Abstract
In Caenorhabditis elegans, the kinase ZYG-1 is required for centrosome duplication. To identify factors that interact with ZYG-1, we used a classical genetic approach and identified 21 szy (suppressor of zyg-1) genes that when mutated restore partial viability to a zyg-1 mutant. None of the suppressors render animals completely independent of zyg-1 activity and analysis of a subset of the suppressors indicates that all restore the normal process of centrosome duplication to zyg-1 mutants. Thirteen of these suppressor mutations confer phenotypes of their own and cytological examination reveals that these genes function in a variety of cellular processes including cell cycle timing, microtubule organization, cytokinesis, chromosome segregation, and centrosome morphology. Interestingly, several of the szy genes play a role in attaching the centrosome to the nuclear envelope. We have found that one such szy gene is sun-1, a gene encoding a nuclear envelope component. We further show that the role of SUN-1 in centrosome duplication is distinct from its role in attachment. Our approach has thus identified numerous candidate regulators of centrosome duplication and uncovered an unanticipated regulatory mechanism involving factors that tether the centrosome to the nucleus.
TO accomplish its various tasks, the microtubule cytoskeleton is constantly reorganized throughout the cell cycle. In mitosis, microtubules are assembled into a bipolar spindle to segregate chromosomes and position the cytokinetic furrow. In interphase they are organized into a radial array that participates in the trafficking of material along the cell's central-peripheral axes. This reorganization is largely under control of the centrosome, the cell's primary microtubule-organizing center. Through its capacity to nucleate and anchor microtubules, the centrosome organizes the radial arrays of interphase cells and the poles of the mitotic spindle (Doxsey 2001).
The morphology of the animal centrosome varies somewhat among species but overall its basic structure is conserved (Azimzadeh and Bornens 2004). It is composed of two parts: an orthogonally aligned pair of centrioles and an associated matrix of pericentriolar material (PCM). Centrioles are cylindrical structures composed of a ninefold symmetric arrangement of microtubules and are important for maintaining a discrete domain of PCM (Bobinnec et al. 1998). Each centrosome contains one “mother” centriole that is at least one cell cycle old and one “daughter” centriole synthesized during the last round of centrosome duplication. The PCM is the site of microtubule nucleation and anchoring. Although its exact molecular composition and structural organization are not known, it contains a high concentration of coiled-coil domain proteins that are thought to provide a scaffold for anchoring the γ-tubulin ring complexes that nucleate microtubules (Doxsey 2001).
Unlike other organelles, the centrosome is not membrane bound but it does maintain a close association with the nuclear envelope. Recently, three proteins have been identified that play a key role in maintaining this close association. Loss of ZYG-12, a Caenorhabditis elegans member of the Hook family of cytoskeletal linker proteins, results in detachment of the centrosome from the nucleus (Malone et al. 2003). Such mutants exhibit spindle defects, chromosome missegregation, and lethality, indicating that at least in the C. elegans embryo, association of the centrosome and nucleus is essential. ZYG-12 localizes to both the nuclear envelope and centrosomes and is thought to maintain anchorage through self-association. ZYG-12 also physically interacts with cytoplasmic dynein (Malone et al. 2003) and loss of dynein activity results in a detached centrosome phenotype (Gonczy et al. 1999a; Yoder and Han 2001). Finally, another conserved component of the nuclear envelope, SUN-1, is required for centrosome–nuclear association. SUN-1, a member of a family of proteins characterized by the presence of a membrane-spanning region and a C-terminal SUN domain (Starr and Fischer 2005), is required for nuclear localization of ZYG-12 (Malone et al. 2003).
Like DNA, centrosomes duplicate precisely once per cell cycle during S phase. Ultrastructural studies have resolved duplication into a few discrete steps (Sluder 2004). The first step involves the separation of mother and daughter centrioles, which move a short distance apart, losing their orthogonal orientation. Centriole synthesis then initiates with the formation of a precursor, or procentriole, next to and at a right angle to each preexisting centriole. Finally, the procentrioles elongate to form complete daughter centrioles. The two resulting centriole pairs ultimately migrate apart. As the cell approaches mitosis, each centrosome “matures” as it accumulates PCM and acquires increased microtubule-nucleating capacity.
Defects in centrosome duplication can result in spindles with an abnormal number of poles. For instance, monopolar spindles can result from duplication failure while multipolar spindles can result from a failure to limit centrosome duplication to one round per cell cycle. Interestingly, many tumor cells contain more than two centrosomes, suggesting that errors in centrosome duplication contribute to genomic instability and cancer (Sankaran and Parvin 2006). Despite the obvious importance of centrosome duplication, little is known about the molecular events that compose this process. In addition, how duplication is limited to one round per cell cycle and how it is temporally coordinated with other cell cycle events are still not well understood.
Work in a variety of organisms over the past decade has led to an expanding inventory of proteins that function in this process. In particular, genetic analysis in C. elegans has led to the identification of five core components of the duplication machinery. These include the kinase ZYG-1 (O'Connell et al. 2001) and four coiled-coil domain-containing proteins: SPD-2, SAS-4, SAS-5, and SAS-6 (Kirkham et al. 2003; Leidel and Gonczy 2003; Dammermann et al. 2004; Delattre et al. 2004; Kemp et al. 2004; Pelletier et al. 2004; Leidel et al. 2005). Loss of any one of these factors leads to a complete block whereby mother and daughter centrioles separate but no new centrioles are formed. In addition, Dammermann et al. (2004) have shown that SPD-5, another coiled-coil domain protein (Hamill et al. 2002), and γ-tubulin also are at least partially required for centriole formation. In the case of SAS-6, a vertebrate ortholog has been identified and demonstrated to have the same function (Leidel et al. 2005). Potential vertebrate orthologs of SPD-2 and SAS-4 exist as well, suggesting that worms and vertebrates utilize the same basic machinery (Leidel and Gonczy 2003; Pelletier et al. 2004).
