Abstract
Functional diversification of duplicated genes can contribute to the emergence of new organ morphologies. Model eudicot plants like Arabidopsis thaliana and Antirrhinum majus have a single PI/GLO gene that together with AP3/DEF regulate petal and stamen formation. Lodicules of grass flowers are morphologically distinct reduced organs occupying the position of petals in other flowers. They serve a distinct function in partial and transient flower opening to allow stamen emergence and cross-pollination. Grasses have duplicated PI/GLO-like genes and in rice (Oryza sativa) one these genes, OsMADS2, controls lodicule formation without affecting stamen development. In this study, we investigate the mechanistic roles played by OsMADS2. We ascribe a function for OsMADS2 in controlling cell division and differentiation along the proximal–distal axis. OsMADS2 is required to trigger parenchymatous and lodicule-specific vascular development while maintaining a small organ size. Our data implicate the developmentally late spatially restricted accumulation of OsMADS2 transcripts in the differentiating lodicule to control growth of these regions. The global architecture of transcripts regulated by OsMADS2 gives insights into the regulation of cell division and vascular differentiation that together can form this highly modified grass organ with important functions in floret opening and stamen emergence independent of the paralogous gene OsMADS4.
“B-FUNCTION” floral-organ-patterning activity requires a pair of related genes in model laboratory plant species. These are APETALA3 (AP3) and PISTILLATA (PI) in Arabidopsis thaliana and DEFICIENS (DEF) and GLOBOSA (GLO) in Antirrhinum majus. In these species, uniform and continued expression of these genes is required for development of petals as dorsiventrally flattened sterile organs (Bowman et al. 1989; Zachgo et al. 1995). Petunia has two AP3/DEF- and two PI/GLO-like genes. Mutational analysis of one of the AP3/DEF-like genes, PhDEF/GREEN PETAL (GP), showed its essential role in petal development alone despite its broader expression in both petal and stamen primordia (Angenent et al. 1992; van der Krol et al. 1993). The other gene, a paleo-AP3-like gene, PhTM6, has functionally diverged from GP and is required only for determining the stamen identity (Rijpkema et al. 2006). Similarly, among the duplicated tomato AP3-like genes, TAP3 has diverged from TM6 to acquire petal-specific functions (de Martino et al. 2006). Functional diversification of duplicated orchid (Dendrobium crumenetum) AP3/DEF genes has also been shown recently by their effects upon overexpression in Arabidopsis and by studying their ability to complement ap3 mutants (Xu et al. 2006). In contrast, the duplicated petunia PI genes, PhGLO1 and PhGLO, have similar expression profiles and redundant activities (Vandenbussche et al. 2004) particularly with respect to petal development.
Studies on B-function gene expression domains in divergent monocots have been informative in understanding floral organ homologies among angiosperms. Florets in grasses, a family of monocot plants, are reduced structures with highly derived organs. Immediately peripheral to the stamens are specialized floret organs called lodicules, external to which are two modified bract-like organs, the lemma and the palea, that completely enclose all internal organs (Schmidt and Ambrose 1998). Lodicules of rice florets are asymmetrically positioned on the second-whorl primordium. The absence of a whorl of lodicules creates the appearance that the palea and lodicules arise from a single whorl. The lodicules are small, fleshy, cup-shaped nongreen organs with a broad base and a narrow apex. Rice florets, as in other grasses, are mostly closed and open in a regulated manner for just 1–2 hr to allow anther emergence. This process requires rapid swelling and shrinking of the lodicules. Grass species have a single AP3/DEF ortholog but have duplicated PI/GLO-like genes (Munster et al. 2001). The rice SPW1 and the maize SILKY1 genes, i.e., the AP3/DEF orthologs, are expressed in lodicules and stamens and regulate the identity of these organs (Ambrose et al. 2000; Nagasawa et al. 2003). Preliminary RNA expression studies of one of these rice PI/GLO paralogs, OsMADS2, localized its expression to lodicules and stamens (Kyozuka et al. 2000; Nandi et al. 2000) but knockdown of its expression impaired only lodicule development, implicating the second PI/GLO paralog, OsMADS4, as sufficient for stamen specification (Prasad and Vijayraghavan 2003). In this study, we report the dynamic expression pattern of OsMADS2 during lodicule development and its unique role in regulating lodicule differentiation and growth. We provide evidence that OsMADS2 functions independently of the other rice PI/GLO paralog, OsMADS4, to control second-whorl cell division and differentiation by regulating genes encoding predicted transcription factors, signaling molecules, and cell cycle regulators.
MATERIALS AND METHODS
In situ hybridizations:
RNA in situ hybridization for OsMADS2 and OsMADS4 transcripts was done using gene-specific RNA probes. Probes for detection of OsMADS2 were prepared as in Nandi et al. (2000). XhoI-linearized pBlueScript KS(+)-OsMADS4 with the 289-bp C-terminal and 3′-UTR cDNA sequences was transcribed with T7 RNA polymerase to make antisense RNAs. The same plasmid linearized with EcoRI was the template for sense-strand probe synthesis with T3 RNA polymerase. To generate gene-specific probes for OsMADS2-regulated downstream genes, PCR-amplified gene-specific regions for each of these genes were cloned in pBlueScript KS(+). The resulting clones for AK064240/TCP, AK106784/YABBY1b, and AK072687/LEUNIG, when linearized with HindIII and transcribed with T7 RNA polymerase, generated antisense RNA probes. The same plasmids linearized with BamHI were transcribed with T3 RNA polymerase to produce sense probes. The AK070518/cyclin B clone in pBlueScript linearized with BamHI was transcribed with T3 RNA polymerase to make antisense RNAs, and the same clone digested with XhoI was transcribed with T7 RNA polymerase to produce the sense RNA probe. Hybridizations were done as in Prasad et al. (2005) and signal was visualized using anti-DIG-AP-conjugated antibodies (Roche) or anti-DIG rhodamine-conjugated antibody (Roche, gift from K. Vijayraghavan; National Center for Biological Science, Bangalore, India).
Microscopy:
For histology, florets fixed in 3% glutaraldehyde were dehydrated in a graded ethanol series and embedded in SPURR resin. Sections (Ultramicrotome Leica, GmBH) of 0.5 μm were stained with 0.05% toleudine blue, air dried, and mounted. All sections were imaged using a Zeiss Axioscope2 Mot Plus microscope (Carl Zeiss, GmBH) and images were processed in Adobe Photoshop version 7.0.
