Abstract
Livestock populations are usually kept in groups. As a consequence, social interactions among individuals affect productivity, health, and welfare. Current selection methods (individual selection), however, ignore those interactions and yield suboptimal or in some cases even negative responses. In principle, selection between groups instead of individuals offers a solution, but has rarely been adopted in practice for two reasons. First, the relationship between group selection theory and common animal breeding concepts, such as the accuracy of selection, is unclear. Second, application of group selection requires keeping selection candidates in groups, which is often undesirable in practice. This work has two objectives. First, we derive expressions for the accuracy of individual and group selection, which provides a measurement of quality for those methods. Second, we investigate the opportunity to improve traits affected by interactions by using information on relatives kept in family groups, while keeping selection candidates individually. The accuracy of selection based on relatives is shown to be an analogy of the classical expression for traits not affected by interactions. Our results show that selection based on relatives offers good opportunities for effective genetic improvement of traits affected by interactions.
NEARLY all living organisms are affected by interactions among individuals (Wilson 1977; Griffing 1989; Moore 1990; Moore et al. 1997; Agrawal et al. 2001; Clutton-Brock 2002; Muir 2005). Such interactions may be due to competition for limited resources, such as daylight or soil nutrients, or due to social behaviors, such as aggression, social dominance, competitive ability, helping behavior, or interactions between mothers and their offspring (maternal effects). Those interactions have received a lot of attention in the field of evolutionary biology (e.g., Hamilton 1964; Frank 1998; Keller 1999; Clutton-Brock 2002), but are also of great importance in domestic populations of animals and plants (Muir 1996, 2005; Denison et al. 2003).
There is clear evidence that interactions among individuals may contribute to the heritable variation in traits (Wade 1976, 1977; Moore 1990; Muir 1996, 2005; Brichette et al. 2001; Wolf 2003; Bijma et al. 2007b). For example, Bijma et al. (2007b) found a total heritable variance for survival time in laying hens equal to 20% of the total phenotypic variance, of which two-thirds originated from interactions among individuals. Furthermore, selection experiments to reduce mortality due to cannibalism in domestic chicken (Muir 1996) and in flour beetle (Wade 1976, 1977) and to increase or decrease leaf area in cress (Goodnight 1985) have demonstrated that heritable interactions can contribute substantially to response to selection.
The inheritance of traits affected by interactions differs from that of classical traits, because trait values are determined in part by heritable effects that originate from other individuals (Moore et al. 1998). As a consequence, response to selection consists of two components (Willham 1963; Griffing 1967). The first component is the usual response in the direct effect of a genotype on the phenotype of the individual itself. The second component is the response in the effect of that genotype on phenotypes of other individuals. Following Griffing (1967), we refer to the effect of a genotype on phenotypes of other individuals as the associative effect of that genotype. With competition among individuals, selection methods that target only the direct effects of genotypes yield a negative correlated response in the associative effects and may yield a negative total response (Griffing 1967). For example, Wade (1976) showed that individual selection for increased population size of Tribolium decreased population size in the next generation. Similar results were also found in other studies (Craig 1982; Goodnight 1985). Genetic improvement of traits affected by interactions, therefore, requires selection methods that aim at both the direct effects of genotypes and at the associative effects of genotypes (Griffing 1967).
Despite the evidence for heritable interactions among individuals, selection methods currently used in livestock genetic improvement, such as mass selection or selection based on information from relatives, consider only the direct effects of genotypes (with the exception of maternal effects). Those methods are, therefore, inadequate for improving traits affected by interactions among individuals. Both theoretical and experimental work shows that selection between groups, where the group is the unit of selection, offers a solution, because group selection simultaneously improves direct and associative effects (Griffing 1967, 1976a; Maynard-Smith 1976; Moore et al. 1997; Wolf et al. 1998, 1999; Agrawal et al. 2001; Muir 2005; Bijma et al. 2007a). Group selection has, however, rarely been adopted in animal breeding practice, primarily for two reasons. First, the theoretical works on group selection have not been written using the usual expression for response to artificial selection, which is the product of intensity of selection, accuracy of selection, and the genetic standard deviation in the trait. This has caused group selection to be not fully understood and accepted in the field of animal breeding. Second, application of group selection requires that the selection candidates are kept in groups. Keeping selection candidates in groups, however, is often undesirable or difficult to apply in practice; first, because it interferes with recording data on an individual basis for important traits such as feed intake and, second, it may increase loss of selection candidates due to both infectious diseases and aggression.
This article has two objectives. First, we derive expressions for the accuracy of individual and group selection, which provides a measurement of quality for those methods. Second, we investigate the opportunity to improve traits affected by interactions by using information on relatives kept in family groups, while keeping selection candidates individually. Finally, we compare selection responses obtained with this strategy to responses obtained with existing strategies that are based on individual and group selection.
THEORY
In artificial breeding, the general expression for response to selection is
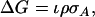 |
(1) |
in which ι is selection intensity, ρ is the correlation between the selection criterion and the breeding value for the trait of interest, usually referred to as the accuracy, and 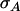 is the additive genetic standard deviation in the trait of interest (Falconer and Mackay 1996). The quality of selection methods is commonly measured by their accuracy, which is easy to interpret because it takes values between zero and one (Falconer and Mackay 1996; Kinghorn et al. 2000). Previous studies have yielded expressions for response to individual and group selection (Griffing 1967, 1976a; Wolf 2003; Bijma et al. 2007a). Those expressions were based on the covariance between the selection criterion and the breeding value, but did not distinguish between accuracy and genetic variance in the trait. Thus the accuracies of individual and group selection are unclear at present.
is the additive genetic standard deviation in the trait of interest (Falconer and Mackay 1996). The quality of selection methods is commonly measured by their accuracy, which is easy to interpret because it takes values between zero and one (Falconer and Mackay 1996; Kinghorn et al. 2000). Previous studies have yielded expressions for response to individual and group selection (Griffing 1967, 1976a; Wolf 2003; Bijma et al. 2007a). Those expressions were based on the covariance between the selection criterion and the breeding value, but did not distinguish between accuracy and genetic variance in the trait. Thus the accuracies of individual and group selection are unclear at present.
In the following, we reformulate existing equations for response to individual and group selection into components of Equation 1 and provide expressions for their accuracy. The results show that expressions for response to selection with interactions among individuals are a generalization of classical expressions for response in the absence of interactions. First, we briefly summarize the basic quantitative genetic theory of interactions among individuals presented in Griffing (1967), Wolf et al. (1998), Muir (2005), and Bijma et al. (2007a).
Model:
In the classical quantitative genetic model, the phenotype of an individual is the sum of its genetic merit or “breeding value” and a residual nonheritable effect (P = A + E). With interactions among individuals, the model needs to be extended to incorporate effects originating from other individuals. When interactions occur among n (number of animals per group) individuals, the phenotype of an individual can be modeled as the sum of its own direct phenotypic effect and the summed associative phenotypic effects of its 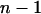 associates:
associates: 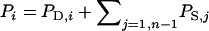 (Griffing 1967) (see Table 1 for notation). Thus, the phenotype of each individual consist of two terms, a direct effect (PD) originating from the genes and the physical environment of the individual itself and the sum of associative effects (PS) originating from each of its
(Griffing 1967) (see Table 1 for notation). Thus, the phenotype of each individual consist of two terms, a direct effect (PD) originating from the genes and the physical environment of the individual itself and the sum of associative effects (PS) originating from each of its 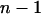 group members. Because each individual has both a direct and an associative effect, the model applies to each of the n individuals. Note that Pi is the observed phenotype, whereas PD and PS may be unobservable. Models used for maternal genetic effects, in which the phenotype of an offspring is the sum of an unobserved direct effect due to the offspring and an unobserved maternal effect due to its dam, can be seen as a specific case of this more general model (Willham 1963).
group members. Because each individual has both a direct and an associative effect, the model applies to each of the n individuals. Note that Pi is the observed phenotype, whereas PD and PS may be unobservable. Models used for maternal genetic effects, in which the phenotype of an offspring is the sum of an unobserved direct effect due to the offspring and an unobserved maternal effect due to its dam, can be seen as a specific case of this more general model (Willham 1963).