Despite a multitude of mutant hunts (Hirsh and Vanderslice 1976; Miwa et al. 1980; Cassada et al. 1981; Kemphues et al. 1988a,b; O'Connell et al. 1998; Gonczy et al. 1999b) and exhaustive genomewide RNAi-based screening (Kamath et al. 2003; Simmer et al. 2003; Sonnichsen et al. 2005), no additional factors have been implicated in centrosome duplication. It is thus unlikely that existing forward and reverse genetic screening strategies will be very effective in identifying many more genes with important roles in this process. Therefore, a more focused screening strategy is needed. One such strategy is a genetic modifier screen whereby one screens for mutations that enhance or suppress the phenotype of an existing mutant. Genes whose products interact in either a positive or a negative manner with the gene of interest can thus be identified. For lethal mutations, the suppressor screen is particularly powerful as it allows one to rapidly and efficiently select for mutations that restore some degree of viability.
Here we have devised a highly sensitive version of the suppressor screen to identify mutations that restore viability to strains carrying a lethal mutation. The design of the screen is such that it can be performed on any scale to rapidly and efficiently identify suppressor mutations of high or low potency, including those suppressor mutations that are deleterious. We have applied this approach to identify suppressor mutations that restore centrosome duplication to a strain compromised for zyg-1 function and have identified 40 independent suppressor mutations that define 21 genes. Many of these genes appear to encode factors with essential functions. Unexpectedly, we identified one of these suppressors as a loss-of-function allele of the sun-1 gene. RNAi of sun-1 in a zyg-1 mutant strain also restores centrosome duplication whereas RNAi of zyg-12 does not, indicating that SUN-1 regulates centrosome duplication independently of its role in centrosome–nuclear attachment. Thus, our approach has identified a large number of candidate regulators of the centrosome duplication pathway and has uncovered an unexpected SUN-1-dependent regulatory pathway.
MATERIALS AND METHODS
Worm strains and culture conditions:
Nematode strains carrying the following markers were derived from the wild-type Bristol strain N2: LGI, dpy-5(e61) unc-13(e1091); LGII, lin-31(n301), dpy-25(e817), zyg-1(it25), zyg-1(or409), bli-2(e768), dpy-10 (e128), unc-4(e120), and unc-53(n569); LGIII, unc-93 (e1500), dpy-17(e164), unc-32 (e189), dpy-18(e364), unc-25(e156), and unc-64(e246); LGV, dpy-11(e224), sma-1(e30), and unc-76(e911); LGX, daf-3(e1376) and lon-2(e678). Genetic analysis was performed using the following gfp-marked balancer chromosomes: hT2[bli-4(e937) qIs48] (I:III), mIn1[dpy-10(e128) mIs14] II, and nT1[qIs51] (IV;V). Each balancer chromosome was marked with the same three fusion constructs: myo-2∷gfp, pes-10∷gfp, and a gut enhancer fused to the gfp gene. An integrated egl-15∷gfp transgene (ayIS2 IV) was used to mark suppressor heterozygotes, and the wild-type Hawaiian variant CB4856 was used for single-nucleotide polymorphism (SNP) mapping.
Worms were cultured on NGM, modified Youngren's, only Bacto-peptone (MYOB) (Church et al. 1995), or high growth media seeded with Escherichia coli strain OP50. Strains were maintained at 16° or 20° and tested for suppression at 23.5°, 24°, or 25°. Incubator temperature was checked periodically with a high precision temperature probe and maintained within 0.2° of the set point. Tests for suppression were carried out with positive and negative controls and, where possible, all controls carried the same morphological markers as the test strains.
Suppressor screen:
Worms were mutagenized on three separate occasions as follows. Mixed-stage cultures of zyg-1(it25) II; daf-3(e1376) lon-2(e678) X worms were washed off plates with M9 buffer and treated in suspension with 40 mm ethyl methanesulfonate (EMS) as described by Brenner (1974). The inclusion of the daf-3 lon-2 chromosome prevented animals from entering dauer diapause and thus allowed us to screen at high worm density. Following EMS treatment, 600 Po L4 larvae were picked, 25 per plate, to 24 100-mm NGM plates and incubated at 16° until the majority of F1 individuals became gravid. Each plate (or pool) was then processed individually as follows. The worms were washed off with water and transferred to a 15-ml conical tube. To determine the number of haploid genomes screened in each pool, a sample of the worm suspension was removed and the number of gravid F1 hermaphrodites counted. From this number we estimated the total number of gravid F1 worms and doubled the number to arrive at the number of haploid genomes. Worms were collected from the remainder of each F1 worm suspension by centrifugation and a pool of F2 eggs was then isolated by treating the worm suspension with 1 ml of 1% hypochlorite, 0.5 m NaOH for 5 min at room temperature in a microfuge tube. After all adults and larvae were dissolved, intact embryos, which are resistant to hypochlorite owing to the presence of an egg shell, were recovered by centrifugation at 4000 rpm for 3 min. The embryos were washed one to two times with 1 ml M9 buffer and then distributed between two 100-mm high growth plates. The embryos were allowed to hatch overnight at 16° and shifted to 24° the next day. Plates were incubated between 3 and 6 weeks and examined periodically for viable lines. To ensure independence, only one suppressor was isolated from each pool. All initial isolates were maintained at 20°, and those that grew reproducibly upon retesting at 23.5° or 24° were selected for further analysis.