Microarray and real-time PCR:
Total RNA from rice panicles (0.5–4 cm), wild type or dsRNAiOsMADS2, was extracted with Tri reagent (Sigma, St. Louis) and purified with RNeasy cleanup kit (QIAGEN, Valencia, CA). Two independent pools of mutant RNAs were compared to a wild-type RNA pool. For the microarray analysis, Agilent Technologies custom rice (22,000) microarrays were hybridized with Cy3- and Cy5-labeled cRNAs according to the manufacturer's instructions. The data are available with Gene Expression Omnibus (http:/www.ncbi.nlm.nih.gov under accession no. GSE7192). The data were analyzed using GeneSpring GX. An average ratio of mutant to wild type of <0.250 for a given gene was the criteria for its differential expression. For real-time quantitative RT–PCR analysis, first-strand cDNA was synthesized from 1 μg of total RNA and Superscript II (Invitrogen, San Diego) enzyme and used in triplicate quantitative PCR (qPCR) reactions using ABI Prism 7000 system. The difference in Ct value between mutant and wild type for the normalized transcript levels was used to calculate fold-down regulation of the deregulated gene. The downregulation of OsMADS2 in dsRNAiOsMADS2 panicles and the organ-specific expression levels of selected potential OsMADS2-regulated genes were analyzed by semiquantitative RT–PCR. The primer sequences are in supplemental Table S2 at http://www.genetics.org/supplemental/.
RESULTS
Dynamic temporal and spatial expression of OsMADS2 RNA during floret development:
The reduced grass floret has well-developed stamens but lacks obvious large petals. The predicted OsMADS2 protein bears domain organization and relatedness to other PI/GLO-like proteins. Further, like other B-function genes, the stable expression of OsMADS2 transcripts is in whorl 2 and 3 primordia and in these developing organs (Kyozuka et al. 2000; Nandi et al. 2000). We have reexamined domains in the young floret meristem where the early activation of OsMADS2 expression occurs and have carefully investigated its expression during second- and third-whorl organ differentiation. This formed the basis for us to decipher the molecular mechanisms underlying the phenotypes created upon its knockdown and its global effects on gene expression.
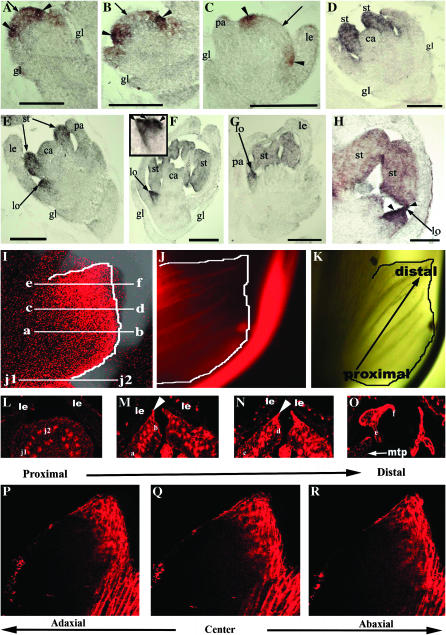
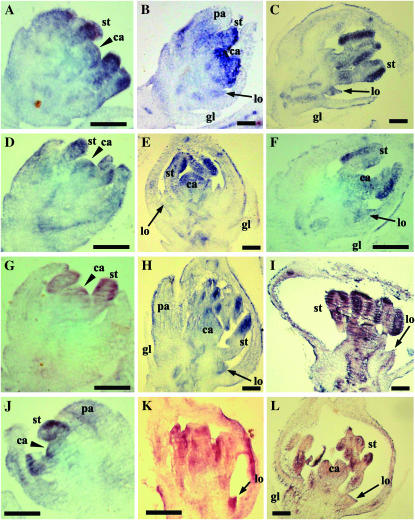
The transcription activation of OsMADS2 to a great extent is localized to the presumptive lodicule and stamen primordia on a newly established floral meristem. In these very young spikelet meristems where only the lemma primordia have been initiated, we note low-level OsMADS2 expression in the central region of the floral meristem that normally forms the carpel anlagen (Figure 1A). Slightly later in development, upon emergence of the palea primordia, this weak expression in the floret meristem center is reduced (Figure 1B). The emerging lemma and palea transiently express low levels of OsMADS2 (Figure 1B), which is eliminated slightly later in development (Figure 1C). This temporally early slightly broad domain of OsMADS2 activation was undetected in previous studies perhaps due to its low levels and transient nature (Kyozuka et al. 2000; Nandi et al. 2000). In fact, the current analyses show that the very early expression profile of OsMADS2 is similar to that of Arabidopsis PI (Goto and Meyerowitz 1994). The temporally later downregulation of OsMADS2 transcripts from the fourth-whorl anlagen is coupled with its high-level expression in the second- and third-whorl anlagen (Figure 1B). At subsequent stages where lemma are well-developed, hood-shaped organs but when lodicule and stamen primordia are yet to initiate, OsMADS2 expression is completely excluded from developing lemma/palea and carpel primordia (Figure 1C). In rice florets, stamen primordia initiate earlier than the lodicule primordia. OsMADS2 is expressed strongly in these early stage stamen primordia (Figure 1D). The lodicule primordia emerge only when stamen primordia begin to differentiate tetralocular anthers. At this stage, high-level OsMADS2 expression is observed in the newly arising second-whorl organs; with similar expression levels in developing lodicules and stamens (Figure 1E). Thus, the timing and early localization of OsMADS2 RNAs are consistent with the documented expression patterns for eudicot “B-function” genes, supporting the analogy between petals and lodicules (Ambrose et al. 2000; Munster et al. 2001; Nagasawa et al. 2003). However, subsequently, during second-whorl organogenesis, expression levels of OsMADS2 deviated from those reported for PI and several of its orthologs. While expression of OsMADS2 remains high in lodicules, its expression is reduced substantially in differentiating stamens (Figure 1, F–H). OsMADS2 transcripts are differentially distributed in the lodicule during its differentiation. They occur at higher levels at the distal lodicule end (Figure 1, F–H). To further examine OsMADS2 transcript distribution along the proximal–distal and the abaxial–adaxial axes, we analyzed its distribution in whole-mount and transverse sections of differentiated lodicules. As seen in longitudinal sections, OsMADS2 expression is greatly reduced in proximal regions of the lodicule (Figure 1, I and L) and expression is at progressively higher levels in distal regions (Figure 1I). Peripheral regions that are most distant from the palea also express high levels of OsMADS2 (Figure 1, H and I and M–O). The high-level expression in distal portions of the lodicule does not vary in the abaxial–adaxial axis (Figure 1, M–O and P–R). The asymmetric distribution of OsMADS2 transcripts on the proximal–distal axis suggests a likely role in the early growth arrest that must occur distally and peripherally in the emerging lodicule, perhaps to regulate its size and shape.
Figure 1.—
Expression pattern of OsMADS2. (A–H) Longitudinal sections through developing rice spikelets probed with digoxigenin-UTP antisense RNAs detected with AP-conjugated anti-DIG antibodies. (A) An incipient floret meristem. Black arrowheads mark the lodicule and stamen anlagen and the black arrow points to the central floret cells, all of which express OsMADS2. (B) Young florets with strong expression in lodicule and stamen anlagen and very low expression in lemma/palea primordia. (C) Developing young florets with emerging lemma/palea. No expression was seen in developing glumes, lemma/palea, and the floral center. (D and E) Equal expression in developing stamen (st) and lodicule (lo) primordia. (F and H) Florets during organogenesis show differential OsMADS2 expression. Black arrowheads point to distal and peripheral regions of a differentiated lodicule (lo). Inset in F is a magnified lodicule. (I–O) RNA in situ hybridization of whole-mount (I and J) and transverse sections (L–O) of mature lodicules detected with anti-DIG-rhodamine-conjugated antibodies. The section planes are indicated in I. Probe in I and L–O is antisense and in J is sense OsMADS2 RNA. (K) Lodicule brightfield image. White arrowheads in M and N mark the lodicule peripheral regions. (P–R) Longitudinal optical sections are arranged abaxially or adaxially with respect to the center of the lodicule. The right (abaxial) and the left (adaxial) sections are 24 μm apart. In both these sections, accumulation of OsMADS2 transcripts is in distal regions and in peripheral regions that grow away from palea. le, lemma; mtp, marginal tissue of palea.