TABLE 1.
Notation key
| Symbol | Meaning |
|---|---|
| Pi | Observed trait value for individual i |
| ΔG | Selection response in observed trait value per generation |
| PD,i, PS,i | Phenotypic direct and associative effect |
| AD,i, AS,i | Direct and associative breeding value (DBV, SBV) |
| TBV | Total breeding value, TBVi = AD,i + (n − 1)AS,i |
| TPV | Total phenotypic value TPVi = PD,i + (n − 1)PS,i |
| n, m | Group size, no. of groups per selection candidate |
| r | Relatedness between selection candidate and its relatives |
| rbr | Relatedness between group members |
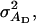 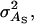 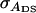
|
Direct genetic variance, associative genetic variance, covariance between direct genetic variance and associative genetic variance |
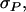 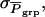 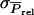
|
Standard deviation among phenotypic values of individuals, among average phenotypic values of groups, and among average phenotypic values of relatives in family groups |
| τ | Intraclass correlation among relatives adjusted for interactions, 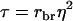
|
| η2 | Heritability adjusted for interactions, 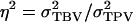
|
i, ρ, 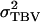
|
Selection intensity, accuracy of selection, variance of TBV |
Both the direct and the associative effect can be divided into an additive genetic (A) and a residual (E) component,
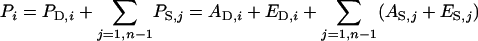 |
(2) |
(Muir 2005; Bijma et al. 2007a), where AD,i is the direct breeding value (DBV) of individual i, ED,i is the nonheritable direct effect of individual i, AS,j is the associative breeding value (SBV) of associate j, and ES,j is the nonheritable associative effect of associate j. Note that DBV and SBV are genetically distinct traits, even though they affect a single phenotype. For example, when interest is in growth rate, the DBV refers to the breeding value of an individual for its own growth rate, whereas the SBV refers to its heritable effect on growth rate of other individuals, which may, for example, be related to aggression or competition for feed. So, the DBV is equivalent to the classical breeding value (Lynch and Walsh 1998), whereas the SBV is a generalization of a breeding value for a maternal effect (Willham 1963).
Each individual expresses its DBV once in its own phenotype and its SBV 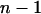 times in the phenotypes of its associates. The heritable contribution of a single individual to total performance of its group, referred to as its total breeding value (TBV), equals therefore TBVi = DBVi + (n − 1)SBVi (Bijma et al. 2007a). Response to selection
times in the phenotypes of its associates. The heritable contribution of a single individual to total performance of its group, referred to as its total breeding value (TBV), equals therefore TBVi = DBVi + (n − 1)SBVi (Bijma et al. 2007a). Response to selection 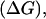 i.e., the genetic change of the mean trait value per generation, equals the per generation increase of the average TBV of the population. Analogous to Equation 1, response to selection can be expressed as
i.e., the genetic change of the mean trait value per generation, equals the per generation increase of the average TBV of the population. Analogous to Equation 1, response to selection can be expressed as
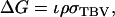 |
(3) |
in which ρ is the accuracy and 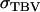 the standard deviation of total breeding values among individuals. In Equation 3, the accuracy is the correlation between the selection criterion and the TBVs of individuals. It measures the quality of selection methods for traits affected by interactions among individuals. The
the standard deviation of total breeding values among individuals. In Equation 3, the accuracy is the correlation between the selection criterion and the TBVs of individuals. It measures the quality of selection methods for traits affected by interactions among individuals. The 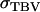 is the square root of the total heritable variation in the trait, which equals
is the square root of the total heritable variation in the trait, which equals
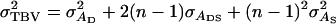 |
(4) |
(Bijma et al. 2007a), where 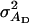 is the direct genetic variance,
is the direct genetic variance, 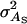 is the associative genetic variance, and
is the associative genetic variance, and 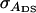 is the covariance between DBVs and SBVs of individuals.
is the covariance between DBVs and SBVs of individuals.
Accuracies of individual and group selection:
In this section we derive the accuracies of individual and group selection, so that response of these selection methods can be expressed as in Equation 3. Table 2 summarizes the selection methods and the corresponding equations for accuracy of selection.
TABLE 2.
Accuracies of selection methods
| Method | Accuracy | Range |
|---|---|---|
| Individual selection | 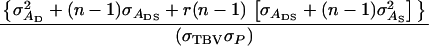 |
−1– +1 |
| Group selection | 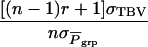 |
0–(0.707–1)a |
| Selection based on relatives |
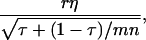 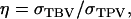 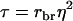
|
|
| Full sib | 0–0.707 | |
| Half sib | 0–0.5 | |
| Half-sib progeny | 0–1.0 |
Depending on group size.
Individual selection:
With individual selection, selection candidates are kept in groups. Individuals with a phenotypic value greater than a chosen threshold value are selected to become parents of the next generation, irrespective of the performance of their group members (Griffing 1960). Response to individual selection in a population consisting of groups of size n equals
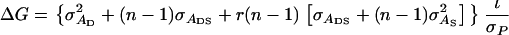 |
(5) |
(Bijma et al. 2007a), in which r denotes the additive genetic relatedness between group members, and 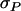 is the standard deviation among phenotypic trait values of individuals, where
is the standard deviation among phenotypic trait values of individuals, where 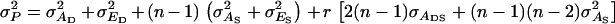 Combining Equations 3 and 5 shows that the accuracy of individual selection equals
Combining Equations 3 and 5 shows that the accuracy of individual selection equals
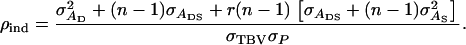 |
(6) |
When there are no interactions among individuals, so that 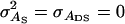 and
and 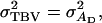 Equation 6 reduces to
Equation 6 reduces to 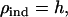 the square root of heritability, and Equation 3 to
the square root of heritability, and Equation 3 to 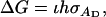 the usual expression for response to mass selection.
the usual expression for response to mass selection.
When there are no interactions, the accuracy is between zero and one (Falconer and Mackay 1996; Kinghorn et al. 2000). Investigation of Equation 6, however, shows that with interactions, the accuracy of individual selection can be negative, which would result in a negative response to selection. With unrelated group members (r = 0), for example, the latter occurs when 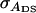 is negative and greater in absolute magnitude than
is negative and greater in absolute magnitude than 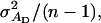 which depends on the genetic correlation between direct and associative effects (rA). When this correlation is negative, individuals with high DBVs have on average negative SBVs, i.e., a negative effect on the phenotypes of their group members. As a consequence, the use of individual selection can result in a negative response to selection (Griffing 1967). With full relatedness (r = 1), i.e., when interactions are among clones, the numerator of Equation 6 becomes equal to
which depends on the genetic correlation between direct and associative effects (rA). When this correlation is negative, individuals with high DBVs have on average negative SBVs, i.e., a negative effect on the phenotypes of their group members. As a consequence, the use of individual selection can result in a negative response to selection (Griffing 1967). With full relatedness (r = 1), i.e., when interactions are among clones, the numerator of Equation 6 becomes equal to 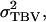 which is positive by definition. Thus relatedness among interacting individuals has the effect of making the correlation between phenotypes and TBVs of individuals a positive value.
which is positive by definition. Thus relatedness among interacting individuals has the effect of making the correlation between phenotypes and TBVs of individuals a positive value.
