasites treated with ActD for the indicated times. Hybridizations were performed with mTS-3′ UTR and SAPA probes. The asterisk indicates the detection of a new band after 1 h of ActD treatment. A β-tubulin probe was used for loading control. (C) Structure of mTS, the new polyadenylated mTS, and the SL-mTS-3′ RNAs generated by trans-splicing. Nucleotides where trans-splicing and polyadenylation events take place are indicated. 3′SS, 3′ trans-splicing sites; pPy, polypyrimidine tract. The drawings are not in scale. One representative sequence is shown. (D) Northern blots of total RNA from wild-type parasites (ctrol.) and parasites transfected with different pTEX constructs (D, TS with a 90-nt deletion including the pPy sequence and two 3′SS, R, TS in which the above-mentioned 90-nt fragment has been replaced by another sequence) without treatment (−) or incubated with ActD for 4 h (4). Hybridizations were performed with mTS-3′ UTR probe. The upper arrow indicates the SL-mTS-gapdh-3′ species. The lower arrow shows the endogenous SL-mTS-3′. (E) Structure of the recombinant mTS-gapdh mRNA and the SL-mTS-gapdh-3′ RNA generated by trans-splicing. 3′SS, acceptor splice site; pPy, polypyrimidine tract.
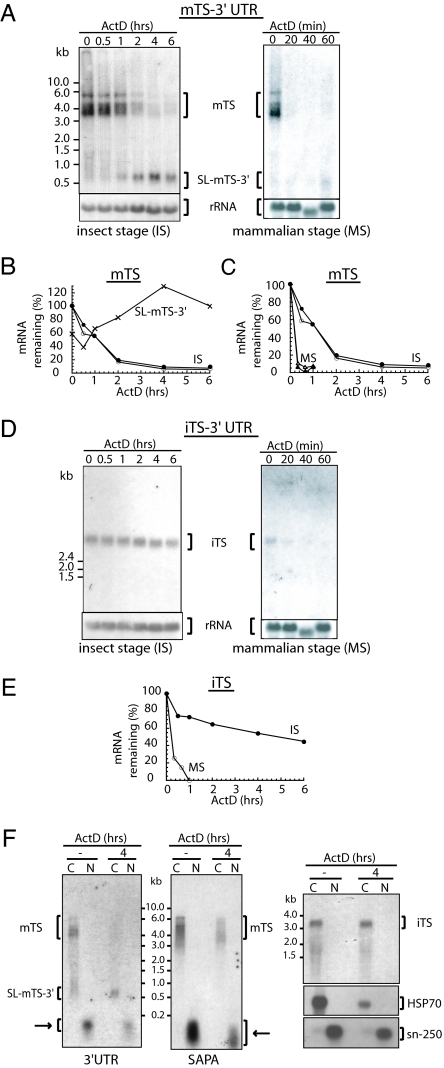
Fig. 4.
Stability and subcellular localization of mTS and iTS mRNAs. (A and D) Northern blots of total RNA from parasites treated with ActD for the indicated times. Hybridizations were performed with mTS-3′ UTR (A) and iTS-3′ UTR (D) probes. For the Northern blot made from mammalian-stage parasites, sequential hybridizations were performed by using the same blot after complete removal of the radioactive signal. Films were scanned, the signals quantified, and the ratio of TS RNAs to rRNA plotted (B and C for mTS and E for iTS). In the case of mTS, the larger RNA bands are indicated with open circles or closed triangles, and the smaller RNA bands are indicated with closed circles or open triangles (B and C). The SL-mTS-3′ RNA is hardly detectable in the mammalian stage, so we were able to quantify this RNA species only in the insect-vector parasite stage (B). (F) Subcellular localization of the different RNAs. Insect-stage parasites after 4 h of ActD treatment or without treatment were used to prepare nuclear (N) and cytoplasmic (C) fractions for RNA extraction. Hybridizations were performed with the indicated probes: 3′ UTR or SAPA probes for mTS and 3′ UTR probe for iTS. HSP70 and sn-250 mRNAs were used as controls for cytoplasmic and nuclear fractions, respectively. Arrows indicate small RNA products in the nuclear fraction.