Effect of OsMADS2 knockdown on second-whorl-specific cell differentiation:
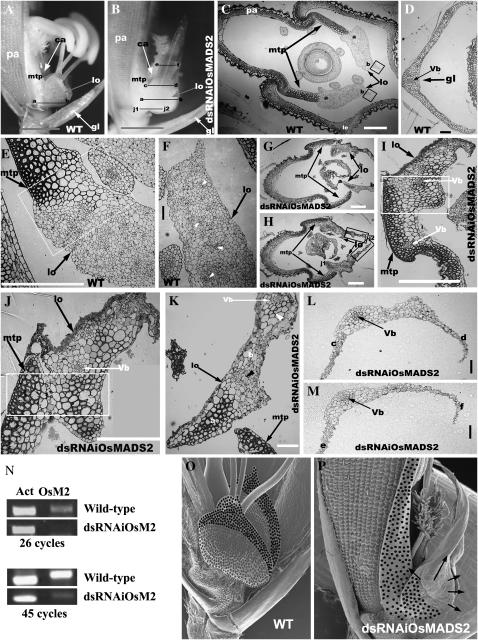
The second-whorl organs of grass florets are small rudimentary fleshy structures due to poor dorsiventral (abaxial–adaxial) flattening coupled with reduced growth along the proximal–distal axis (Figure 2A). The knockdown of OsMADS2 results in continued growth of the distal region, forming an extended bract-like structure (Figure 2B; Prasad and Vijayraghavan 2003). The basal or proximal portion of the transformed lodicule is also significantly larger with abnormal, severe dorsiventral flattening (compare Figure 2C vs. 2H). Because OsMADS2 is expressed at high levels in the presumptive lodicule anlagen even before outgrowth of these second-whorl cells from the floral primordia (Figure 1B), we examined its role in triggering lodicule-specific cell division and differentiation by serial ultrathin-section anatomical analysis of the entire transformed lodicule in OsMADS2 knockdown transgenic spikelets (Prasad and Vijayraghavan 2003). Anatomical features of the wild-type glume, lemma, and palea are distinctive (Figure 2, C and D). The rice palea has a characteristic marginal tissue, which is absent in the lemma (Figure 2C). This marginal tissue of palea (mtp) differs from rest of the palea in its unique smooth upper epidermis without epicuticular thickenings (Figure 2A) and in the underlying two to three layers of fibrous schlerenchymatous cells (Figure 2, C and E). The mtp schlerenchymatous cells differ from those in the glume with regard to their cell-wall thickness (compare Figure 2D vs. 2E). Unlike the palea, the wild-type lodicule contains a dense population of parenchymatous cells interspersed with vascular trachea elements (Figure 2, E and F). Another striking difference among glume, lemma/palea, and lodicules is that, while vascular tissues are organized as bundles in the glume (Figure 2D) and in the lemma/palea, in the lodicule they are dispersed throughout the organ (Figure 2F). This pattern of vascular differentiation is perhaps necessary for the uniform rapid swelling of the lodicule. We studied transverse sections through the proximal (basal) region of the lodicule that is closely juxtaposed with the marginal tissue of palea and found no identifiable border cells demarcating these two organs (Figure 2, C and E). Loss of OsMADS2 causes unique lodicule cell differentiation defects, notable of which is the transformation of cells at the proximal regions to cells typical of the immediately adjacent organ, the marginal tissue of palea (Figure 2, G–J). On the other hand, regions of the transformed lodicule positioned distant from marginal tissue of palea show a mixture of schlerenchyma and parenchyma cells (region b in Figure 2, B and K). And further, the extended distal portions of the transformed lodicule contain schlerenchymatous cells typical of the glume and not the mtp of the palea (Figure 2, L and M). We note that, in addition to these cellular transformations, the lodicule basal parts are flattened, comprising very few cell layers and only two to three vascular bundles. These vascular bundles are similar to those of the glume, lemma, or palea (Figure 2, I–M). Additionally, the continued growth of the lodicule margins is consistently seen in multiple transgenic lines due to which the normal cup-shaped fleshy lodicule structure is always lost (compare Figure 2C vs. 2H). Thus, the normal differentiation program of wild-type lodicule is entirely disrupted in these transformed lodicules, with the cells committing to different fates along the proximal–distal axis, and we obtained cleistogamous rice lines where spikelets remain unopened. The extent of downregulation of the endogenous OsMADS2 transcripts in dsRNAiOsMADS2 panicles was determined by semiquantitative RT–PCR (Figure 2N). Transcript levels are extremely low and are barely detected after 45 cycles of PCR amplification, indicating that near-null phenotypes are generated in these transgenic lines. Together, we reason that the likely role for the early expression of OsMADS2 in lodicule primordia is to delineate the boundary between palea and lodicule at its proximal end and to trigger second-whorl-specific cell differentiation throughout the lodicule (Figure 2, O and P). The temporally later expression of OsMADS2 in specific regions of the lodicule perhaps serves to initiate growth arrest distally and toward the lodicule periphery (Figure 2, O and P).
Figure 2.—
Ultrathin transverse sections show cell proliferation and differentiation defects in OsMADS2 knockdown florets. (A) Partially dissected wild type. (B) OsMADS2 knockdown spikelet. The transverse section planes are marked by lines (j1–j2, a–b, c–d, e–f). (C) Overview of wild-type lemma (le), palea (pa), marginal tissue of palea (mtp), and lodicule (lo) cell types. Boxed areas indicate the lodicule peripheral regions. (D) Wild-type glume histology. (E) Magnified view of the juxtaposed wild-type mtp and lodicule at the floret base. (F) Parachymatous cells and vascular elements (open arrowheads) of a wild-type lodicule. (G and H) Overview of OsMADS2 knockdown floret histology. The boxed areas mark the overgrown peripheral regions of the transformed lodicule (lo). (I and J) Magnified view of the base of a transformed lodicule in an OsMADS2 knockdown spikelet; compare with E. (K–M) Sections of the transformed lodicule through plane a–b in K, c–d in L, and e–f in M. Bar, 1.0 cm in A and B; 200 μm in C, E, and G–J; 50 μm in D, F, and K–M. (N) RT–PCR showing extent of downregulation of endogenous OsMADS2 transcripts in dsRNAiOsMADS2 panicles. Actin transcript levels are used for normalization. PCR amplification was done for 26 and 45 cycles. (O and P) Representation of the differential distribution of OsMADS2 transcripts and local effects upon its knockdown. (O) Lodicule distal and peripheral regions with high transcript levels have solid dots; regions with low expression have shaded dots. (P) Early OsMADS2 expression in developing lodicule (not shown here) may delineate the boundary between the schlerenchymatous cells of the mtp (solid dots) and the lodicule cells. Solid arrows mark the regions that overgrow upon OsMADS2 knockdown.