Group selection:
With group selection, individuals in groups with an average phenotypic value greater than a given value are selected to become parents of the next generation. Thus the entire group is either selected or rejected solely on the basis of the mean phenotypic value of the entire group, 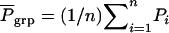 (Griffing 1967). Response to group selection is given by Bijma et al. (2007a),
(Griffing 1967). Response to group selection is given by Bijma et al. (2007a),
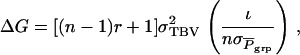 |
(7) |
in which 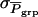 is the standard deviation among group means,
is the standard deviation among group means, 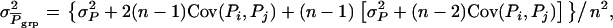 with
with 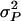 given above Equation 6, and
given above Equation 6, and 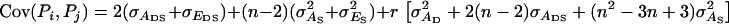 (see example in Bijma et al. 2007a). Combining Equations 3 and 7 shows that the accuracy of group selection equals
(see example in Bijma et al. 2007a). Combining Equations 3 and 7 shows that the accuracy of group selection equals
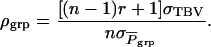 |
(8) |
Because both the numerator and the denominator of Equation 8 are positive, the accuracy and response to group selection are always positive. Furthermore, as indicated by the term 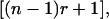 the accuracy of group selection is greater with family groups than with groups of unrelated individuals, which agrees with expressions for response to selection obtained by Griffing (1976a,b).
the accuracy of group selection is greater with family groups than with groups of unrelated individuals, which agrees with expressions for response to selection obtained by Griffing (1976a,b).
Selection based on relatives:
When selection candidates are housed individually, their phenotypes provide no information on their SBVs. In that case, information for selection methods aiming to improve the population average TBV needs to come from relatives of the candidate, which are kept in groups. These relatives of the selection candidates are assumed to be present at the same time as the selection candidates themselves. The phenotypic value of a relative, say j, consists of the direct effect of that relative and the summed associative effects of its group members, denoted by k: 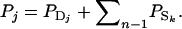 If the group members of the relative are unrelated to the candidate (rik = 0), then the phenotype of the relative provides information only on the direct effect of the candidate, not on its associative effect. This is because
If the group members of the relative are unrelated to the candidate (rik = 0), then the phenotype of the relative provides information only on the direct effect of the candidate, not on its associative effect. This is because 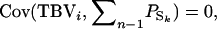 so that
so that 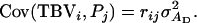 To capture the entire TBV of the selection candidate, relatedness between the candidate and the group members of its relatives needs to be equal to relatedness between the candidate and its relatives, rik = rij, so that
To capture the entire TBV of the selection candidate, relatedness between the candidate and the group members of its relatives needs to be equal to relatedness between the candidate and its relatives, rik = rij, so that 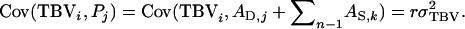 A situation with rik = rij is obtained by keeping relatives in family groups. For example, when selection is based on sib information, groups may consist of full sibs of the candidate, so that rik = rij = 0.5. In the following, therefore, we consider selection based on relatives of the candidate that are kept in family groups.
A situation with rik = rij is obtained by keeping relatives in family groups. For example, when selection is based on sib information, groups may consist of full sibs of the candidate, so that rik = rij = 0.5. In the following, therefore, we consider selection based on relatives of the candidate that are kept in family groups.
The accuracy of selection based on the mean phenotypic value of relatives kept in family groups can be expressed analogous to the situation with traits not affected by interactions (see appendix a for the derivation). In the absence of interactions, the accuracy of selection based on relatives is commonly formulated in terms of relatedness between the candidate and its relatives, r, the square root of heritability, h, and the intraclass correlation t between the relatives,
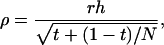 |
(9) |
in which t = rbrh2, the product of relatedness between the relatives and heritability, and N is the number of relatives (Falconer and Mackay 1996; Cameron 1997; Lynch and Walsh 1998). Hence, we distinguish between relatedness r between the candidate and its relatives and mutual relatedness rbr between the relatives. For example, for half-sib progeny of the candidate, we would have 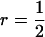 and
and 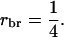 The analogy of Equation 9 for traits affected by interactions is
The analogy of Equation 9 for traits affected by interactions is
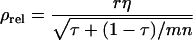 |
(10) |
(appendix a). In Equation 10, we used Greek symbols to denote heritability and intraclass correlation adapted to account for interactions: 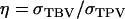 is an analogy of the square root of heritability,
is an analogy of the square root of heritability, 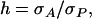
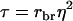 is an analogy of the intraclass correlation between relatives
is an analogy of the intraclass correlation between relatives 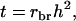 and mn is the number of relatives in m groups consisting of n individuals each. The η and τ account for interactions among individuals and, therefore, depend on the TBV and on the total phenotypic value (TPV) contributed by an individual. The TPV is an analogy of the TBV. The TPV of individual i represents the phenotypic effect on the population mean that originates from individual i, which equals its direct phenotypic value plus n − 1 its associative phenotypic value,
and mn is the number of relatives in m groups consisting of n individuals each. The η and τ account for interactions among individuals and, therefore, depend on the TBV and on the total phenotypic value (TPV) contributed by an individual. The TPV is an analogy of the TBV. The TPV of individual i represents the phenotypic effect on the population mean that originates from individual i, which equals its direct phenotypic value plus n − 1 its associative phenotypic value, 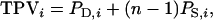 so that
so that 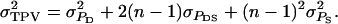 Note that TPVi differs from the observed phenotypic value of individual i,
Note that TPVi differs from the observed phenotypic value of individual i, 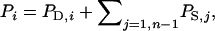 which contains associative effects of the group members j of i, whereas TPVi contains the associative effect of i itself. Thus, the TPV measures the total effect of an individual on performance of its group, the TBV is the heritable component of the TPV, and
which contains associative effects of the group members j of i, whereas TPVi contains the associative effect of i itself. Thus, the TPV measures the total effect of an individual on performance of its group, the TBV is the heritable component of the TPV, and 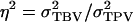 is the proportion of the variance of the TPV that is heritable, fully analogous to the classical heritability. The intraclass correlation τ equals the correlation between TPVs of relatives, analogous to the classical intraclass correlation t, which equals the correlation between phenotypes of relatives for traits not affected by interactions. In conclusion, Equation 10 shows that response to selection based on relatives kept in family groups can be obtained from the classical expression for response to selection based on relatives, when replacing heritability by
is the proportion of the variance of the TPV that is heritable, fully analogous to the classical heritability. The intraclass correlation τ equals the correlation between TPVs of relatives, analogous to the classical intraclass correlation t, which equals the correlation between phenotypes of relatives for traits not affected by interactions. In conclusion, Equation 10 shows that response to selection based on relatives kept in family groups can be obtained from the classical expression for response to selection based on relatives, when replacing heritability by 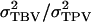 and the intraclass correlation between relatives by
and the intraclass correlation between relatives by 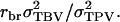 In the absence of interactions, η reduces to h, τ reduces to t, and
In the absence of interactions, η reduces to h, τ reduces to t, and 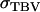 reduces to
reduces to 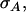 so that Equation 10 reduces to Equation 9.
so that Equation 10 reduces to Equation 9.