Genetic analysis:
Before analysis, strains were backcrossed at least twice to the original zyg-1(it25) line to remove any extraneous mutations produced during EMS treatment. To quantify suppression, L4 hermaphrodites from each backcrossed line were picked to individual 35-mm MYOB plates and incubated at either 23.5° or 24°. Approximately 24 hr later, hermaphrodites were removed and the plates were returned to 24°. Live (larvae) and dead (unhatched eggs) were counted the next day. For each strain, the progeny of 4–10 hermaphrodites were analyzed. In an identical manner, we tested zyg-1(it25) animals heterozygous for each suppressor. Suppressor heterozygotes were generated by mating suppressor-bearing zyg-1(it25) hermaphrodites to zyg-1(it25); ayIs2 IV males. Outcross L4 hermaphrodite progeny [genotype zyg-1(it25); szy/+; ayIs2/+] were identified on the basis of the presence of the egl-15∷gfp marker. For sun-1(bs12) and szy-20(bs52), we found that embryonic viability was higher during the second 24-hr period at elevated temperature. We report these values in Figure 1C and Table 1.
Figure 1.—
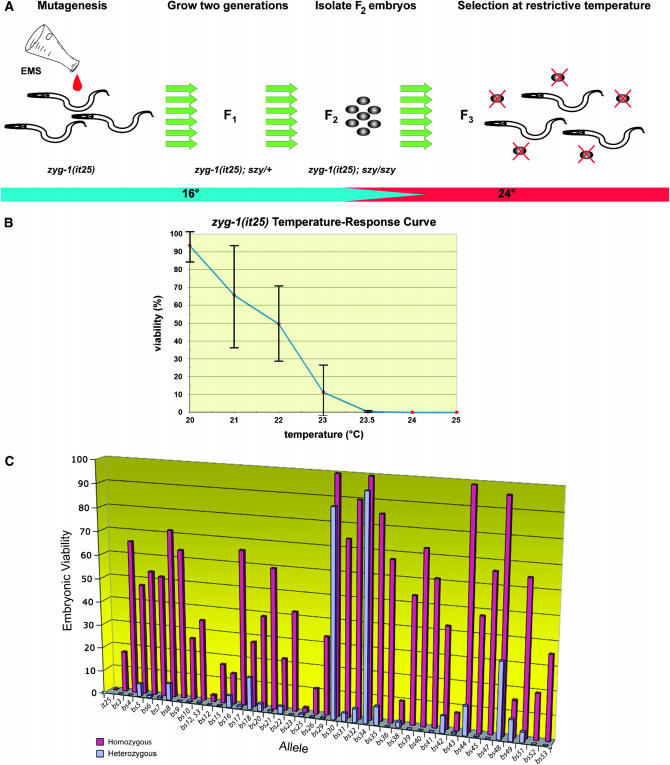
Suppressor screen. (A) To identify mutations that restore centrosome duplication and viability to a strain compromised for zyg-1 function, animals carrying the temperature-sensitive mutation zyg-1(it25) were treated with EMS. At the permissive temperature, EMS-induced germ-line szy mutations are transmitted to the F1 generation in the heterozygous state (szy/+) and to the F2 generation in the homozygous state (szy/szy). To identify szy-bearing individuals, F2 embryos were isolated, allowed to complete larval development at permissive temperature, and then shifted to restrictive temperature for 3–6 weeks. szy-bearing lines give rise to multiple generations over this time period while all other individuals die off. The columns of green arrows indicate that individual pools of mutagenized lines were processed in parallel. To ensure independence, only one suppressor was retained from each pool. (B) Percentage of embryonic viability of a zyg-1(it25) strain as a function of temperature. Each data point represents the average percentage of viability among the offspring of four to five individuals shifted as L4 larvae to the indicated temperature for 24 hr. The vertical bars indicate the standard deviation. Note that the largest standard deviations are observed at the intermediate temperatures of 21°–23°. (C) Potency of szy alleles. Each szy allele in the homozygous (red) or heterozygous (blue) state was assayed for suppression of zyg-1(it25) embryonic lethality. Shown is the average percentage of viable offspring. Numerical values including standard deviations and the number of embryos scored are listed in Table 1 along with assay conditions and complete strain genotypes.
TABLE 1.
Genetics of suppression
|
szy/szy
|
szy/+
|
||||||
|---|---|---|---|---|---|---|---|
| Strain | Temperature | Averagea | SD | Nb | Averagea | SD | Nb |
| Potency of szy alleles in homozygous and heterozygous state | |||||||
| zyg-1(it25) | 23.5° | 0 | 0 | 194 | |||
| zyg-1(it25) | 24° | 0 | 0 | 1852 | |||
| zyg-1(bs8 it25) | 24° | 71.5 | 6.9 | 240 | 7.2 | 7 | 458 |
| zyg-1(bs18 it25) | 24° | 27.6 | 9 | 342 | 13.8 | 13.8 | 321 |
| zyg-1(bs30 it25) | 24° | 99.7 | 0.7 | 253 | 87.5 | 6.6 | 282 |
| zyg-1(bs34 it25) | 24° | 99.6 | 0.8 | 224 | 94.6 | 2.6 | 286 |
| zyg-1(bs44 it25) | 24° | 98.8 | 1.8 | 298 | 12.4 | 8.2 | 267 |
| zyg-1(bs48 it25) | 24° | 95.8 | 2.7 | 272 | 32 | 9 | 208 |
| zyg-1(it25); sun-1(bs12)c | 24° | 2.8d | 2.7 | 1307 | 0d | 0 | 1501 |