Timing and localization of transcripts for the second rice PI/GLO paralog OsMADS4:
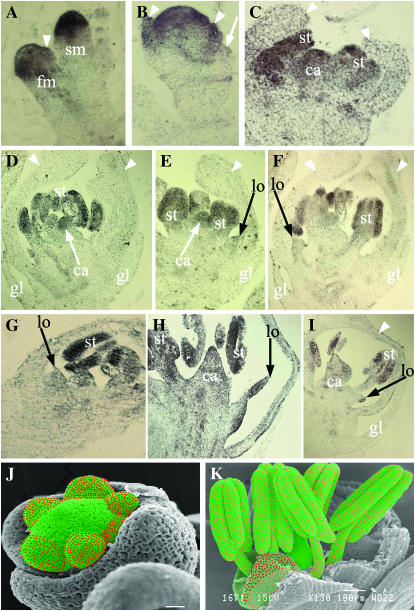
OsMADS4, together with the sole rice AP3 homolog SUPERWOMAN (SPW1), specifies lodicules and stamens (Kang et al. 1998; S. Lee et al. 2003; Nagasawa et al. 2003) as indicated from the phenotypic similarities of spw1 flowers and florets with OsMADS4 downregulation. To determine the OsMADS4 expression pattern, we monitored the spatial and temporal distribution of its transcripts by RNA in situ hybridization. We found that OsMADS4 transcription activation occurs very early, and uniformly, during spikelet meristem initiation prior to the emergence of even the first floret organ primordial, i.e., the lemma primordia (Figure 3A). The lemma and palea primordia express OsMADS4 from their outgrowth with continuing expression in the anlagen for the second-, third-, and fourth-whorl organs (Figure 3B). Since these transcripts are excluded from glumes, OsMADS4 expression is specific to floret organs (Figure 3B). Thus, the timing and domain of OsMADS4 transcription activation deviates from that of OsMADS2 from very early stages of floral meristem specification. During floret organogenesis high levels of OsMADS4 expression occur in stamen and carpel whorls (Figure 3, C and D) with reduced expression in the differentiating lodicules (Figure 3, E and F) and are completely excluded from lemma and palea (Figure 3, D–F). Importantly, from this stage onward, despite reduced OsMADS4 expression, the distribution in the lodicule is uniform (Figure 3, E and F) and no expression is detectable in the differentiated lemma or palea (Figure 3, D–F). Unlike OsMADS2, strong expression of OsMADS4 is maintained in differentiating fourth-whorl primordia (Figure 3, D and E). These data show that all three rice class B-genes share a common conserved expression domain (whorls 2 and 3) as is typical of many B-function genes. But they differ in the onset of their expression in the floret meristem and in their transcript levels and distribution within whorls 2 and 3. These rice paralogous genes also differ in their expression patterns in other floret whorls.
Figure 3.—
RNA in situ expression analysis of OsMADS4 during spikelet development. (A) Strong expression of OsMADS4 in spikelet meristem (sm) and floral meristem (fm). (B) Expression of OsMADS4 in young floral primordia and in lemma/palea primordia is marked by white arrowheads. White arrow denotes emerging glume. (C) OsMADS4 is expressed in stamen (st) and carpel (ca) primordia and is completely excluded from developing lemma and palea (arrowheads). (D–F) OsMADS4 expression patterns in near mature spikelets. High level of OsMADS4 expression continues in stamens (st) and carpel (ca) but is drastically reduced in lodicules (lo). Note the reduced but uniform expression of OsMADS4 throughout the lodicule (lo). (G–I) OsMADS4 expression in differentiating OsMADS2 knockdown spikelets. Spikelet in G represents slightly earlier developmental stage than shown in H where lodicule is completely transformed. (I) Similar to H and showing only a small portion of the proximal region of the transformed lodicule. Whorl-specific expression of OsMADS4 is retained in OsMADS2 knockdown spikelets. (J and K) OsMADS2 and OsMADS4 expression patterns in developing floral organ primordia (J) and in differentiated floral organs (K). OsMADS2 expression domain is denoted by red dots while the green-colored region represents the OsMADS4 expression domain. Pink dots and light green color represent reduced levels of expression.
Because knockdown of OsMADS2 completely perturbs lodicule differentiation, we examined if there are any changes in the expression profile of OsMADS4 in the transformed lodicules of Ubi-dsRNAiOsMADS2 transgenic florets (Prasad and Vijayraghavan 2003). RNA in situ hybridization shows that OsMADS4 expression is retained in the second whorl of these knockdown florets in addition to its normal expression in whorl 3 and whorl 4 (Figure 3, G–I). In fact, OsMADS4 transcripts are slightly upregulated in overgrown distal regions of transformed lodicules (Figure 3H). These data suggest that whorl-specific expression of OsMADS4 is retained in transgenic spikelet and that the abundant OsMADS4 transcripts cannot rescue the second-whorl-specific defects caused by the loss of OsMADS2.
Potential targets for OsMADS2 during lodicule differentiation:
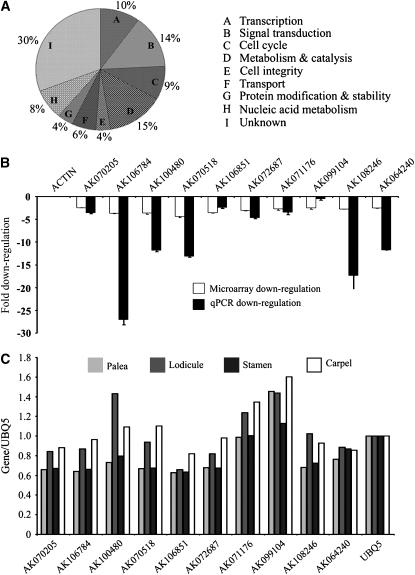
To gain mechanistic insights into OsMADS2 functions, we have employed global expression profiling to identify downstream genes regulated by OsMADS2. RNAs from two independent biological pools of dsRNAiOsMADS2 transgenic inflorescences, at various stages of organ specification and differentiation, were compared to a wild-type RNA pool. These transgenic RNA pools were used in two competitive hybridizations with the wild-type RNA pool on rice microarrays (materials and methods). A third hybridization was performed with reciprocally dye-labeled wild-type RNA and one of the mutant RNAs. We first identified transcripts downregulated in both the mutant RNA pools and then further screened for transcripts with more than twofold downregulation even after dye-swap labeling of the wild-type and mutant RNAs. These transcripts were manually inspected for their predicted or known expression profiles in available databases (KOME at http://cdna01.dna.affrc.go.jp/cDNA/ and our unpublished data) to arrive at a list of 385 affected genes whose expression was independently documented in developing wild-type florets. On the basis of the occurrence of protein domains, we ascribed functional categories to these differentially expressed genes. We found a preponderance of transcription factors and signaling molecules (10 and 14%) among the transcripts affected by the loss of OsMADS2 (Figure 4A). In addition, a large set of genes (9%) functioning in diverse aspects of cell division control; including cyclins, histones, and replication factors, are deregulated (Figure 4A; supplemental Table S1 at http://www.genetics.org/supplemental/). These observations are interesting in light of the global expression profiling data from Arabidopsis pi and ap3 mutants where the affected downstream genes do not include such a large proportion of cell division genes (Zik and Irish 2003; Wellmer et al. 2004). On the other hand ∼8% of the potential target genes of Antirrhinum DEF are factors affecting cell cycle and DNA processing (Bey et al. 2004).