Investigation of the accuracy for large numbers of observations, i.e., for 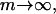 shows that the limiting accuracy equals
shows that the limiting accuracy equals 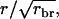 which is 0.5 for half-sib information, 0.707 for full-sib information, and 1 for half-sib progeny of the selection candidate. These results are the same as those for classical selection based on relatives in the absence of interactions. Hence, limiting accuracies for traits affected by interactions among individuals are the same as those for classical traits. For example, it is possible to obtain an accuracy approaching unity by using information on a large number of half-sib progeny kept in groups consisting of half sibs.
which is 0.5 for half-sib information, 0.707 for full-sib information, and 1 for half-sib progeny of the selection candidate. These results are the same as those for classical selection based on relatives in the absence of interactions. Hence, limiting accuracies for traits affected by interactions among individuals are the same as those for classical traits. For example, it is possible to obtain an accuracy approaching unity by using information on a large number of half-sib progeny kept in groups consisting of half sibs.
APPLICATION
So far, we have presented expressions for responses to selection and accuracies of individual selection, group selection, and selection based on relatives. In this section we numerically illustrate the accuracy for the three selection methods for a varying degree of competition. The magnitude of interactions was varied by varying the associative phenotypic variance 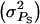 Large
Large 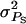 causes large differences in SBVs of individuals and thus reflects a situation with large interactions. The type of interactions was varied by varying the genetic correlation (rA) between direct and associative effects. Negative rA corresponds to competition among individuals, zero rA corresponds to neutral interactions, and positive rA corresponds to cooperation. For example, large
causes large differences in SBVs of individuals and thus reflects a situation with large interactions. The type of interactions was varied by varying the genetic correlation (rA) between direct and associative effects. Negative rA corresponds to competition among individuals, zero rA corresponds to neutral interactions, and positive rA corresponds to cooperation. For example, large 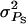 together with a strongly negative rA represents strong competition, whereas small
together with a strongly negative rA represents strong competition, whereas small 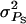 together with a slightly positive rA represents mild cooperation. Table 3 summarizes the default values of genetic parameters used. The default value of
together with a slightly positive rA represents mild cooperation. Table 3 summarizes the default values of genetic parameters used. The default value of 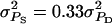 together with group size (n) of 4 implies that ∼50% of the total phenotypic variance is due to associative effects. Larger values of
together with group size (n) of 4 implies that ∼50% of the total phenotypic variance is due to associative effects. Larger values of 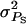 indicate strong interactions and vice versa. The following selection methods were compared: individual selection with groups consisting of either unrelated individuals (r = 0) or full sibs (
indicate strong interactions and vice versa. The following selection methods were compared: individual selection with groups consisting of either unrelated individuals (r = 0) or full sibs (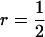 ), group selection with groups of full sibs (
), group selection with groups of full sibs (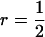 ), and selection based on relatives with groups of half sibs (
), and selection based on relatives with groups of half sibs (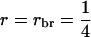 ), full sibs (
), full sibs (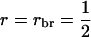 ), or half-sib progeny (
), or half-sib progeny (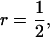
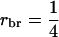 ).
).
TABLE 3.
Default values used to compare selection methods
| Parameter | Abbreviation | Default value |
|---|---|---|
| No. of animals per group | n | 4 |
| No. of groups per selection candidate | m | 1 |
| Heritability of direct effecta | 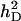 |
0.10 |
| Heritability of associative effectb | 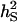 |
0.10 |
| Direct phenotypic variance | 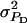 |
1 |
| Associative phenotypic variance | 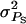 |
0.33 |
| Genetic, environmental, and phenotypic correlation between direct and associative effect | rA, rE, rP | 0 |
Obtained as 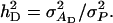
Obtained as 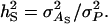
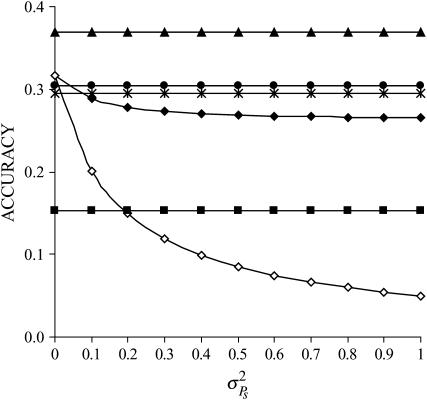
Figure 1 illustrates the impact of the magnitude of interactions (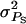 ) for the case where interactions are neutral (rA = 0) and for a single group of relatives (m = 1). The accuracy of group selection and selection based on relatives was not affected by the magnitude of interactions. This equivalence results from the fact that the heritabilities of direct and associative effects were equal and that the genetic and environmental correlations between direct and associative effects were equal also (Table 3). In all situations, group selection with groups of full sibs yielded the highest accuracy, even when associative effects were absent. This is because the heritability was low (0.10), so that information on relatives is more important than one's own information. When heritability was high (0.5), individual selection had higher accuracy in the absence of associative effects than selection based on groups of full sibs (results not shown). As expected, selection based on full sibs or progeny yielded higher accuracy than selection based on half sibs. Individual selection with groups of unrelated individuals performed well in the absence of interactions (
) for the case where interactions are neutral (rA = 0) and for a single group of relatives (m = 1). The accuracy of group selection and selection based on relatives was not affected by the magnitude of interactions. This equivalence results from the fact that the heritabilities of direct and associative effects were equal and that the genetic and environmental correlations between direct and associative effects were equal also (Table 3). In all situations, group selection with groups of full sibs yielded the highest accuracy, even when associative effects were absent. This is because the heritability was low (0.10), so that information on relatives is more important than one's own information. When heritability was high (0.5), individual selection had higher accuracy in the absence of associative effects than selection based on groups of full sibs (results not shown). As expected, selection based on full sibs or progeny yielded higher accuracy than selection based on half sibs. Individual selection with groups of unrelated individuals performed well in the absence of interactions (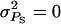 ), but poorly when the magnitude of interactions increased. Individual selection with groups of full sibs performed well over the entire range of
), but poorly when the magnitude of interactions increased. Individual selection with groups of full sibs performed well over the entire range of 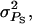 although somewhat less well than selection based on full sibs or progeny and group selection.
although somewhat less well than selection based on full sibs or progeny and group selection.
Figure 1.—
Accuracy of selection methods as a function of the associative phenotypic variance (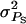 ) (n = 4; m = 1;
) (n = 4; m = 1; 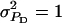 ;
; 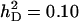 ;
; 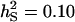 ; rA = rE = 0). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•).
; rA = rE = 0). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•).
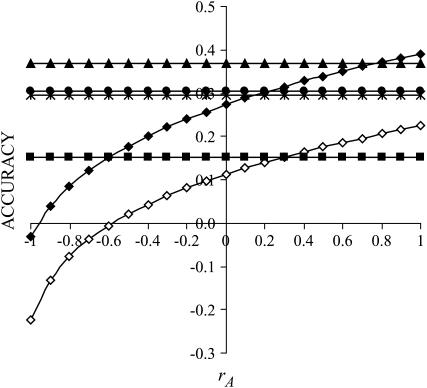
Figure 2 illustrates the impact of the type of interactions (rA) for the case where interactions contributed 50% of the phenotypic variance (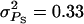 ) and for a single group of relatives (m = 1). The accuracy of group selection and selection based on relatives was not affected by rA and was always positive. The accuracy of individual selection was highest with strong cooperation, but decreased strongly when interactions became more competitive (lower rA) and became negative with strong competition. Together, Figures 1 and 2 illustrate that, in contrast to individual selection, group selection and selection based on relatives are robust against variation in the magnitude and type of interactions.