The presence of the SL indicates that this small transcript, named SL-mTS-3′ (from SL mTS-3′ UTR), is the result of a trans-splicing event occurring in the 3′ UTR of the mTS transcript (Fig. 3C). The 5′ end of 49 different SL-mTS-3′ clones, from nine independent RT-PCRs, was sequenced. Seven of these clones were RT-PCR products from parasites that were not treated with ActD, confirming that this RNA is also produced without ActD treatment. There were at least four possible trans-splicing acceptor sites within a region of ≈70 nt (Fig. 3C), from which two were the most frequently used. A polypyrimidine tract required for both trans-splicing and polyadenylation (7) is located between ≈20 and 40 nt upstream from the main 3′ splice sites (Fig. 3C). The acceptor site in trans-splicing is usually an AG (22). However, the 3′ splice site of SL-mTS-3′ RNAs could be either the canonical AG or a noncanonical GG site. The presence of both AG and GG acceptor sites was confirmed by the analysis of products from parasites transfected with recombinant DNA expressing a single TS gene (see below). RT-PCR of RNA extracted from the mammalian stage of the parasite also showed that SL-mTS-3′ RNA is produced in this trypanosomal stage (see below).
Because trans-splicing and polyadenylation are coupled processes, we devised a 3′ RACE to search for the putative polyadenylated form of the mTS coding region. Such a product was identified and was shown to be polyadenylated at three different sites, between 75 and 93 nt upstream from the polypyrimidine tract (Fig. 3C). This process generated a polyadenylated mTS mRNA bearing a shorter 3′ UTR (≈185 nt) (Fig. 3C), such as the intermediate product indicated by an asterisk in the Northern blot with the SAPA probe in Fig. 3B. This result implies that the generation of SL-mTS-3′ RNA is associated with a new polyadenylation event within the 3′ end of the coding mTS mRNA. Thus, mTS mRNAs processing resembles what we initially observed for the TcUBP dicistron in that a preRNA is further processed into monocistrons by trans-splicing/polyadenylation.
The SL-mTS-3′ contains a small ORF that varies in size between the different 3′ UTR sequences from the mTS family. We identified and synthesized a peptide that was common to all of the predicted ORFs, and polyclonal antibodies were raised against this peptide. The antiserum recognizes the synthetic peptide but was unable to detect any band of the expected size (4–8 kDa) in Western blots using total protein extracts from the insect or mammalian stages of the parasite (results not shown). Thus, this observation makes it unlikely that the SL-mTS-3′ small RNA is translated into a protein product.
A Single Recombinant TS Transcript Recapitulates the Process of Endogenous mRNA Processing.
The in vivo analysis indicated that the mTS transcript previously considered to be the mature TS mRNA is in fact a precursor, which can be processed under natural conditions. However, a drawback of these results is that TS is encoded by a large gene family, and thus the analysis was performed on several different transcripts simultaneously. To determine how a single transcript is being processed, we obtained the complete TS coding sequence together with 310 bp of the 5′ UTR and 733 bp of the 3′ UTR. This DNA was cloned in the trypanosome pTEX expression vector (pTEX-TS), and transient transfections in the insect parasite stage were analyzed. This vector contains GAPDH sequences surrounding the multiple cloning site that provide the cloned sequence with all of the required trans-splicing and polyadenylation signals. Approximately 450 nt are expected to be incorporated at the 3′ end of the TS 3′ UTR as a result of polyadenylation/trans-splicing events. We used these GAPDH sequences as a tag that should give rise to a SL-mTS-3′ species (called SL-mTS-gapdh-3′) longer than the endogenous one. A small RNA product of the expected size was indeed detectable in Northern blots of poly(A)+ RNA [see upper arrow in Fig. 3D (TS)], and this product was present even in the absence of ActD treatment. The GAPDH sequences increased the size of the small transcript to ≈1.2 kb. This RNA species is polyadenylated and contains the SL sequence added at three different acceptor sites (Fig. 3E). SL addition and polyadenylation sites were mapped exactly to the same sites used by the endogenous SL-mTS-3′ product. Some of the trans- splicing sites identified also included the noncanonical GG site, confirming that a GG site can act as a 3′ splice acceptor site. Altogether, these results confirm that a single TS transcript is trans-spliced at the 3′ UTR, and that several trans-splicing sites can be used during the processing.