Figure 4.—
Global expression analysis of OsMADS2-regulated genes. (A) Transcripts downregulated in OsMADS2 knockdown florets categorized on the basis of their predicted functions. (B) Comparison and verification of expression data retrieved from microarray experiments with quantitative real-time PCR analysis of expression levels. The bar graphs compare fold downregulation in wild type vs. dsRNAiOsMADS2 florets for 10 transcripts. Two biological replicates for the RNAs and at least triplicate qPCRs were performed to compute mean and standard deviation for expression levels determined by qRT–PCR. (C) Normalized expression levels for a few selected potential OsMADS2 target genes in the wild-type floret organs palea, lodicule, stamen, and carpel.
From the subset of potential OsMADS2-regulated genes, we chose 10 genes for validation of their downregulation in the absence of OsMADS2. These genes (Table 1) include factors with predicted functions in signaling and cell proliferation (GDSL lipase, F-box factor, cyclin B) and transcriptional regulators (YABBY domain factors, the TCP family of regulators, LEUNIG). Their expression levels, in wild-type panicle RNAs and in transgenic dsRNAiOsMADS2 panicles, were determined by qRT–PCR. The fold downregulation seen in qRT–PCR was compared with the data from microarray analysis (Figure 4B). Expression of all but one of these genes was dependent on OsMADS2 as seen by both assays. The expression levels of these genes in various wild-type floret organs were examined by semiquantitative RT–PCR specifically in the palea, lodicule, stamens, and carpels. From comparison of the normalized expression level, we found that all of these genes are expressed in wild-type lodicules (Figure 4C) and are expressed at comparable levels in carpels. The expression levels in stamens and palea are somewhat lower.
TABLE 1.
Ten selected genes downregulated in OsMADS2 knockdown floret
| Accession nos.a | Annotation/descriptionb | Closest Arabidopsis hit |
|---|---|---|
| AK070205 | Axial regulator YABBY1 | At2g45190 |
| AK106784 | Axial regulator YABBY1 | At2g45190 |
| AK100480 | GDSL-motif lipase | At1g09390 |
| AK070518 | Cyclin, similar to B-like cyclin | At1g76310 |
| AK106851 | Cellulase, identical to ATCEL2 | At1g02800 |
| AK072687 | Transcriptional corepressor LEUNIG | At4g32551 |
| AK071176 | SET-domain transcription regulator | At2g17900 |
| AK099104 | F-box family protein | At1g53320 |
| AK108246 | ZF-HD homeobox family protein | At4g24660 |
| AK064240 | TCP family transcription factor | At1g69690 |
Accession numbers according to KOME (http://cdna01.dna.affrc.go.jp/cDNA).
Annotation/description as in KOME (http://cdna01.dna.affrc.go.jp/cDNA).
The rice drooping leaf-superman (dl-sup) mutant forms ectopic stamens at the expense of carpel tissues, while lodicules are unaffected (Nagasawa et al. 2003). We observed that the expression domain and localization of OsMADS2 transcripts to specific regions of the lodicule in dl-sup flowers remained unaffected (supplemental Figure S1, A–C, at http://www.genetics.org/supplemental/), corroborating the dynamic regulation of OsMADS2 in lodicules of mutant plants of other genotypes. Similarly, expression of SPW1 is normal in dl-sup lodicules (Nagasawa et al. 2003), suggesting that OsMADS2 and SPW1 may suffice to trigger lodicule differentiation. Since lodicule morphology and OsMADS2 expression domains within the lodicule are normal in dl-sup flowers, we used this rice mutant as a control to verify the expression levels of genes regulated by OsMADS2. We found that the transcription factors AK106784/Yabby1b, AK071176/ZF-SET, and AK064240/TCP and the signaling or cell division factors such as AK070518/cyclin B, AK099104/F-box factor, and AK100480/GDSL lipase are expressed at normal or near-normal levels in differentiated florets of dl-sup panicles (supplemental Figure S1D at http://www.genetics.org/supplemental/). Together, these data show these genes to be regulated by OsMADS2. As a first approximation in identifying genes that might be direct targets of OsMADS2, we inspected genomic sequences up to 1 kb upstream of all 385 deregulated genes for the presence of cis-acting CArG elements that are binding sites for MADS domain proteins (supplemental Table S1 at http://www.genetics.org/supplemental/). We found that 47% of these genes have CArG elements in this segment of their genomic loci and, notably, of these, ∼37% of the genes have multiple such elements (supplemental Table S1 at http://www.genetics.org/supplemental/).
Spatial transcript distribution for genes regulated by OsMADS2 in developing florets:
Four of the genes regulated by OsMADS2—encoding three transcription regulators and a cell division regulator—were further examined by RNA in situ hybridization to determine their expression domains in developing floral organs. Transcripts for all of these genes were detected in the lodicule right from its emergence; expression continues during its development and persists to varying extents in the fully developed lodicules (Figure 5). This profile is in general similar to that of OsMADS2 (Figure 1, E–H). However, not unexpectedly, the expression of these genes in other floral organs differs from that of OsMADS2. For example, their expression in the fourth whorl is maintained during carpel development as is expression in the mature stamens (compare Figure 5 and Figure 1). These genes also differ from each other in their relative expression in stamens, the carpel, and mature lodicules.
Figure 5.—
Spatial expression profile of OsMADS2-regulated genes in wild-type florets. (A–C) Localization of AK064240/TCP transcripts. (D–F) Expression profile of AK070518/cyclin B. (G–I) Spatial distribution AK106784/Yabby1b transcripts. (J–L) Expression pattern of AK072687/LEUNIG. Spikelets in A, D, G, and J have initiated stamen differentiation but lodicules have yet to initiate. In B, E, H, and K, spikelets are at the early stage of lodicule development. C, F, I, and L show florets at late stages of lodicule development. Arrowheads point to carpel primordia and arrows point to lodicules. gl, outer glume; pa, palea; lo, lodicule; st, stamen; ca, carpel. Bar in B, C, E, H, I, and L, 50 μm; 100 μm in A, D, F, G, J, and K.