) and for a single group of relatives (m = 1). The accuracy of group selection and selection based on relatives was not affected by rA and was always positive. The accuracy of individual selection was highest with strong cooperation, but decreased strongly when interactions became more competitive (lower rA) and became negative with strong competition. Together, Figures 1 and 2 illustrate that, in contrast to individual selection, group selection and selection based on relatives are robust against variation in the magnitude and type of interactions.
Figure 2.—
Accuracy of selection methods as a function of the genetic correlation (rA) when rA = rE (n = 4; m = 1; 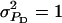 ;
; 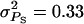 ;
; 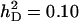 ;
; 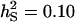 ). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•).
). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•).
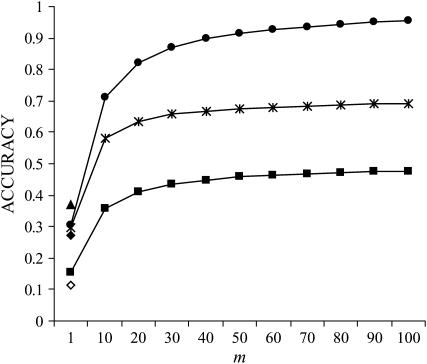
In contrast to group and individual selection, selection based on relatives offers the opportunity to use information on multiple groups of individuals. For example, males may be selected on the mean performance of 10 groups of offspring, instead of on the performance of a single group. Figure 3 illustrates the impact of the number of groups of relatives. The accuracy of selection based on relatives was independent of rA and 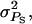 so that results in Figure 3 apply to any magnitude and type of interactions (as long as rA = rE and
so that results in Figure 3 apply to any magnitude and type of interactions (as long as rA = rE and 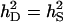 ). The accuracies of selection based on relatives increased substantially with the number of groups of relatives. The relationship between accuracy and the number of groups in Figure 3 was similar to that between accuracy and the number of relatives for classical traits not affected by interactions (Falconer and Mackay 1996). For comparison, Figure 3 also shows accuracies of individual and group selection for neutral interactions (rA = 0). Those accuracies were substantially smaller than values obtained with selection based on multiple groups of relatives.
). The accuracies of selection based on relatives increased substantially with the number of groups of relatives. The relationship between accuracy and the number of groups in Figure 3 was similar to that between accuracy and the number of relatives for classical traits not affected by interactions (Falconer and Mackay 1996). For comparison, Figure 3 also shows accuracies of individual and group selection for neutral interactions (rA = 0). Those accuracies were substantially smaller than values obtained with selection based on multiple groups of relatives.
Figure 3.—
Accuracy of selection methods as a function of the number of groups per selection candidate (m) (n = 4; 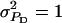 ;
; 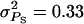 ;
; 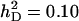 ;
; 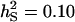 ; rA = rE = 0). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives, where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•). When m = 1, selection based on full sibs and selection based on half-sib offspring received the same symbol, because the accuracy is, respectively, 0.29 and 0.31.
; rA = rE = 0). The accuracy is shown for individual selection when the animals in a group are full sibs (♦) or unrelated (⋄); for group selection with groups of full sibs (▴); and for selection based on relatives, where relatives can be half sibs (▪), full sibs (Û), or half-sib offspring (•). When m = 1, selection based on full sibs and selection based on half-sib offspring received the same symbol, because the accuracy is, respectively, 0.29 and 0.31.
DISCUSSION
In this article, we derived expressions for the accuracy of individual and group selection and investigated opportunities for selection based on information from relatives kept in family groups. This work rests primarily on the foundational work of Willham (1963) and Griffing (1967, 1976a). Unfortunately, the work of Griffing (1967, 1976a) has had relatively little impact in the field of livestock genetic improvement, which is mainly due to the difficulty of derivations and the treatment of distinct situations as special cases. In contrast to most previous work on interactions among individuals (e.g., Griffing 1967, 1976a; Wade 1978; Wolf et al. 1998), our results are expressed in terms familiar to animal breeders, such as intensity of selection, accuracy of selection, and the standard deviation of (total) breeding values. We expect that this way of expressing results will stimulate the acceptance of quantitative genetic theory of interactions among individuals and its application in livestock genetic improvement. They also provide insight into the prospects for development of better selection strategies; i.e., low accuracies indicate substantial prospects for improvement of the selection strategy, whereas values near unity indicate little prospects for improvement.
Our results show that selection based on relatives kept in family groups acts directly on the TBVs of selection candidates and always yields a positive response to selection. Selection based on relatives kept in family groups can be interpreted as an analogy of selection based on relatives for classical traits, on the condition that the definition of heritability and intraclass correlation between relatives are extended to account for interactions. This analogy suggests that, analogous to classical traits, selection on the mean performance of relatives kept in family groups is the optimum way to use information from relatives for traits affected by interactions. This hypothesis is supported by the fact that asymptotic accuracies obtained with large numbers of relatives are identical to values for classical traits not affected by interactions.
The added value of selection based on relatives compared to individual or group selection depends on the genetic parameters of the trait, the consequences for parameters of the breeding scheme (intensity of selection and generation interval), and costs involved in keeping different numbers of animals. The advantage is largest with strong competition and low heritability. With cooperation (rA > 0), individual selection yields a positive response, so that group selection or selection based on relatives is not required to ensure a positive response (Griffing 1967). However, when multiple groups of relatives can be used, selection based on relatives yields substantially higher accuracy, particularly when heritability is low. Group selection is robust in the sense that it always yields a positive response (Griffing 1976a), but requires keeping selection candidates in groups, which is often undesirable. Moreover, group selection cannot be used for traits that require killing the individuals providing the information, such as meat percentage in chickens or pigs, because it would require killing the selection candidate.
At present there is very little information on the genetic parameters underlying traits affected by interactions ( ; Wolf 2003; Muir 2005; Bijma et al. 2007b). The selection method developed here can be implemented without knowledge of the genetic parameters in that population, but the efficiency will depend on the parameters used. Genetic parameters of traits affected by interactions can be estimated from livestock populations, but the amount of data required to accurately estimate those parameters is substantially larger than that for classical traits (Bijma et al. 2007b). For example, Bijma et al. (2007b) used information on ∼3800 individuals with a full-sib–half-sib pedigree structure and groups composed of unrelated individuals and obtained an estimated genetic correlation between direct and associative effects of +0.28. This value did, however, not differ significantly from zero. With knowledge of the genetic parameters, individuals could be selected on the basis of an index of group and individual performance (Griffing 1969). Furthermore, when information is available on the genetic parameters, the use of best linear unbiased predictions (BLUP) of breeding values with an animal model allows combining information from different types of relatives into a single estimate of the breeding value (Henderson 1984; Muir 2005; Bijma et al. 2007b). The accuracy of that breeding value will exceed the accuracy obtained with selection based on relatives. When accurate information on genetic parameters is not available, however, selection based on relatives kept in family groups provides a robust solution that is relatively easy to implement.