To confirm that the mapped sequence is indeed acting as a trans-splicing signal, we deleted ≈90 nt, including the polypyrimidine tract and the first two of the three possible trans-splicing acceptor sites (referred to as deletion mutant D; Fig. 3D). In addition, we made a construct that has the natural 90-nt pyrimidine-rich region replaced by a purine-rich tract, completely lacking any 3′ trans-splicing sites (referred to as replacement mutant R; Fig. 3D). After transfection of the insect-stage parasites with these constructs, the SL-mTS-gapdh-3′ product was undetectable in both cases, indicating that the trans-splicing/polyadenylation events were completely abolished. These results confirm the importance of the above 90-nt pyrimidine-rich sequence for the posttranscriptional processing of the unspliced TS transcript.
Processing of TS 3′ UTR Is Faster in the Developmental Stage of the Parasite Where the Transcript Is Translated.
mTS transcripts are translated in the mammalian stage of the parasite but not in the insect vector stage, where transfection experiments can be performed, because this is the replicative stage of the trypanosome. To further analyze whether the rate of trans-splicing processing might play a role in gene-expression and/or translation regulation, we treated parasites from both (mammalian and insect) stages with ActD and analyzed the half-life and the processing of TS transcripts. Surprisingly, the half-life of these transcripts is <10 min in the mammalian stage of the trypanosome, the stage where translation of the transcript occurs, and ≈90 min in the insect parasite stage in which the transcript is not translated (Fig. 4 A and C). These results also confirmed that trans-splicing and polyadenylation take place in both stages of the parasite (Fig. 4A), in agreement with our previous finding by RT-PCR. More importantly, mTS-3′ UTR processing is much faster in the parasite stage, where translation occurs, suggesting that 3′ UTR cleavage by trans-splicing and polyadenylation might be related to expression.
We also asked whether iTS mRNAs, which are translated only in the insect stage of the parasite, are also subject to endogenous cleavage within the 3′ UTR. The iTS gene family has a completely different 3′ UTR sequence (A.V.J., R. Muiá, O. Campetella, and A.C.F., unpublished data), allowing the design of a 3′ UTR-specific probe. This probe detected a single band of ≈3.5 kb, the expected size for the iTS transcript family (Fig. 4D). Experiments done with ActD showed that these transcripts have a long half-life (≈5 h) in the insect- vector stage, whereas their stability was much shorter (≈10 min) in the mammalian stage (Fig. 4E). These results are in agreement with the concept that trypanosomes use mRNA stabilization/destabilization mechanisms to promote/prevent gene expression. In contrast to the processing of mTS, no small RNA product was generated during iTS transcript processing (Fig. 4D). Thus, only one of the two groups of TS transcripts is processed by trans-splicing and polyadenylation in its 3′ UTR.
We next analyzed the subcellular localization of the different mRNAs products by Northern blots of RNA fractions from insect-stage parasites treated or not with ActD for 4 h (Fig. 4F). As nuclear and cytoplasmic markers, the sn-250 RNA (23) and the heat-shock protein 70 (HSP70) mRNA were used, respectively. Hybridization experiments showed that iTS transcripts are cytoplasmic, as might be expected for an mRNA that is actively translated in this parasite stage. mTS transcripts were also located in the cytoplasm, including the full-length form as well as the SL-mTS-3′ small species (Fig. 4F). In the nuclear fractions, we detected small RNA products of ≈150–200 nt that hybridized with both the mTS-3′ UTR and SAPA probes (see arrows in Fig. 4F), suggesting that mTS transcripts might be degraded in the nucleus. These RNA degradation products are specific for mTS because no fragments were detected with other probes, including sn-250 transcripts.
In Silico and Experimental Studies Indicate the Presence of Putative Splicing Signals Within TcUBP and the mTS UTRs.