The AK064240/TCP and AK070518/cyclin gene transcripts in the lemma, palea, and emerging stamens of young florets are at equal levels (Figure 5, A and D). The expression in the carpel primordia is somewhat lower (Figure 5, A and D). While the expression in the lemma and palea is shut down during their later differentiation, high-level expression is seen in differentiating stamens, particularly in anthers (Figure 5, B, C, E, and F). Expression levels in the lodicules are maintained uniformly throughout its development (Figure 5, B, C, E, and F). The transcripts for AK106784/Yabby1b and AK072687/LEUNIG appear to be temporally regulated in differentiating floral organs. They occur at higher abundance early during lodicule development and are at lower levels later during lodicule development (Figure 5, H, I, K, and L). The expression of both these genes in stamens is maintained throughout stamen differentiation and in later stages is particularly high in anthers (Figure 5, G–L). The recently reported expression levels of YABBY1b (Toriba et al. 2007), as determined by quantitative RT–PCR, are consistent with our RNA in situ hybridization data (Figure 5, G–I) of developing flowers.
DISCUSSION
Developmental innovations by gene duplication:
Duplication and diversification of evolutionarily conserved transcription factors can bring about morphological variation between closely related organisms. Mechanistically, such functional differences between duplicated genes could arise from changes in the transcription factors themselves, or from their altered expression, or from changes in how they regulate their target genes (reviewed in Irish and Litt 2005). We demonstrate that, of the two rice PI-like genes, OsMADS2 controls development of the rice lodicule, a floret organ critical for regulated opening of the rice floret and for cross-pollination. Its unique regulatory role is evident from the specific effects, obtained upon OsMADS2 knockdown, on the fate of lodicule cells along the proximal vs. distal axis. These distinct functions are not seen with downregulation of its paralog OsMADS4 (Kang et al. 1998). The functional diversification of OsMADS4 with respect to second-whorl organ development has been shown using a heterologous plant system, tobacco, as a model (Kang and An 2005). Unlike the effect of Antirrhinum GLO overexpression, the ectopic overexpression of OsMADS4 in tobacco did not cause any alteration in first-whorl organs (Kang and An 2005). Together with our evidence that OsMADS4 expression in second-whorl organs of dsRNAiOsMADS2 florets does not rescue second-whorl lodicule formation, we can conclude that OsMADS4 alone cannot confer the unique second-whorl organ fate in rice florets. Such effects may arise from alterations in the conserved MADS domain or from changes in the functionally important PI motif (supplemental Figure S2 at http://www.genetics.org/supplemental/; Lamb and Irish 2003) or even from changes in the profile within the conserved B-function expression domains. Interestingly, the early transcriptional activation of OsMADS2 in whorl 2 and 3 primordia is a pattern similar to that of many eudicot PI genes, but the later subdomains of OsMADS2 expression differ from that known for Arabidopsis PI and Antirrhinum GLO (Trobner et al. 1992; Goto and Meyerowitz 1994). In whorl 2 organs, high-level OsMADS2 transcripts are predominantly localized toward their distal end. We ascribe a function for this expression in regulating growth as this region overgrows upon OsMADS2 knockdown. Recent analysis of several Arabidopsis gene-trap lines reveals that these petals also have underlying differences in gene expression, which are reflected in reporter profiles along the proximal–distal axis (Nakayama et al. 2005). However, these molecular signatures are distinct from the expression pattern of Arabidopsis B-class genes or their regulators. The restricted late expression of OsMADS2 to specific regions of the lodicule bears similarity to the expression of some lower eudicot PI-like genes of the Ranunculid family where expression is limited to petal edges (Kramer and Irish 1999). Such localized expression may serve specific functions in petal development in these species.
The expression pattern of OsMADS2 not only is distinct from higher eudicot B-class genes but also diverges from its paralog, OsMADS4, in local regions within the second-and third-whorl. High-level OsMADS2 expression is restricted to specific regions of lodicules and expression is generally downregulated in stamens. In contrast, OsMADS4 expression is downregulated uniformly in all lodicule cells and its strong uniform expression is maintained in stamens, supporting its critical role in defining the stamen identity (Kang et al. 1998; S. Lee et al. 2003). Additionally, OsMADS4 acquires expression in the fourth whorl, a pattern not seen for OsMADS2. These observations also explain the need for repression of SPW1 in fourth whorl (Nagasawa et al. 2003). We deduce that the continued fourth-whorl expression of OsMADS4 throughout floret development necessitates the repression of its interacting partner SPW1 to prevent the formation of ectopic fourth-whorl stamens (S. Lee et al. 2003). Changes in SPW1 affect OsMADS4 transcripts reciprocally without any changes in OsMADS2 transcript levels (Xiao et al. 2003). Therefore, rice B-function activity provided by OsMADS4/SPW1 heterodimer may auto-regulate its expression, as has been reported for the Arabidopsis PI/AP3 and Antirrhinum GLO/DEF heterodimers (Trobner et al. 1992; Krizek and Meyerowitz 1996). All of these data suggest divergence in the regulation of the two rice PI/GLO paralogs.
Triggering differentiation of the lodicule, a highly derived grass-specific second-whorl organ:
One of the most intriguing effects of OsMADS2 knockdown is the perturbation of lodicule cell differentiation in a context-dependent fashion. In these knockdown florets, lodicule cells in close proximity to the marginal tissue of palea (mtp) acquire cellular features of the mtp. On the other hand, the proximal tissues of the transformed lodicule at a distance from mtp acquire a mixed fate of parenchyma and schlerenchyma cells. Further still, the distal portions of the transformed lodicule overgrow but acquire an exclusively schlerenchymatous cell fate. This suggests direct cell–cell communication in regions of the floret where the distinctive cells of the mtp and lodicules are juxtaposed. We speculate that the primary role of OsMADS2 in this proximal region is to delineate the boundary between mtp and lodicule, perhaps an early event during commitment of floret meristem cells to the lodicule fate. Our data also show that OsMADS2 expression promotes second-whorl-specific cell differentiation throughout the lodicule as the transformed organs have dramatic alterations in vascular differentiation. Our report represents a novel example of a class B gene exerting context-dependent local effects on growth and differentiation.
Our studies provide possible explanations for the conversion of lodicules to mtp and not a fully differentiated lemma or palea in the spw1 mutant (Nagasawa et al. 2003). Loss-of-B-function mutants in Arabidopsis and Antirrhinum have petals transformed to the adjacent peripheral organ, i.e., the sepals. In the rice floret, the mtp is placed adjacent to whorl 2 organs; therefore we may interpret the transformation of lodicules to mtp seen in rice B-function mutants as being similar to the phenotypic transformations of Arabidopsis mutants. These homeotic organ transformations support the notion that lodicules are petal homologs as suggested from studies in maize (Ambrose et al. 2000; Whipple et al. 2004).