; Wolf 2003; Muir 2005; Bijma et al. 2007b). The selection method developed here can be implemented without knowledge of the genetic parameters in that population, but the efficiency will depend on the parameters used. Genetic parameters of traits affected by interactions can be estimated from livestock populations, but the amount of data required to accurately estimate those parameters is substantially larger than that for classical traits (Bijma et al. 2007b). For example, Bijma et al. (2007b) used information on ∼3800 individuals with a full-sib–half-sib pedigree structure and groups composed of unrelated individuals and obtained an estimated genetic correlation between direct and associative effects of +0.28. This value did, however, not differ significantly from zero. With knowledge of the genetic parameters, individuals could be selected on the basis of an index of group and individual performance (Griffing 1969). Furthermore, when information is available on the genetic parameters, the use of best linear unbiased predictions (BLUP) of breeding values with an animal model allows combining information from different types of relatives into a single estimate of the breeding value (Henderson 1984; Muir 2005; Bijma et al. 2007b). The accuracy of that breeding value will exceed the accuracy obtained with selection based on relatives. When accurate information on genetic parameters is not available, however, selection based on relatives kept in family groups provides a robust solution that is relatively easy to implement.
An example of a trait affected by interactions among individuals is survival in laying hens. Survival in laying hens is affected by cannibalistic interactions among cage members, which has made it difficult to improve survival using conventional selection methods (Muir 1996). Bijma et al. (2007b) estimated variance components for survival time in a commercial line of laying hens with intact beaks. In this line, mean survival till 80 weeks of age was 54%, with a mean survival time of 454 days (SD 122 days). The estimated genetic parameters were 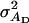 = 960 days2,
= 960 days2, 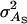 = 132 days2, rA = 0.28,
= 132 days2, rA = 0.28, 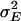 = 12,369 days2, n = 4, and
= 12,369 days2, n = 4, and 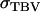 = 52.4 days. With a selection intensity of unity (ι = 1), predicted responses for this population are 10.7 days for individual selection with groups of unrelated individuals, 16.6 days for individual selection with groups of full sibs, and 22.8 days for group selection with groups of full sibs. With selection based on a single group of relatives, predicted responses are 9.6 days for half sibs, 18.3 days for full sibs, and 19.2 days for half-sib progeny. With selection based on 10 groups of relatives, predicted responses are 20.4 days for half sibs, 32.4 days for full sibs, and 40.8 days for half-sib progeny. These results show that, even with moderate cooperation, selection based on information from relatives kept in family groups enables substantial response to selection.
= 52.4 days. With a selection intensity of unity (ι = 1), predicted responses for this population are 10.7 days for individual selection with groups of unrelated individuals, 16.6 days for individual selection with groups of full sibs, and 22.8 days for group selection with groups of full sibs. With selection based on a single group of relatives, predicted responses are 9.6 days for half sibs, 18.3 days for full sibs, and 19.2 days for half-sib progeny. With selection based on 10 groups of relatives, predicted responses are 20.4 days for half sibs, 32.4 days for full sibs, and 40.8 days for half-sib progeny. These results show that, even with moderate cooperation, selection based on information from relatives kept in family groups enables substantial response to selection.
The responses to selection mentioned above refer to the total genetic improvement due to the combination of direct and associative effects. There may also be interest, however, in the response to selection for the direct and the associative effect separately. For group selection, responses for direct and associative effects follow from results presented in Bijma et al. (2007a). For selection based on relatives response in direct effects equals 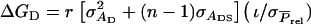 and response in associative effects equals
and response in associative effects equals 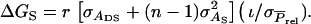 Using the estimated genetic parameters of Bijma et al. (2007b), selection based on half sibs yielded a response for the direct effect of 4.4 days and a response for the associative effect of 1.7 days, that based on full sibs yielded, respectively, 8.4 days and 3.3 days, and that based on half-sib offspring yielded responses of, respectively, 8.8 days and 3.5 days. When judging these results, it is important to realize that the contribution of the associative effects to the total response equals the response in associative effects multiplied by (n − 1).
Using the estimated genetic parameters of Bijma et al. (2007b), selection based on half sibs yielded a response for the direct effect of 4.4 days and a response for the associative effect of 1.7 days, that based on full sibs yielded, respectively, 8.4 days and 3.3 days, and that based on half-sib offspring yielded responses of, respectively, 8.8 days and 3.5 days. When judging these results, it is important to realize that the contribution of the associative effects to the total response equals the response in associative effects multiplied by (n − 1).
Commercial conditions:
For all three selection methods we have to keep in mind that the housing conditions of the relatives kept in groups (selection based on relatives) or the selection candidates (individual and group selection) should accurately reflect the commercial conditions under which the animals will be reared by the farmer. Especially group size can have large effects on the impact of social interactions and thus on response to selection. Furthermore, in this article only homogeneous groups of the same type of relatives have been assumed, e.g., only full sibs or only half sibs in one group. Under commercial conditions, however, groups can consist of a mix of full sibs, half sibs, and unrelated individuals. For both group selection and selection based on relatives it is possible to use an average r and rbr to estimate the accuracy and response to selection.
Index selection:
When the genetic parameters of direct and associative effects are known, selection of individuals using a selection index is an alternative to selection between groups or based on relatives. With interactions among individuals, the goal parameter of such an index, usually referred to as the “aggregate genotype,” would be the TBV of an individual. In matrix notation, the TBV of individual i is given by 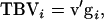 in which
in which 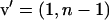 and
and 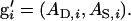 The direct effect of an individual is expressed in its own phenotype, whereas its associative effect is expressed in its group members. Hence, a simple index aiming to maximize response by simultaneous improvement of direct and associative effects can be composed of the phenotype of the individual itself, Pi, and the average phenotype of its n − 1 group members,
The direct effect of an individual is expressed in its own phenotype, whereas its associative effect is expressed in its group members. Hence, a simple index aiming to maximize response by simultaneous improvement of direct and associative effects can be composed of the phenotype of the individual itself, Pi, and the average phenotype of its n − 1 group members, 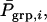
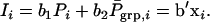 |
(11) |
It follows from selection index theory that optimum index weights are b = P−1Gv, where P is the 2 × 2 (co)variance matrix of information sources in xi, and G is the 2 × 2 matrix of covariances between information sources in xi and true breeding values for direct and associative effects in g (Hazel 1943). Elements of P and G are 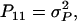
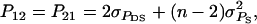
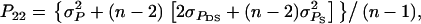
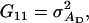
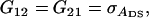 and
and 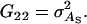 From selection index theory, response to selection equals
From selection index theory, response to selection equals
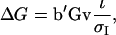 |
(12) |
in which 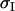 is the standard deviation of the index,
is the standard deviation of the index, 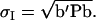
To compare response with index selection to other selection methods, we calculated response for the genetic parameters of Bijma et al. (2007b) given above. Predicted response from Equation 12 was 11.7 days. This is only a little more than the 10.7 days of response from individual selection with groups of unrelated individuals, but substantially less than the 16.6 days of response for individual selection with groups of full sibs. Thus optimum index selection using groups of unrelated individuals performs worse than individual selection using groups of relatives. This result indicates that using groups composed of relatives contributes more to the accuracy than optimizing index weights. The index calculations can be extended to apply to groups composed of relatives, but the derivations become complex in that case. Selection based on groups composed of relatives, in contrast, is simple and robust. It does not require knowledge of the genetic parameters, and its accuracy does not depend on the real values of the genetic parameters (Figures 1 and 2).
Acknowledgments
E.E. thanks Han Mulder for stimulating discussions during the development of this work. Johan van Arendonk and Roel Veerkamp are acknowledged for helpful comments on earlier versions of the manuscript. This research is part of a joint project of Hendrix Genetics, Nutreco, and Wageningen University on “Genetics of robustness in laying hens,” which is financially supported by SenterNovem.