| zyg-1(it25); sun-1(bs12) szy-18(bs53) | 24° | 34.6 | 12.5 | 277 | 0.4 | 1.3 | 415 |
| szy-1(bs3); zyg-1(it25) | 24° | 17.2 | 19.7 | 150 | 0 | 0 | 207 |
| zyg-1(it25); szy-2(bs4) | 24° | 65.5 | 11.8 | 283 | 0 | 0 | 427 |
| zyg-1(it25) szy-3(bs5) | 24° | 47.1 | 21.1 | 216 | 5.5 | 5.9 | 301 |
| zyg-1(it25) szy-4(bs6) | 24° | 53.3 | 22.1 | 261 | 0.9 | 1 | 270 |
| zyg-1(it25) szy-4(bs17) | 24° | 65.7 | 6.9 | 214 | 1.1 | 2.1 | 266 |
| zyg-1(it25) szy-4(bs23) | 24° | 42.2 | 31.8 | 210 | 0 | 0 | 211 |
| szy-5(bs7); zyg-1(it25) | 24° | 51.5 | 18.9 | 193 | 0.7 | 1.4 | 284 |
| zyg-1(it25) szy-6(bs9) | 24° | 63.5 | 10.7 | 311 | 0 | 0 | 263 |
| zyg-1(it25) szy-7(bs10) | 24° | 26.4 | 17.7 | 271 | 0.4 | 1.2 | 573 |
| zyg-1(it25) szy-7(bs41) | 24° | 61.1 | 11.2 | 251 | 0 | 0 | 310 |
| zyg-1(it25); szy-8(bs15) | 24° | 16.6 | 10.4 | 176 | 0.21 | 0.66 | 503 |
| zyg-1(it25) szy-9(bs20) | 24° | 38.9 | 14.5 | 343 | 3.1 | 1.2 | 288 |
| zyg-1(it25) szy-9(bs25) | 24° | 2.1 | 1.5 | 290 | 0.8 | 1 | 243 |
| zyg-1(it25) szy-9(bs26) | 24° | 10.8 | 4.3 | 332 | 0.2 | 0.6 | 949 |
| zyg-1(it25) szy-9(bs32) | 24° | 89.9 | 10.1 | 165 | 5.8 | 9 | 337 |
| zyg-1(it25) szy-9(bs40) | 24° | 72.8 | 10 | 405 | 0 | 0 | 299 |
| zyg-1(it25) szy-9(bs45) | 24° | 47.9 | 14.4 | 265 | 0.3 | 0.6 | 350 |
| zyg-1(it25) szy-10(bs21) | 24° | 59.4 | 11.3 | 207 | 0.7 | 0.8 | 295 |
| zyg-1(it25); szy-11(bs22) | 24° | 22 | 24.2 | 494 | 3.1 | 5.2 | 591 |
| zyg-1(it25); szy-12(bs16) | 23.5° | 13.1 | 6 | 376 | 5 | 3 | 543 |
| szy-13(bs29) zyg-1(it25) | 24° | 33.1 | 15.4 | 239 | 0.4 | 0.8 | 285 |
| zyg-1(it25) szy-14(bs31) | 24° | 74 | 12.7 | 279 | 3.5 | 5.4 | 528 |
| zyg-1(it25) szy-14(bs38) | 24° | 9.7 | 10.9 | 404 | 2.5 | 2 | 264 |
| szy-15(bs35); zyg-1(it25) | 23.5° | 85 | 13.2 | 187 | 7.8 | 9.3 | 350 |
| zyg-1(it25) szy-16(bs36) | 24° | 67.3 | 16.9 | 242 | 0 | 0 | 278 |
| zyg-1(it25); szy-17(bs39) | 24° | 53.5 | 25.5 | 177 | 0 | 0 | 303 |
| zyg-1(it25); szy-17(bs42) | 24° | 42.5 | 21 | 340 | 7.2 | 4.3 | 181 |
| zyg-1(it25); szy-17(bs43) | 24° | 7.4 | 7.8 | 266 | 0.7 | 0.9 | 676 |
| zyg-1(it25); szy-17(bs47) | 24° | 66 | 27 | 390 | 0 | 0 | 308 |
| zyg-1(it25); szy-17(bs51) | 23.5° | 64.8 | 17.4 | 414 | 4.5 | 2.7 | 520 |
| zyg-1(it25); szy-18(bs53)e | 24° | 35.3 | 5.6 | 295 | 0 | 0 | 584 |
| zyg-1(it25) szy-19(bs49) | 24° | 15.3 | 7.1 | 368 | 9 | 2 | 269 |
| zyg-1(it25) szy-20(bs52) | 24° | 19.2d | 14.5 | 552 | 0d | 0 | 484 |
| Suppression on control plates
|
Suppression on RNAi plates
|
||||||
| Strain | Temperature | Averagea | SD | Nb | Averagea | SD | Nb |
| Test for zyg-1 bypass suppression of szy alleles | |||||||
| zyg-1(it25) | 24° | 0.04 | 0.2 | 4258 | 0 | 0 | 4038 |
| zyg-1(it25); sun-1(bs12) szy-18(bs53) | 24° | 45.7 | 5.9 | 583 | 0 | 0 | 438 |
| szy-1(bs3); zyg-1(it25) | 24° | 32.5 | 20.5 | 276 | 0 | 0 | 301 |
| zyg-1(it25); szy-2(bs4) | 24° | 71.3 | 28.7 | 606 | 0 | 0 | 536 |
| zyg-1(it25) szy-3(bs5) | 24° | 6.0 | 4.2 | 456 | 0 | 0 | 565 |
| zyg-1(it25) szy-4(bs6) | 24° | 93.8 | 4.7 | 339 | 0 | 0 | 540 |
| zyg-1(it25) szy-4(bs17) | 23.5° | 98.5 | 2.0 | 381 | 0 | 0 | 193 |
| zyg-1(it25) szy-4(bs23) | 24° | 46.6 | 29.1 | 3950 | 0 | 0 | 132 |
| szy-5(bs7); zyg-1(it25) | 24° | 16.9 | 11.5 | 238 | 0 | 0 | 504 |
| zyg-1(it25) szy-6(bs9) | 24° | 42.7 | 7.1 | 548 | 0 | 0 | 554 |
| zyg-1(it25) szy-7(bs10) | 24° | 60.0 | 11.0 | 358 | 0 | 0 | 307 |
| zyg-1(it25) szy-7(bs41) | 24° | 26.0 | 18.1 | 363 | 0 | 0 | 439 |
| zyg-1(it25); szy-8(bs15) | 24° | 36.2 | 13.1 | 198 | 0 | 0 | 197 |
| zyg-1(it25) szy-9(bs20) | 24° | 17.2 | 9.5 | 429 | 0 | 0 | 482 |