Target genes regulated by OsMADS2 encode putative cell proliferation, differentiation, and signaling factors:
OsMADS2 controls the shape and fleshy characteristics of the second-whorl floret organ by regulating cell proliferation and by maintaining a unique differentiation pattern. Several genes deregulated upon knockdown of OsMADS2 are those predicted to regulate cellular growth and differentiation and ultimately organ shape. The transcription activation of these genes must be independent of the OsMADS4/SPW1 heterodimer as the normal whorl-specific expression of OsMADS4 does not rescue the phenotype of OsMADS2 knockdown. OsMADS2-regulated genes with potential growth regulatory functions, as well as those genes needed for the development of highly vascularized parenchymatous cells, is particularly interesting. Both these features distinguish the lodicule from other sterile whorl organs. Of the 49 rice cyclins identified (La et al. 2006), we found that six cyclins are deregulated in the absence of OsMADS2. Three of these are B-type cyclins, two are A-type cyclins, and another is a D-type cyclin (Table 1; supplemental Table S1 at http://www.genetics.org/supplemental/). We have validated the downregulation of AK070518 (OsCycB2-2) upon loss of OsMADS2. Intriguingly, there are three CArG elements, potential binding sites for MADS domain proteins, positioned within 1-kb sequences upstream to the predicted OsCycB2-2 transcription start site. Further, its spatial expression domain overlaps with OsMADS2. These observations make it one of the candidate genes that may be directly regulated by OsMADS2I; investigations on this line are ongoing. Importantly, independent studies have shown that this cyclin is nuclear localized and that its overexpression causes increased root growth, most likely by promoting cell division (J. Lee et al. 2003). We speculate that, as a potential target of OsMADS2, perhaps OsCycB2-2 promotes proliferation of parenchymatous cells in the lodicule. The importance of regulated cell proliferation during lodicule development is further reinforced by our discovery of two members of the TCP family of transcription regulators deregulated in the absence of OsMADS2, a characteristic not seen among potential targets of AP3 or PI (Zik and Irish 2003; Wellmer et al. 2004). Among 25 predicted rice TCP factors (http://ricetfdb.bio.uni-potsdam.de/v2.1/), we found that AK064240, an uncharacterized class I factor similar to PCF1, and another PCF7 (AK058570), a class II factor, are deregulated on OsMADS2 knockdown. The AK064240/TCP is expressed immediately following lodicule emergence, with expression continuing during its differentiation. Further sequences upstream to the predicted transcription start site have one CArG element. The class II TCP family includes the well-studied Antirrhinum CIN and CYC proteins, the maize TB1, and the rice OsTB1 proteins (Doebley et al. 1997; Luo et al. 1999; Takeda et al. 2003; Crawford et al. 2004). Functional studies on CIN and CYC show them to regulate growth of lateral organs (Nath et al. 2003; Crawford et al. 2004; Costa et al. 2005). Similar functions are seen for the Lotus japonicus TCP-box gene LjCYC2 that regulates asymmetric growth of petals and maintains its shape (Feng et al. 2006). The first mechanistic link between TCP factors and cell proliferation came from in vitro studies showing that rice PCF1 and PCF2 bind cis elements in the PCNA gene, a key protein of the DNA replication complex (Kosugi and Ohashi 1997, 2002). Another link identified in a recent study shows that Arabidopsis TCP20 can bind elements in the promoter of CycB1 in addition to promoters of a few of the ribosomal protein genes (Li et al. 2005). Notably, an opposite effect of the repression of cell proliferation by a class I TCP domain factor is exemplified by the interaction between Antirrhinum TIC and CUPULIFORMIS, a NAC-domain protein (Weir et al. 2004). This study provides the attractive hypothesis that localized expression of TCP and NAC domain transcription factors can repress cell division to establish boundaries between lateral organs (Weir et al. 2004), a scenario that may apply in lodicule development as four NAC-domain-containing factors are deregulated in the absence of OsMADS2 (data not shown). Thus multiple alternative mechanisms operate to link this class of transcription regulators with cell proliferation. Together, these lines of evidence provide hypotheses to test the impact of OsMADS2 in regulating cell division.
Our global transcript profiling shows how OsMADS2 can trigger lodicule-specific differentiation. The Arabidopsis transcription coregulators SUESS and LEUNIG control petal blade cell number and petal internal vasculature (Franks et al. 2006). Similar functions for the Antirrhinum homologs of LUG and SEU, i.e., STY, AmSEU1, AmSEU2, and AmSEU3, in organ initiation, laminar growth, and venation have been found (Navarro et al. 2004). STY has also been shown to physically interact with these Antirrhinum SEU-like proteins as well as with the Antirrhinum FIL ortholog, GRAMINIFOLIA (GRAM). While SEU and LUG are transcription regulators, DNA-binding activity is likely provided by their partner, FIL1/YAB1 (Navarro et al. 2004). In another study, SEU has been shown to mediate interaction of LUG with MADS box proteins AP1 and SEP3 and thereby acquire DNA-binding specificity (Sridhar et al. 2006). LUG-SEU has also been shown to acquire DNA-binding specificity from the AP1-SVP and AP1-AGL24 MADS protein dimers (Gregis et al. 2006). It is striking that the closest rice homologs for these factors (AK072687-OsLEU, AK070205-OsYAB1a, AK106784-OsYAB1b, and AK100227, a rice MADS gene) are deregulated in the absence of OsMADS2, hinting at a role for OsMADS2 in regulating vascular development through the actions of a homologous complex. Both OsLEU and OsYAB1b are expressed during lodicule development. While none of the above-mentioned rice factors are by themselves functionally characterized, it is noteworthy that a related YABBY domain factor, DL-SUP, is required for leaf midvein development (Yamaguchi et al. 2004) and for carpel development. DL-SUP is not expressed in lodicules, but the YABBY genes identified here are expressed in both lodicules and carpels. Functional studies of these rice genes, including understanding their direct vs. indirect regulation by OsMADS2, are required to elucidate their specific roles in lodicule development.
Our preliminary comparative survey of genes deregulated on knockdown of OsMADS2, potential targets of Arabidopsis AP3 and PI or potential targets of Antirrhinum DEF (Zik and Irish 2003; Bey et al. 2004; Wellmer et al. 2004), reveals some commonalities and some important differences in the global profile of affected genes. Many commonly affected genes in these diverse species are expressed late in petal differentiation, such as genes for lipases, hydrolases, fatty acid elongation, and lipid transfer proteins. Another similarity in the global profile of genes regulated by B-function genes is the occurrence of predicted receptor-like kinases and signaling factors. It is noteworthy that a sizable fraction of the genes affected upon loss of OsMADS2 are those with functions in cell division, a global profile not seen in targets of PI (Zik and Irish 2003; Wellmer et al. 2004). These studies suggest that, while some target genes of higher eudicot and grass class B genes many encode molecules with similar functions, many downstream genes of OsMADS2 are distinct from those identified thus far for PI or DEF. This likely reveals the differences in the cascade of genes regulated by B-function activity in these diverse species that have distinct petal morphologies and where petals serve different functions.
Acknowledgments
We are grateful to Y. Nagato for providing the dl-sup mutant. U.V. acknowledges grant support from the Department of Biotechnology, Government of India, for rice functional genomics and expression profiling. Infrastructure support from Indian Council of Medical Research, Government of India, to the Department of Microbiology and Cell Biology is acknowledged. A Senior Research Fellowship from Council of Scientific and Industrial Research, Government of India, provided support to S.R.Y.