APPENDIX A: DERIVATION OF EQUATION 10
With selection based on relatives kept in family groups, the selection criterion for individual i equals the mean phenotypic value of its relatives, 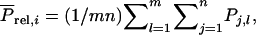 in which
in which 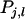 is the phenotypic trait value of relative j in group l, and m is the number of family groups of selection candidate i, each consisting of n individuals. The
is the phenotypic trait value of relative j in group l, and m is the number of family groups of selection candidate i, each consisting of n individuals. The 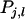 has components as indicated by Equation 2:
has components as indicated by Equation 2: 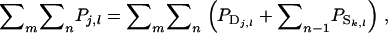 in which k denotes group members of j. Response to selection is obtained as the regression coefficient of the TBV of the selection candidates on the selection criterion,
in which k denotes group members of j. Response to selection is obtained as the regression coefficient of the TBV of the selection candidates on the selection criterion, 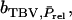 multiplied by the selection differential S. Thus, response to selection equals
multiplied by the selection differential S. Thus, response to selection equals 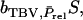 with
with 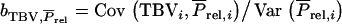 and
and 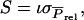 so that
so that 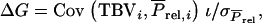 in which
in which 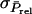 is the standard deviation of the selection criterion. The term
is the standard deviation of the selection criterion. The term 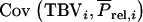 can be split into mn covariances, giving
can be split into mn covariances, giving 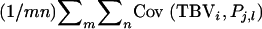 =
= 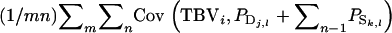 =
= 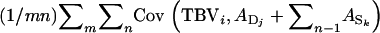 When all relatives have the same relatedness with the selection candidate, i.e.,
When all relatives have the same relatedness with the selection candidate, i.e., 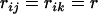 for all i, j, and k, then all covariances have the same value, giving
for all i, j, and k, then all covariances have the same value, giving 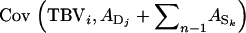 =
= 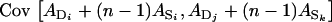 =
= 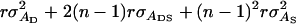 =
= 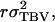 in which r denotes additive genetic relatedness between the selection candidate and its relatives kept in family groups. Response to selection based on relatives equals, therefore,
in which r denotes additive genetic relatedness between the selection candidate and its relatives kept in family groups. Response to selection based on relatives equals, therefore,
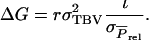 |
(A1) |
Combining Equations 1 and A1 shows that the accuracy of selection based on relatives kept in family groups equals
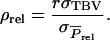 |
(A2) |
Although Equation A2 provides an expression for the accuracy, it is not easy to use, because it depends on 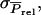 which is not a common genetic parameter. In the following, therefore, we reformulate Equation A2 in terms of Equation 9, starting with the derivation of
which is not a common genetic parameter. In the following, therefore, we reformulate Equation A2 in terms of Equation 9, starting with the derivation of 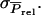 To derive
To derive 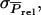 it is convenient to split
it is convenient to split 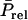 into a component that is common to all relatives of the selection candidate, plus a remaining term that no longer contains genetic relationships between individuals. For example, when considering half sibs, the common term would be half the TBV of the sire, so that
into a component that is common to all relatives of the selection candidate, plus a remaining term that no longer contains genetic relationships between individuals. For example, when considering half sibs, the common term would be half the TBV of the sire, so that 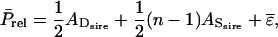 in which
in which 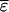 is the remainder of
is the remainder of 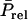 after subtracting
after subtracting 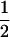 TBVsire. The ɛ is specific to each individual. In general, the common term equals
TBVsire. The ɛ is specific to each individual. In general, the common term equals 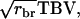 in which rbr is mutual relatedness between the relatives. In the derivation of
in which rbr is mutual relatedness between the relatives. In the derivation of 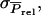 distinguishing between
distinguishing between 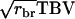 and
and 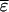 is convenient, because we do not have to consider averaging for the
is convenient, because we do not have to consider averaging for the 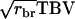 term, we can ignore relatedness among individuals for
term, we can ignore relatedness among individuals for 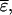 and both terms are mutually independent (when mating is at random). Thus
and both terms are mutually independent (when mating is at random). Thus 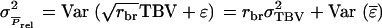
For the derivation of 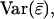 it is convenient to group the direct and associative components in
it is convenient to group the direct and associative components in 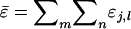 according to the individual from which they originate, instead of according to the individual in whose phenotype they are expressed. Each individual expresses its direct effect once and its associative effect n − 1 times. Therefore, we can write that
according to the individual from which they originate, instead of according to the individual in whose phenotype they are expressed. Each individual expresses its direct effect once and its associative effect n − 1 times. Therefore, we can write that 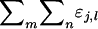 =
= 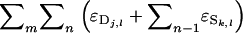 =
= 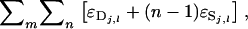 in which
in which 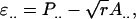 indicating the remainder when subtracting
indicating the remainder when subtracting 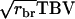 from
from 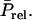 In the last summation, direct and associative effects are grouped according to the individual j from which they originate. Next,
In the last summation, direct and associative effects are grouped according to the individual j from which they originate. Next, 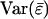 =
= 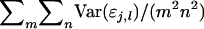 =
= 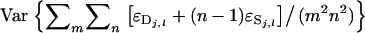 =
= 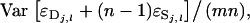 because the
because the 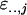 are independent after subtraction of the common part. Finally, using
are independent after subtraction of the common part. Finally, using 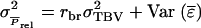 shows that
shows that
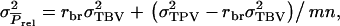 |
(A3) |
in which TPV denotes the total phenotypic value of an individual, 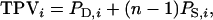 and
and 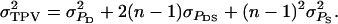 The TPV is an analogy of the TBV (see main text). In Equation A3, the term
The TPV is an analogy of the TBV (see main text). In Equation A3, the term 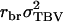 is the variance of the part of
is the variance of the part of 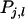 that is common to all relatives, which is not averaged, and the term
that is common to all relatives, which is not averaged, and the term 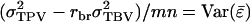 is the variance of the average value of the mn parts of
is the variance of the average value of the mn parts of 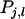 that are specific to each relative. Combining Equations A2 and A3 gives an expression for the accuracy of selection. First, we rewrite Equation A3 as
that are specific to each relative. Combining Equations A2 and A3 gives an expression for the accuracy of selection. First, we rewrite Equation A3 as 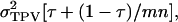 in which
in which 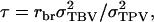 which is the covariance between TPVs of relatives expressed as a proportion of the variance of TPVs, so that τ is a so-called intraclass correlation among relatives adjusted to account for interactions among individuals. Substituting this result into Equation A2 gives Equation 10.
which is the covariance between TPVs of relatives expressed as a proportion of the variance of TPVs, so that τ is a so-called intraclass correlation among relatives adjusted to account for interactions among individuals. Substituting this result into Equation A2 gives Equation 10.