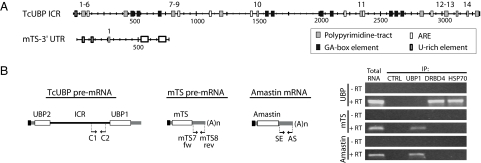
Using bioinformatics approaches, we focused on the detection of putative cis-elements present in the regulatory regions of both unspliced RNAs described here. A Blast search against UTRdb and EST databases revealed several GA- and AU-rich elements that are conserved among different trypanosomatid transcripts [see details in supporting information (SI) Table 1]. Particularly in the ICR of the TcUBP pre-mRNA dicistron, we identified AUUUA pentamers reported to be target sites for the HSP70 (24). In addition, we found various polypyrimidine tracts within the ICR. Among them, a variety of target sites for the polypyrimidine tract-binding protein (PTB), such as UCUUC or CUCUCU (25), were recognized (see Fig. 5A).
Fig. 5.
Functional cis-acting elements in the noncoding regions of TcUBP dicistron interact in vivo with DRBD4 and HSP70. (A) Scheme of cis-acting elements predicted in the ICR and mTS-3′ UTR using bioinformatics tools. Polypyrimidine tracts, GA-rich elements, AU-rich elements, U-rich elements, and PTB sites are indicated. The numbers indicate putative PTB sites. The following numbers indicate the beginning and end positions of PTB sites within the ICR: UCUUC, 103–107, 106–110, 109–113, 817–821, 977–981, 1490–1494, 2356–2360, 3052–3056, 3055–3059, and 3169–3173; and CUCUCU, 4–9, 78–83, 80–85, 82–87, 795–800, and 797–802. Within the mTS-3′ UTR, the only putative PTB-binding site found was UCUUC, 288–292. (B) Agarose gel showing the RT-PCR products of TcUBP, mTS-3′ UTR, and Amastin from total RNA, coimmunoprecipitated in vivo with control rabbit serum (immunoprecipitation control, CTRL), anti-UBP1, anti-DRBD4, or -HSP70 antibodies. RT was performed with (+) or without (−) SuperScript II enzyme. For mTS-3′ UTR cDNA synthesis, we used oligod(T)18. For TcUBP, the internal specific primer NH2/AS-ubp1 (see SI Table 3) was used. Schemes of mRNAs with 3′ UTRs and position of primers used in PCRs are shown at the left. RT, reverse transcriptase enzyme.
The mTS-3′ UTR contains two short U-rich motifs, two conserved long GU-rich tracts located in the 3′ end of the molecule that are putative UBP1-binding sites, and a single polypyrimidine tract that is responsible for the trans-splicing event. Comparison of the dicistron and mTS sequences indicated that >10 putative target sites for the splicing repressor PTB were detected along the ICR of TcUBP (see above), whereas only one putative PTB site (UCUUC) is present in mTS-3′ UTR located 5 nt downstream of the polypyrimidine tract (Fig. 5A). Furthermore, the AUUUA pentamers for HSP70 binding are absent in the mTS-3′ UTR.
To test whether the double RNA-binding Domain 4 (DRBD4), the trypanosome homolog of the splicing regulator PTB (13), and the chaperone HSP70 interact with the putative sites identified in silico, we performed immunoprecipitation (IP) of the ribonucleoprotein complexes present in vivo with polyclonal antibodies against each of the proteins mentioned above. The IP samples were analyzed by RT-PCR for the presence of the different RNAs, TcUBP dicistron, and mTS transcripts. We found that the TcUBP RNA can be coimmunoprecipitated with both anti-TcDRBD4 and -HSP70 antibodies, whereas no product was amplified in IP experiments performed with antibodies against TcUBP1 or control serum (Fig. 5B). The mTS transcript was detectable in the TcUBP1 immunoprecipitates (Fig. 5B), together with the Amastin transcript [a known target of TcUBP1 (15)], used as an experimental control. These findings suggest that the TcUBP dicistron interacts with both DRBD4 and HSP70, whereas the mTS and Amastin transcripts do not likely because they possess a single putative binding site for PTB (see Discussion).
