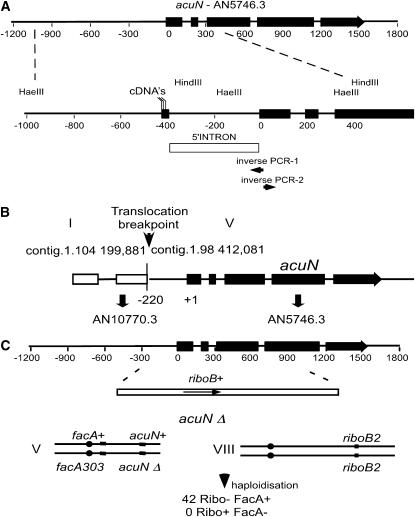
Figure 2.—
Characterization of the acuN356 mutation. (A) Schematic of the acuN gene corresponding to AN5746.3 in the A. nidulans database. The positions of exons are shown as thick lines and the coordinates are relative to the position of the ATG start codon. An expanded view of the 5′-end of the gene shows the position of the HindIII and HaeIII restriction sites used to map the acuN356 translocation breakpoint by Southern blotting of genomic DNA. The position of the intron in the 5′ untranslated region is shown (open box) and the position of 5′ endpoints of sequenced cDNAs is indicated, as are the positions of primers (inverse PCR-1 and -2) used for cloning the acuN356 translocation region. (B) Schematic of the acuN356 translocation breakpoint showing the contig coordinates on chromosomes 1 and 5 and the position of the breakpoint relative to the genes AN10770.3 and AN5746.3 (acuN). (C) Deletion of the acuN gene by replacement with the A. nidulans riboB gene in a riboB2 homozygous diploid strain. The relevant genotypes on linkage groups V and VIII of the resulting diploid are shown and phenotypes of haploids are recovered by haploidizing this diploid. All haploids have the riboB2 mutation (linkage group VIII) but none were phenotypically Ribo+ and all were FacA+, indicating that no haploids with the acuNΔ were recovered.