APPENDIX B: EXAMPLE OF CALCULATION
This example illustrates the calculation of accuracy and response to selection for selection based on information of a single group of half-sib offspring (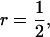
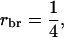 and m = 1). Estimated genetic parameters for survival time in a commercial line of laying hens are taken from Bijma et al. (2007b) and given in Table B1. The accuracy follows from Equation 10,
and m = 1). Estimated genetic parameters for survival time in a commercial line of laying hens are taken from Bijma et al. (2007b) and given in Table B1. The accuracy follows from Equation 10, 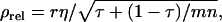 in which
in which 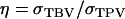 and
and 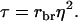 Thus, calculation of the accuracy requires calculating the variance of total breeding values,
Thus, calculation of the accuracy requires calculating the variance of total breeding values, 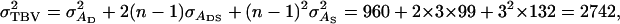 and of the variance of total phenotypic values,
and of the variance of total phenotypic values, 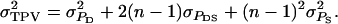 The statistical data analyses, however, do not provide estimates of the phenotypic variances of direct and associative effects, meaning that we cannot calculate
The statistical data analyses, however, do not provide estimates of the phenotypic variances of direct and associative effects, meaning that we cannot calculate 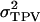 directly. For this reason,
directly. For this reason, 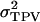 is calculated as the sum of
is calculated as the sum of 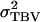 and the variance of the total environmental values of individuals,
and the variance of the total environmental values of individuals, 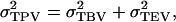 in which
in which 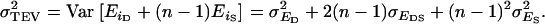 The data analyses provide an estimate of the residual variance,
The data analyses provide an estimate of the residual variance, 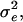 and an estimate of the correlation between residuals of group members, denoted ρ in Bijma et al. (2007b).
and an estimate of the correlation between residuals of group members, denoted ρ in Bijma et al. (2007b). 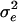 and ρ can be combined into an estimate of
and ρ can be combined into an estimate of 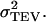 Bijma et al. (2007b) showed that
Bijma et al. (2007b) showed that 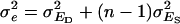 and
and 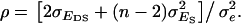 Therefore, it follows that
Therefore, it follows that 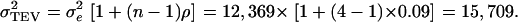 Next,
Next, 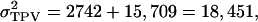 so that
so that 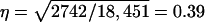 and
and 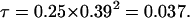 Thus the accuracy of selection based on a single group of half-sib offspring equals
Thus the accuracy of selection based on a single group of half-sib offspring equals 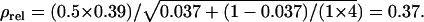 When selection is by truncation with a selection intensity of unity (ι = 1) (Falconer and Mackay 1996), response to selection is
When selection is by truncation with a selection intensity of unity (ι = 1) (Falconer and Mackay 1996), response to selection is 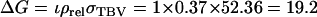 days.
days.
TABLE B1.
Genetic parameters for the example
| Parameter | Estimate |
|---|---|
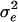 |
12,369 |
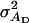 |
960 |
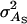 |
132 |
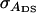 |
99 |
| ρ | 0.09 |
References
- Agrawal, A. F., E. D. Brodie III and M. J. Wade, 2001. On indirect genetic effects in structured populations. Am. Nat. 158: 308–323. [DOI] [PubMed] [Google Scholar]
- Bijma, P., W. M. Muir and J. A. M. Van Arendonk, 2007. a Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics 175: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P., W. M. Muir, E. D. Ellen, J. B. Wolf and J. A. M. Van Arendonk, 2007. b Multilevel selection 2: estimating the genetic parameters determining inheritance and response to selection. Genetics 175: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichette, I., M. I. Reyero and C. García, 2001. A genetic analysis of intraspecific competition for growth in mussel cultures. Aquaculture 192: 155–169. [Google Scholar]
- Cameron, N. D., 1997. Selection Indices and Prediction of Genetic Merit in Animal Breeding. CAB International, Oxon, UK.
- Clutton-Brock, T., 2002. Breeding together: kin selection and mutualism in cooperative vertebrates. Science 296: 69–72. [DOI] [PubMed] [Google Scholar]
- Craig, D. M., 1982. Group selection versus individual selection: an experimental analysis. Evolution 36: 271–282. [DOI] [PubMed] [Google Scholar]
- Denison, R. F., E. T. Kiers and S. A. West, 2003. Darwinian agriculture: When can humans find solutions beyond the reach of natural selection? Q. Rev. Biol. 78: 145–168. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Pearson Education, Harlow, England.
- Frank, S. A., 1998. Foundations of Social Evolution. Princeton University Press, Princeton, NJ.
- Goodnight, C. J., 1985. The influence of environmental variation on group and individual selection in a cress. Evolution 39: 545–558. [DOI] [PubMed] [Google Scholar]
- Griffing, B., 1960. Theoretical consequences of truncation selection based on the individual phenotype. Aust. J. Biol. Sci. 13: 307–343. [Google Scholar]
- Griffing, B., 1967. Selection in reference to biological groups I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 20: 127–139. [PubMed] [Google Scholar]
- Griffing, B., 1969. Selection in reference to biological groups IV. Application of selection index theory. Aust. J. Biol. Sci. 22: 131–142. [PubMed] [Google Scholar]
- Griffing, B., 1976. a Selection in reference to biological groups. V. Analysis of full-sib groups. Genetics 82: 703–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing, B., 1976. b Selection in reference to biological groups. VI. Use of extreme forms of nonrandom groups to increase selection efficiency. Genetics 82: 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing, B., 1989. Genetic analysis of plant mixtures. Genetics 122: 943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D., 1964. The genetical evolution of social behaviour I. J. Theor. Biol. 7: 1–16. [DOI] [PubMed] [Google Scholar]
- Hazel, L. N., 1943. The genetic basis for constructing selection indexes. Genetics 28: 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, C. R., 1984. Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, ON, Canada.
- Keller, L., 1999. Levels of Selection in Evolution. Princeton University Press, Princeton, NJ.
- Kinghorn, B., J. Van Der Werf and M. Ryan, 2000. Animal Breeding, Use of New Technologies. The Post Graduate Foundation in Veterinarian Science of the University of Sydney, Sydney.
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Maynard-Smith, J., 1976. Group selection. Q. Rev. Biol. 51: 277–283. [Google Scholar]
- Moore, A. J., 1990. The inheritance of social dominance, mating behaviour and attractiveness to mates in male Nauphoeta cinerea. Anim. Behav. 39: 388–397. [Google Scholar]
- Moore, A. J., E. D. Brodie III and J. B. Wolf, 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51: 1352–1362. [DOI] [PubMed] [Google Scholar]
- Moore, A. J., J. B. Wolf and E. D. Brodie III, 1998. The influence of direct and indirect genetic effects on the evolution of behavior: social and sexual selection meet maternal effects, pp. 22–41 in Maternal Effects as Adaptations, edited by T. A. Mousseau and C. W. Fox. Oxford University Press, Oxford.
- Muir, W. M., 1996. Group selection for adaptation to multiple-hen cages: selection program and direct responses. Poult. Sci. 75: 447–458. [DOI] [PubMed] [Google Scholar]
- Muir, W. M., 2005. Incorporation of competitive effects in forest tree or animal breeding programs. Genetics 170: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J., 1976. Group selection among laboratory populations of Tribolium. Proc. Natl. Acad. Sci. USA 73: 4604–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J., 1977. An experimental study of group selection. Evolution 31: 134–153. [DOI] [PubMed] [Google Scholar]
- Wade, M. J., 1978. Kin selection: a classical approach and a general solution. Proc. Natl. Acad. Sci. USA 75: 6154–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willham, R. L., 1963. The covariance between relatives for characters composed of components contributed by related individuals. Biometrics 19: 18–27. [Google Scholar]
- Wilson, D. S., 1977. Structured demes and the evolution of group advantageous traits. Am. Nat. 111: 157–185. [Google Scholar]
- Wolf, J. B., 2003. Genetic architecture and evolutionary constraint when the environment contains genes. Proc. Natl. Acad. Sci. USA 100: 4655–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B., E. D. Brodie III, J. M. Cheverud, A. J. Moore and M. J. Wade, 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13: 64–69. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B., E. D. Brodie III and A. J. Moore, 1999. Interacting phenotypes and the evolutionary process. II. Selection resulting from social interactions. Am. Nat. 153: 254–266. [DOI] [PubMed] [Google Scholar]