To see whether DRBD4 could interact with the TcUBP dicistron in the cytoplasm, where this transcript is localized (see Fig. 1C), we analyzed the intracellular localization of DRBD4 in the insect-stage parasite. This experiment revealed that the protein is localized in both the nucleus and the cytoplasm (see SI Fig. 7), thus exhibiting a different localization pattern in comparison to other nuclear proteins of trypanosomes, which are totally excluded from the cytoplasm (A.C., J.G.D.G., and A.C.F., unpublished work).
Discussion
Transcriptional regulation is the main mechanism by which gene expression is controlled in most cells. Kinetoplastid parasites show an important variation on this eukaryotic paradigm because these cells have polycistronic transcription coupled with the apparent absence of classic RNA polymerase II promoters, indicating that transcriptional regulation is not relevant (26, 27). Trypanosomatids are also different from other organisms having transcriptomes composed of multigene transcription units, such as Drosophila and Caenorhabditis elegans. Operons from fruit flies are translated in that form, whereas in nematodes, all polycistrons so far described are processed into translatable monocistrons (28). We propose that, during trypanosome RNA processing, trans-splicing/polyadenylation sites are masked, resulting in unspliced RNAs. Once a trypanosome pre-mRNA is transcribed by the action of a RNA polymerase II complex, all of the monocistrons contained in that precursor are likely to be generated by RNA cleavage. However, we show here that transcription-coupled polycistronic RNA processing is not a rule. In some cases, signals within intergenic or UTR regions facilitate the skipping of trans-splicing/polyadenylation signals. This mechanism generates intermediate RNAs, like dicistrons and mRNAs with long 3′ UTRs, which can be further processed by conventional trans-splicing and polyadenylation, thus giving rise to monocistronic mRNAs.
Two examples of intermediate RNAs have been described here. The first example is a TcUBP dicistronic RNA that encodes two functionally related RNA-recognition motif-type RBPs, which are evolutionarily conserved among different protozoan species, such as Trypanosoma brucei and Leishmania major (13). Second, we characterized a 3′ UTR trans-splicing event taking place in the members of one TS transcript family (20). This event leads to the removal of 3′ UTR sequences that can be followed during RNA processing (Fig. 4B). Interestingly, this occurs faster in the parasite stage, where translation takes place. Thus, we postulate that this mechanism could be used to remove translation inhibitory sequences/structures contained in 3′ UTRs. Curiously, the TcUBP dicistron is strongly expressed in all parasite life-cycle stages but encodes one protein that is developmentally regulated, suggesting that the dicistron itself is not a functional mRNA. Similarly, the mTS mRNA is expressed in the insect-vector and mammalian-host stages, but, as mentioned, the protein is translated only in the latter stage, where trans-splicing occurs faster. Altogether, these findings indicate that regulation of trans-splicing of partially processed RNAs might be crucial to generate mature monocistrons for protein synthesis.
In silico identified splicing signals are likely to be functional. For instance, the trans-acting factor DRBD4 interacts in vivo with TcUBP RNA in the cytoplasm, arguing for a role for this protein in RNA processing. As mentioned, DRBD4 is homologous to the mammalian PTB, which is involved in several aspects of mRNA metabolism, including mRNA localization, polyadenylation, translation, and regulation of alternative splicing (29). Because the processing of TcUBP pre-mRNA occurs with very low efficiency, our observations point to the presence of weak or masked trans-splicing signals probably because of the presence of numerous PTB sites that facilitate DRBD4 binding. However, other proteins might also be necessary, and PTB by itself might not be enough to prevent precursor RNA processing. In fact, the mTS-3′ UTR unspliced RNA and Amastin transcripts possess a single putative binding site for PTB, and the protein is not in complex with these transcripts in vivo (Fig. 5B).
The mechanism used by trypanosomes to produce unspliced RNAs, as reported here, is likely to occur in other transcripts (30, 31). Previously, two dicistrons encoding RBPs have been reported in African trypanosomes, although it is not known whether they can be further processed (8, 32). Moreover, we found in the literature that several protein-coding genes related to mRNA metabolism in trypanosomes are expressed as transcripts longer than expected from the prediction of their cDNA length, e.g., poly(A)-binding protein 1 (PABP1) and TSR1 (see SI Table 2). Altogether, these findings suggest that masking of trans-splicing/polyadenylation signals might be a common trait for the control of gene expression.