| zyg-1(it25) szy-9(bs25) | 24° | 32.4 | 21.8 | 330 | 0 | 0 | 445 |
| zyg-1(it25) szy-9(bs26) | 24° | 9.6 | 8.6 | 355 | 0 | 0 | 522 |
| zyg-1(it25) szy-9(bs32) | 24° | 38.6 | 8.5 | 273 | 0 | 0 | 343 |
| zyg-1(it25) szy-9(bs40) | 24° | 21.6 | 6.2 | 236 | 0 | 0 | 412 |
| zyg-1(it25) szy-9(bs45) | 24° | 29.7 | 7.9 | 384 | 0 | 0 | 612 |
| zyg-1(it25) szy-10(bs21) | 24° | 4.9 | 6.7 | 483 | 0 | 0 | 218 |
| zyg-1(it25); szy-11(bs22) | 23.5° | 86.8 | 7.6 | 301 | 0 | 0 | 436 |
| zyg-1(it25); szy-12(bs16) | 24° | 31.3 | 9.8 | 222 | 0 | 0 | 403 |
| szy-13(bs29) zyg-1(it25) | 24° | 49.3 | 21.6 | 374 | 0 | 0 | 501 |
| zyg-1(it25) szy-14(bs31) | 24° | 37.3 | 16.1 | 449 | 0.5 | 0.9 | 412 |
| zyg-1(it25) szy-14(bs38) | 24° | 23.0 | 25.1 | 907 | 0 | 0 | 472 |
| szy-15(bs35); zyg-1(it25) | 24° | 44.8 | 22.2 | 353 | 0 | 0 | 481 |
| zyg-1(it25) szy-16(bs36) | 24° | 24.1 | 1.6 | 414 | 0 | 0 | 297 |
| zyg-1(it25); szy-17(bs39) | 24° | 24.1 | 12.4 | 268 | 0 | 0 | 187 |
| zyg-1(it25); szy-17(bs42) | 24° | 14.5 | 10.4 | 401 | 0 | 0 | 308 |
| zyg-1(it25); szy-17(bs43) | 23.5° | 11.4 | 9.9 | 568 | 0.4 | 0.8 | 497 |
| zyg-1(it25); szy-17(bs47) | 24° | 22.8 | 1.8 | 472 | 0 | 0 | 404 |
| zyg-1(it25); szy-17(bs51) | 24° | 18.2 | 5.2 | 413 | 0 | 0 | 484 |
| zyg-1(it25) szy-19(bs49) | 24° | 13.2 | 8.0 | 455 | 0 | 0 | 588 |
| zyg-1(it25) szy-20(bs52) | 24° | 57.9 | 44.9 | 282 | 0 | 0 | 265 |
Average percentage of embryonic viability.
Number of embryos tallied.
The complete genotype of the strain tested was zyg-1(it25); sun-1(bs12) szy-18(bs53) +/sun-1(bs12) + unc-76(e911). The congenic strain zyg-1(it25); sun-1(bs12) szy-18(bs53) unc- 76(e911)/+ + + served as a negative control and produced 0 ± 0 viable progeny (n = 1501).
Measurement made 24–48 hr after shift to restrictive temperature. All other values reported in the top were measured during the first 24 hr after the shift to restrictive temperature.
The complete genotype of the strain tested was zyg-1(it25); sma-1(e30) + szy-18(bs53)/+ sun-1(bs12) szy-18(bs53). The strain zyg-1(it25); sun-1(bs12) szy-18(bs53) unc-76(e911)/+ + + served as a negative control and produced 0 ± 0 viable progeny (n = 1501).
To assign each suppressor to a chromosome, we first tested for linkage to zyg-1 (chromosome II) using standard genetic methodology (Brenner 1974). Suppressors that did not show linkage to zyg-1 were mapped to one of the other chromosomes using the snip–SNP mapping technique (Wicks et al. 2001). To SNP map suppressors, we created OC118, a zyg-1(it25) Hawaiian congenic strain by backcrossing the zyg-1(it25) line to strain CB4856 10 successive times. Analysis of SNPs in OC118 demonstrated that all chromosomes were of Hawaiian origin except for a small region surrounding the zyg-1 locus. To map, we crossed OC118 males with zyg-1(it25); szy hermaphrodites. F1 outcross hermaphrodites were allowed to produce an F2 generation and F2 progeny were scored individually for the presence of the suppressor. Equal numbers of Szy-positive and Szy-negative F2 individuals were then analyzed for a set of 18 SNP markers (three per chromosome: left arm, right arm, and center) by bulked segregant analysis (Wicks et al. 2001). Suppressors were further localized to a specific genetic interval on each chromosome using standard three-factor mapping techniques (Brenner 1974). As many of the morphological markers used for mapping made scoring for suppression difficult, we often tested for the presence of a recessive suppressor by backcrossing marked recombinants to the original zyg-1(it25); szy line and scoring unmarked F1 for suppression.