References
- Ambrose, B. A., D. R. Lerner, P. Ciceri, C. M. Padilla, M. F. Yanofsky et al., 2000. Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579. [DOI] [PubMed] [Google Scholar]
- Angenent, C. C., M. Busscher, J. Franken, J. N. Mol and A. J. Van Tunen, 1992. Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4: 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey, M., K. Stuber, Z. Fellenberg, H. Schwarz-Sommer, H. Sommer et al., 2004. Characterization of Antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16: 3197–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. L., D. R. Smyth and E. M. Meyerowitz, 1989. Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M. M., S. Fox, A. I. Hanna, C. Baxter and E. Coen, 2005. Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–5101. [DOI] [PubMed] [Google Scholar]
- Crawford, B. C., U. Nath, R. Carpenter and E. S. Coen, 2004. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino, G., I. Pan, E. Emmanuel, A. Levy and V. F. Irish, 2006. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18: 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Feng, X., Z. Zhao, Z. Tian, S. Xu, Y. Luo et al., 2006. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 103: 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, R. G., Z. Liu and R. L. Fischer, 2006. SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224: 801–811. [DOI] [PubMed] [Google Scholar]
- Goto, K., and E. M. Meyerowitz, 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gregis, V., A. Sessa, L. Colombo and M. M. Kater, 2006. AGL24, SHORT VEGETATIVE PHASE and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish, V. F., and A. Litt, 2005. Flower development and evolution: gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15: 454–460. [DOI] [PubMed] [Google Scholar]
- Kang, H. G., and G. An, 2005. Morphological alterations by ectopic expression of the rice OsMADS4 gene in tobacco plants. Plant Cell Rep. 24: 120–126. [DOI] [PubMed] [Google Scholar]
- Kang, H. G., J. S. Jeon, S. Lee and G. An, 1998. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38: 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Y. Ohashi, 1997. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Y. Ohashi, 2002. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348. [DOI] [PubMed] [Google Scholar]
- Kramer, E. M., and V. F. Irish, 1999. Evolution of genetic mechanisms controlling petal development. Nature 399: 144–148. [DOI] [PubMed] [Google Scholar]
- Krizek, B. A., and E. M. Meyerowitz, 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22. [DOI] [PubMed] [Google Scholar]
- Kyozuka, J., T. Kobayashi, M. Morita and K. Shimamoto, 2000. Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41: 710–718. [DOI] [PubMed] [Google Scholar]
- La, H., J. Li, Z. Ji, Y. Cheng, X. Li et al., 2006. Genome-wide analysis of cyclin family in rice (Oryza sativa L.). Mol. Genet. Genomics 275: 374–386. [DOI] [PubMed] [Google Scholar]
- Lamb, R. S., and V. F. Irish, 2003. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100: 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., A. Das, M. Yamaguchi, J. Hashimoto, N. Tsutsumi et al., 2003. Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J. 34: 417–425. [DOI] [PubMed] [Google Scholar]
- Lee, S., J. S. Jeon, K. An, Y. H. Moon, S. Lee et al., 2003. Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217: 904–911. [DOI] [PubMed] [Google Scholar]
- Li, C., T. Potuschak, A. Colon-Carmona, R. A. Gutierrez and P. Doerner, 2005. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D., R. Carpenter, L. Copsey, C. Vincent, J. Clark et al., 1999. Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376. [DOI] [PubMed] [Google Scholar]
- Munster, T., L. U. Wingen, W. Faigl, S. Werth, H. Saedler et al., 2001. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic ‘B-function’ genes of grasses. Gene 262: 1–13. [DOI] [PubMed] [Google Scholar]
- Nagasawa, N., M. Miyoshi, Y. Sano, H. Satoh, H. Hirano et al., 2003. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718. [DOI] [PubMed] [Google Scholar]
- Nakayama, N., J. M. Arroyo, J. Simorowski, B. May, R. Martienssen et al., 2005. Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. Plant Cell 17: 2486–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, A. K., K. Kushalappa, K. Prasad and U. Vijayraghavan, 2000. A conserved function for Arabidopsis SUPERMAN in regulating floral-whorl cell proliferation in rice, a monocotyledonous plant. Curr. Biol. 10: 215–218. [DOI] [PubMed] [Google Scholar]
- Nath, U., B. C. Crawford, R. Carpenter and E. Coen, 2003. Genetic control of surface curvature. Science 299: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Navarro, C., N. Efremova, H. F. Golz, R. Rubiera, M. Kuckenberg et al., 2004. Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131: 3649–3659. [DOI] [PubMed] [Google Scholar]
- Prasad, K., and U. Vijayraghavan, 2003. Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, K., S. Parameswaran and U. Vijayraghavan, 2005. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43: 915–928. [DOI] [PubMed] [Google Scholar]
- Rijpkema, A. S., S. Royaert, J. Zethof, G. Van Der Weerden, T. Gerats et al., 2006. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R., and B. A. Ambrose, 1998. The blooming of grass flower development. Curr. Opin. Plant Biol. 1: 60–67. [DOI] [PubMed] [Google Scholar]
- Sridhar, V. V., A. Surendrarao and Z. Liu, 2006. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166. [DOI] [PubMed] [Google Scholar]
- Takeda, T., Y. Suwa, M. Suzuki, H. Kitano, M. Ueguchi-Tanaka et al., 2003. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33: 513–520. [DOI] [PubMed] [Google Scholar]
- Toriba, T., K. Harada, A. Takamura, H. Nakamura, H. Ichikawa et al., 2007. Molecular characterization of YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genomics 277: 457–468. [DOI] [PubMed] [Google Scholar]
- Trobner, W., L. Ramirez, P. Motte, I. Hue, P. Huijser et al., 1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., J. Zethof, S. Royaert, K. Weterings and T. Gerats, 2004. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol, A. R., A. Brunelle, S. Tsuchimoto and N. H. Chua, 1993. Functional analysis of Petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7: 1214–1228. [DOI] [PubMed] [Google Scholar]
- Weir, I., J. Lu, H. Cook, B. Causier, Z. Schwarz-Sommer et al., 2004. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131: 915–922. [DOI] [PubMed] [Google Scholar]
- Wellmer, F., J. L. Riechmann, M. Alves-Ferreira and E. M. Meyerowitz, 2004. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple, C. J., P. Cicero, C. M. Padilla, B. A. Ambrose, S. L. Bandong et al., 2004. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131: 6083–6091. [DOI] [PubMed] [Google Scholar]
- Xiao, H., Y. Wang, D. Liu, W. Wang, X. Li et al., 2003. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 52: 957–966. [DOI] [PubMed] [Google Scholar]
- Xu, Y., L. L. Teo, P. P. Kumar and H. Yu, 2006. Floral organ identity genes in orchid Dendrobium crumenatum. Plant J. 46: 54–68. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., N. Nagasawa, S. Kawasaki, M. Matsuoka, Y. Nagato et al., 2004. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachgo, S., A. Silva Ede, P. Motte, W. Trobner, H. Saedler et al., 1995. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121: 2861–2875. [DOI] [PubMed] [Google Scholar]
- Zik, M., and V. F. Irish, 2003. Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]