SL trans-splicing is a phylogenetically widespread process that has been discovered in several unrelated metazoans, including flatworms, nematodes, and tunicates (33). Nevertheless, no regulated splicing has been detected in unicellular organisms (34). In the case of the intermediate RNAs described here, they could be produced as a consequence of a regulated process. Whether trans-splicing of these intermediate RNAs takes place in the nucleus or the cytoplasm is unknown at present. It might be postulated that long pre-mRNAs are reimported to the nucleus for further processing when the cell needs to generate monocistrons for translation. However, it has recently been described that splicing might occur in the cytosol of some types of anucleate cells and in neuronal dendrites having a functional cytosolic spliceosome complex (35, 36).
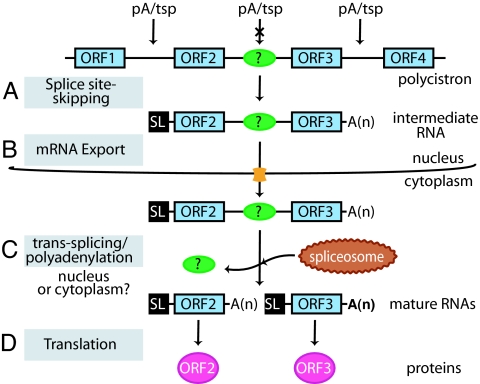
In summary, an intermediate RNA is formed in the nucleus as a consequence of splice site-specific skipping (Fig. 6A), and this is then exported to the cytoplasm, in some cases through a CRM1-dependent pathway (37) (Fig. 6B). This unspliced RNA is then processed into monocistrons by a trans-splicing/polyadenylation event (Fig. 6C) that gives rise to a functional mRNA ready for translation (Fig. 6D) or degradation. Some yet-unidentified signals/elements determine the precise moment of RNA processing, according to the requirements of the cell. Our understanding of the trans-splicing regulation requires elucidation of additional key molecular events involved in RNA metabolism. Future studies will be necessary to determine the location where the unspliced RNAs are stored and stabilized until they are further processed. In this context, we have recently found that poly(A)+ RNAs are stored within cytoplasmic granules together with several RBPs (A.C., J.G.D.G., and A.C.F., unpublished work). In any case, these results further support the idea that trypanosomatids are interesting model cells to analyze the use of peculiar posttranscriptional mechanisms regulating gene expression.
Fig. 6.
Model of intermediate RNA maturation in T. cruzi. Two steps of trans-splicing and polyadenylation processing generate functional monocistronic mRNAs (see Discussion). In some cases, the second monocistron might not be translated into protein, as is the case for SL-mTS-3′ small RNA. ORF1, ORF2, ORF3, and ORF4, ORFs; pA, polyadenylation site; tsp, trans-splicing site.
Methods
Culture conditions for T. cruzi CL-Brener cloned stock (38), parasite transfections parameters, and hybridization experiments were according to Di Noia et al. (19), with modifications described in SI Text. Oligonucleotide sequences used in this work are listed in SI Table 3. Gene cloning, RACE experiments, subcellular fractionation for RNA extraction, microscopic analysis, and bioinformatics were performed as described in SI Text. RNA extractions from ribonucleoprotein complexes were done as described (15). Further details are in SI Text.
Acknowledgments
We are indebted to Berta Franke de Cazzulo, Liliana Sferco, and Agustina Chidichimo for parasite cultures, and to Alan Frankel for critical reading of the manuscript. We thank J. M. Kelly for providing pTEX vector and R. Hernández for pRIBOTEX vector. We also thank to Henri van Luenen, Saara Vainio, and Piet Borst for helpful editorial comments. We apologize to people whose work could not be included in the reference list because of space limitations. This work was supported by National Institutes of Health Grant AI060645-01 and by a grant from the Agencia Nacional de Promoción Científica y Tecnológica (Argentina) (to A.C.F.). The research of A.C.F. was also supported in part by an International Research Scholars Grant from the Howard Hughes Medical Institute. J.G.D.G. is a research fellow and A.C.F. is a researcher from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina.