In the course of our mapping experiments, we discovered that one of our lines carried two genetically linked suppressors: sun-1(bs12) and szy-18(bs53). In the zyg-1(it25) strain, the sun-1(bs12) szy-18(bs53) chromosome conferred robust suppression and, in an otherwise wild-type background, a temperature-sensitive embryonic lethal phenotype. At a cytological level, this chromosome conferred two defects: loss of close association between the centrosome and the nucleus and an S-phase delay. On the basis of the genetic map position of the suppressor and the detached centrosome phenotype we decided to sequence the sun-1 locus on this chromosome and identified a single-base substitution. As loss of sun-1 activity does not cause an S-phase delay (Malone et al. 2003), we postulated the existence of a second suppressor. Fortuitously, the sun-1(bs12) mutation was found to disrupt an AciI site and thus could be followed in crosses using snip–SNP technology. To separate the two suppressors, we isolated Sma-nonUnc and Unc-nonSma recombinants from a parental strain of genotype zyg-1(it25) II; sma-1(e30) + + unc-76(e911)/+ sun-1(bs12) szy-18(bs53) +. Analysis of the recombination data confirmed the existence of two suppressors and revealed that the szy-18(bs53) mutation segregated with relatively strong suppression and embryonic lethality marked by the S-phase delay while the sun-1(bs12) mutation segregated with weaker suppression, no embryonic lethality, and the detached centrosome phenotype.
Complementation tests were employed to determine if recessive and weakly semidominant mutations that mapped to a common genetic interval were allelic. In all tests, males of genotype zyg-1(it25); szy were mated to zyg-1(it25); szy hermaphrodites carrying a morphological marker that conferred a Dpy, Unc, or Egl phenotype. For each test, four unmarked F1 progeny were picked to a 35-mm plate and incubated at 24° for 2 days. Hermaphrodites were removed and the next day the number of live progeny on the plate was counted. All appropriate zyg-1(it25); szy parental controls were tested in parallel. Two suppressors were scored as noncomplementing when the number of viable progeny on the test plate was equal to or greater than the number of viable progeny found on either parental control plate.
To remove the zyg-1(it25) mutation from the suppressor lines, we used one of the following strategies. For unlinked suppressors, we crossed males heterozygous for one of the gfp-marked balancer chromosomes with zyg-1(it25); szy hermaphrodites to create F1 progeny of genotype zyg-1(it25)/zyg-1(+); szy/balancer∷gfp. From these hermaphrodites, two or more zyg-1(+); szy/balancer∷gfp lines were identified. Non-green (szy/szy) progeny were then isolated from each line. To separate suppressors that mapped to the right of dpy-10 on chromosome II, we crossed zyg-1(it25) szy hermaphrodites with dpy-10(e128) unc-4(e120)/++ males and selected zyg-1(it25) + szy +/+dpy-10(e128) + unc-4(e120) F1 progeny. These were picked individually to 35-mm plates and allowed to produce progeny. From this generation we picked Dpy-non-Unc recombinants and isolated lines homozygous for the recombinant chromosome [possible genotypes dpy-10(e128) or dpy-10(e128) szy]. As the probability of recovering a line carrying a given szy mutation was <100%, we isolated and analyzed at least six independent recombinant lines per suppressor. Where possible, suppressor lines were examined for the presence of a number of different phenotypes at 16°, 20°, and 25°. These phenotypes include embryonic lethality, larval lethality, and a high incidence of males (Him) phenotype, as well as anatomical defects. Lines marked with the dpy-10(e128) mutation could not be scored at 16° due to a partially penetrant cold-sensitive embryonic lethal phenotype associated with this morphological marker.
To test for allele specificity among the unlinked suppressors, we crossed males of genotype zyg-1(or409) unc-4(e120)/++; balancer∷gfp/+ with szy hermaphrodites. Several F1 Gfp-positive progeny were picked to individual plates and allowed to produce progeny. From these, zyg-1(or409) unc-4(e120)/++; szy/balancer∷gfp hermaphrodites were identified and several F2 Gfp-negative Unc progeny [genotype zyg-1(or409) unc-4(e120); szy] were picked and used to establish a line. As a control, in parallel we reintroduced the same suppressor mutations into a zyg-1(it25) unc-4(e120) strain using the same strategy. To test for suppression, four L4 larvae from each strain were transferred in parallel to individual 35-mm plates and the plates were incubated at 24° for 2 days. Each plate was then scored for the presence of viable progeny.
Bypass suppression was tested by removing any residual zyg-1 activity in each zyg-1(it25) szy strain using an RNAi feeding protocol (Timmons and Fire 1998). For each zyg-1(it25) szy line, L2 larvae were transferred to MYOB plates containing 1 mm IPTG and 25 mg/ml carbenicillin and seeded with E. coli HT115(DE3) pCK13. These plates were incubated at 20° until larvae reached the L4 stage at which time the plates were transferred to 24°. To assess the ability of each szy allele to bypass zyg-1, we determined the percentage of viable progeny produced during the period of maximal inactivation of zyg-1 (48–72 hr after initial exposure to dsRNA). Controls not subjected to RNAi were processed in parallel.
Molecular biology:
To create the zyg-1(RNAi) plasmid pCK13, we amplified a 732-bp fragment of zyg-1 cDNA with the primers 5′-gaagatctaaaggtggattcggcgttgta-3′ and 5′-gaagatctagtgttctctcgagaagattaccgc-3′. The amplified fragment was digested with BglII and cloned into the BglII site of the RNAi feeding vector L4440 (Timmons and Fire 1998). For RNAi of zyg-12 and sun-1, we used the corresponding clones from the RNAi feeding library (MRC Gene Service). Amplified genomic DNA was analyzed by automated fluorescent dye-terminator sequencing on an ABI Prism 3730xl sequencer (Seqwright).