Abbreviations
- RBP
RNA-binding protein
- TcUBP
T. cruzi U-rich binding protein
- SL
spliced leader
- ActD
actinomycin D
- TS
trans-sialidase
- mTS
mammalian-stage TS
- iTS
insect-stage TS
- PTB
polypyrimidine tract-binding protein
- ICR
intercistronic region.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611125104/DC1.
References
- 1.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keene JD. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haile S, Estevez AM, Clayton C. RNA. 2003;9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton CE. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Orso I, De Gaudenzi JG, Frasch AC. Trends Parasitol. 2003;19:151–155. doi: 10.1016/s1471-4922(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 6.Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. Lancet. 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 7.Liang XH, Haritan A, Uliel S, Michaeli S. Eukaryot Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radwanska M, Couvreur B, Dumont N, Pays A, Vanhamme L, Pays E. Gene. 2000;255:43–50. doi: 10.1016/s0378-1119(00)00306-1. [DOI] [PubMed] [Google Scholar]
- 9.Van der Ploeg LH, Liu AY, Michels PA, De Lange T, Borst P, Majumder HK, Weber H, Veeneman GH, Van Boom J. Nucleic Acids Res. 1982;10:3591–3604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boothroyd JC, Cross GA. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 11.Borst P. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 12.D'Orso I, Frasch AC. J Biol Chem. 2001;276:34801–34809. doi: 10.1074/jbc.M102120200. [DOI] [PubMed] [Google Scholar]
- 13.De Gaudenzi J, Frasch AC, Clayton C. Eukaryot Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Orso I, Frasch AC. J Biol Chem. 2002;277:50520–50528. doi: 10.1074/jbc.M209092200. [DOI] [PubMed] [Google Scholar]
- 15.De Gaudenzi JG, D'Orso I, Frasch AC. J Biol Chem. 2003;278:18884–18894. doi: 10.1074/jbc.M301756200. [DOI] [PubMed] [Google Scholar]
- 16.Coughlin BC, Teixeira SM, Kirchhoff LV, Donelson JE. J Biol Chem. 2000;275:12051–12060. doi: 10.1074/jbc.275.16.12051. [DOI] [PubMed] [Google Scholar]
- 17.Irmer H, Clayton C. Nucleic Acids Res. 2001;29:4707–4715. doi: 10.1093/nar/29.22.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furger A, Schurch N, Kurath U, Roditi I. Mol Cell Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Noia JM, D'Orso I, Sanchez DO, Frasch AC. J Biol Chem. 2000;275:10218–10227. doi: 10.1074/jbc.275.14.10218. [DOI] [PubMed] [Google Scholar]
- 20.Frasch AC. Parasitol Today. 2000;16:282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 21.Cazzulo JJ, Frasch AC. FASEB J. 1992;6:3259–3264. [PubMed] [Google Scholar]
- 22.Nilsen TW. Trends Genet. 2001;17:678–680. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 23.Roberts TG, Sturm NR, Yee BK, Yu MC, Hartshorne T, Agabian N, Campbell DA. Mol Cell Biol. 1998;18:4409–4417. doi: 10.1128/mcb.18.8.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. J Biol Chem. 2001;276:44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 25.Spellman R, Smith CW. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Campbell DA, Thomas S, Sturm NR. Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Palenchar JB, Bellofatto V. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Blumenthal T. Brief Funct Genomic Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- 29.Wagner EJ, Garcia-Blanco MA. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimmapuram J, Duan H, Liu L, Schuler MA. RNA. 2005;11:128–138. doi: 10.1261/rna.7114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savard J, Marques-Souza H, Aranda M, Tautz D. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 32.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaili N, Goringer U, Benne R, Pays E. J Biol Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 33.Hastings KE. Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Ast G. Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 35.Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. Proc Natl Acad Sci USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuevas IC, Frasch AC, D'Orso I. Mol Biochem Parasitol. 2005;139:15–24. doi: 10.1016/j.molbiopara.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Zingales B, Pereira ME, Oliveira RP, Almeida KA, Umezawa ES, Souto RP, Vargas N, Cano MI, da Silveira JF, Nehme NS, et al. Acta Trop. 1997;68:159–173. doi: 10.1016/s0001-706x(97)00088-0. [DOI] [PubMed] [Google Scholar]