Antibodies, immunostaining, and microscopy:
DM1A, an anti-α-tubulin monoclonal antibody, was obtained from Sigma (St. Louis). The affinity-purified rabbit anti-SPD-2 polyclonal antibody was described previously (Kemp et al. 2004). The ZYG-1 antibody was raised in rabbits against a purified fusion protein consisting of ZYG-1 amino acids 201–402 fused to maltose-binding protein. ZYG-1-specific antibodies were affinity purified using a second fusion protein consisting of the same portion of ZYG-1 fused to glutathione S-transferase. This antibody detects ZYG-1 at centrioles throughout the cell cycle with highest levels at anaphase in agreement with published results (O'Connell et al. 2001; Dammermann et al. 2004; Delattre et al. 2006). The specificity of this antibody in immunofluorescence microscopy experiments was confirmed by staining embryos depleted of ZYG-1 by RNAi. Such embryos showed a reproducible and significant reduction in centrosome staining (data not shown). All primary antibodies were used at either 1:500 or 1:1000 dilutions. Alexa fluor 488 goat anti-rabbit and 568 goat anti-mouse secondary antibodies were from Molecular Probes (Eugene, OR). Each was used at a 1:1000 dilution. Immunostaining, spinning-disk confocal microscopy, and four-dimensional differential interference contrast (4D-DIC) microscopy were as described previously (O'Connell 2000; O'Connell et al. 2000; Kemp et al. 2004) and utilized a Plan Apo 100× 1.4-NA lens. For confocal microscopy we used a Nikon Eclipse E800 microscope equipped with a Perkin-Elmer (Norwalk, CT) UltraVIEW spinning disk unit and a Hamamatsu C9100-12 EM-CCD camera. Confocal images were acquired using Openlab software and gain, offset, and gamma adjustments were made with Photoshop. For 4D-DIC microscopy, we used IPLab software to acquire images from a Zeiss Axiovert 200M microscope equipped with a Hamamatsu ORCA-ER camera.
RESULTS
Identification of zyg-1 suppressors:
In the C. elegans embryo, loss of zyg-1 gene activity results in a failure of centrosome duplication (O'Connell et al. 2001). As a consequence, bipolar spindles are not assembled, DNA is not properly segregated, cytokinesis fails, and the embryos die. ZYG-1 is distantly related to vertebrate PLK4, a kinase that is also required for centriole replication (Bettencourt-Dias et al. 2005; Habedanck et al. 2005). As key regulators of duplication, the activity of these kinases is likely stringently controlled. To identify factors that interact with zyg-1 to regulate centrosome duplication we designed a sensitive genetic suppressor screen to identify mutations that restore normal centrosome duplication to an embryo deficient in zyg-1 activity (Figure 1A). Animals homozygous for the temperature-sensitive partial loss-of-function allele zyg-1(it25) appear wild type at 16° but exhibit a fully penetrant embryonic lethal phenotype at 25° (Kemphues et al. 1988a). The mutant form of the protein encoded by zyg-1(it25) contains a single-amino-acid substitution (P442L) in the nonkinase portion of ZYG-1 (Figure 3A), but still localizes to centrosomes (Figure 4A). To identify suppressors, we used EMS to induce germ-line mutations in a population of zyg-1(it25) animals, grew this population for two generations at permissive temperature to allow any suppressor mutations to become homozygous, and then shifted the population to the restrictive temperature to select for those suppressor-bearing individuals. There were three key features of our experimental design. First, to make screening as efficient as possible, we sought to minimize the number of animals screened without reducing the complexity of the pool. We reasoned that since each F1 mother produces many progeny carrying the same EMS-induced mutation, we needed to assay only a small fraction of progeny. Thus, we treated the F1 population with hypochlorite to kill all animals except for the small clutch of F2 eggs present in each uterus. Second, to make our method as sensitive as possible we screened at the lowest possible temperature at which 100% of zyg-1(it25) embryos die. This temperature was found to be 24° (Figure 1B). At this temperature, the mutants still possess residual zyg-1 activity but this level falls just short of that necessary to sustain viability. Third, we selected for suppressors over an extended period of time (the equivalent of 6–12 life cycles). We reasoned that this should reduce the number of false positives as suppressor-bearing lines would be required to survive over multiple generations. This feature would also allow us a greater chance of identifying suppressors with a growth defect, due either to weak suppression or to a deleterious effect caused by the suppressor mutation itself.
Figure 3.—
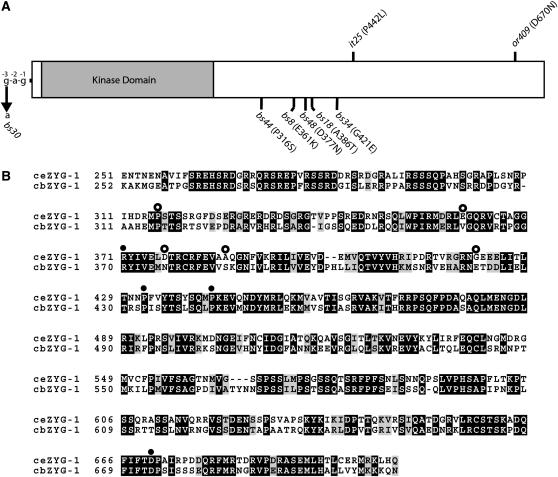
Intragenic zyg-1 suppressors. (A) Schematic of the ZYG-1 protein showing the positions of the zyg-1(it25) and zyg-1(or409) mutations and the six intragenic suppressors. Five suppressor mutations are within the coding region and the presumptive amino acid substitutions are indicated. The bs30 mutation is a G-to-A transition 3 bp upstream of the initiator methionine codon. (B) Amino acid sequence alignment of the nonkinase portions of ZYG-1 from C. elegans and C. briggsae. Identical and similar amino acid residues are shown as solid and shaded areas, respectively. Solid circles indicate the positions of the four known loss-of-function mutations. Open circles indicate the positions of the five suppressor mutations that produce an amino acid substitution.
Figure 4.—
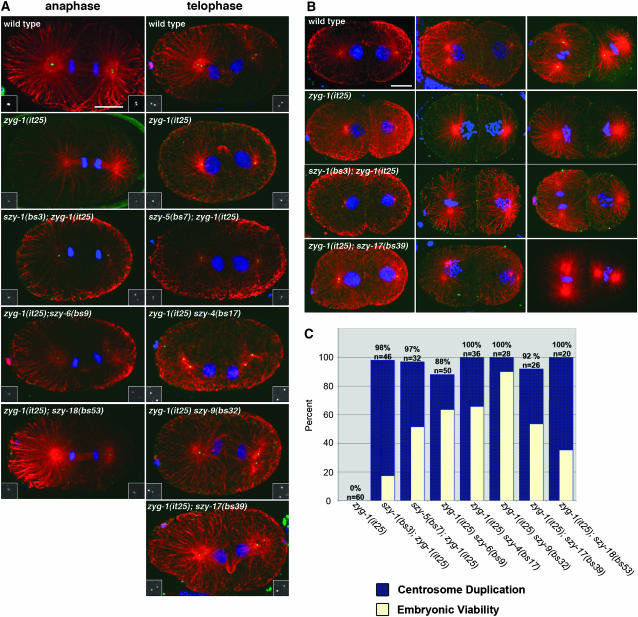
Centrosome duplication is restored in zyg-1(it25); szy mutants. (A and B) Images of wild-type, zyg-1(it25), and zyg-1(it25); szy double mutants stained for tubulin (red), DNA (blue), and ZYG-1 (green). (A) Embryos are in anaphase (left column) or telophase (right column). Insets show ZYG-1 staining at spindle poles. Note that at telophase zyg-1(it25) embryos have only a single dot of ZYG-1 at each pole while all other strains clearly have two, indicating successful centrosome duplication. Bar, 10 μm. (B) Microtubule-organizing activity is similar during the second cell cycle in the wild-type and zyg-1(it25); szy double mutants. Shown are images of wild-type, zyg-1(it25), and zyg-1(it25); szy embryos at early (left column), mid (center column), and late (right column) points of the second cell cycle. In wild-type and zyg-1(it25); szy double mutants microtubule-organizing centers appear similar in size, number, and position, indicating that the normal process of centrosome duplication has been restored. Bar, 10 μm. (C) Quantification of centrosome duplication in zyg-1(it25) and zyg-1(it25); szy double mutants. Centrosome duplication was assessed for each strain by scoring two-cell stage blastomeres in late prophase for the presence of two centrosomes. Blue columns indicate the percentage of successful duplication events. The actual value is shown above the column with the number of duplication events scored. For comparison, the level of embryonic viability of each strain is shown in yellow.
In a screen of an estimated 314,000 haploid genomes, we isolated 39 independent mutant lines that could reproducibly grow at 24° despite carrying the zyg-1(it25) mutation. One of these lines contained two genetically linked suppressor mutations that we ultimately separated and characterized independently (see materials and methods). Thus we identified a total of 40 independent suppressors. After backcrossing, each of the suppressor-bearing zyg-1(it25) lines was assayed for the ability to grow at restrictive temperature. We found that these mutations differed markedly in their potency of suppression (Figure 1C and Table 1). A few backcrossed lines exhibited weak suppression and were assayed at 23.5° (noted in Table 1), but most of the mutant lines exhibited robust levels of viability (>20%) at 24°, including four (bs30, bs34, bs44, and bs48) that exhibited wild-type levels of viability. None of the isolated lines, however, contained a reversion of the zyg-1(it25) mutation; when challenged to grow at 25° all of the lines exhibited significant levels of embryonic lethality (our unpublished data). In fact, we were surprised to find that for most suppressors just a 1° increase in temperature (from 24° to 25°) resulted in a significant reduction in suppression. For instance, in the case of zyg-1(it25) animals carrying the bs7 suppressor, ∼50% of the offspring survived at 24° while none survived at 25°. We conclude that the restrictive temperature employed in the screen is a key determinant of stringency and can profoundly affect the results.
Genetic properties of zyg-1 suppressors:
To determine which of our suppressors are dominant and which are recessive, we determined the percentage of viable zyg-1(it25) embryos produced by strains heterozygous for each suppressor. Twenty-one of the 40 suppressors were found to be recessive, although about half of these heterozygotes do allow an occasional embryo to survive. However, in all cases this amounts to <2.0% of the level seen in the corresponding homozygote and thus we deemed this level of suppression insignificant. Of the remaining 19 suppressors, 15 were found to be semidominant; as heterozygotes, these mutations afforded levels of suppression that range between 4.7 and 38.2% of the corresponding homozygous levels. Just four of the suppressors—bs18, bs30, bs34, and bs49—were found to be truly dominant. When heterozygous, these mutations are ≥50% as effective as when homozygous. Thus, the design of our screen allowed the identification of a genetically diverse set of suppressors.
Suppressors can work through a variety of mechanisms. Bypass suppressors work by rewiring the process under study so that the gene being suppressed is no longer needed. Other suppressors work by restoring activity to the suppressed gene or, conversely, by lowering the requirement for that gene. With respect to the present study, bypass suppressors would work in a manner that would render centrosome duplication (and suppression) completely independent of zyg-1. For example, some suppressors might activate a centrosome-independent spindle assembly pathway as described in vertebrates (Khodjakov et al. 2000). Alternatively, nonbypass suppressors would work to restore the normal process of duplication utilizing the residual zyg-1 activity present in the zyg-1(it25) mutant. To determine by which mechanism each of our suppressors work, we used RNAi to remove the residual zyg-1 activity present in each suppressor strain and then assayed for suppression. Strikingly, none of these strains were able to grow when residual zyg-1 activity was eliminated (Table 1). Thus, none of the suppressors identified in this screen bypass zyg-1. The most likely explanation for this result is that the ZYG-1-dependent centrosome duplication pathway is indispensable for proper embryonic cell division. These results also indicate that all szy suppressors work by increasing the residual activity of the zyg-1(it25) allele or conversely by reducing the level of zyg-1 needed for successful centrosome duplication.
zyg-1 activity is regulated by a large number of szy genes:
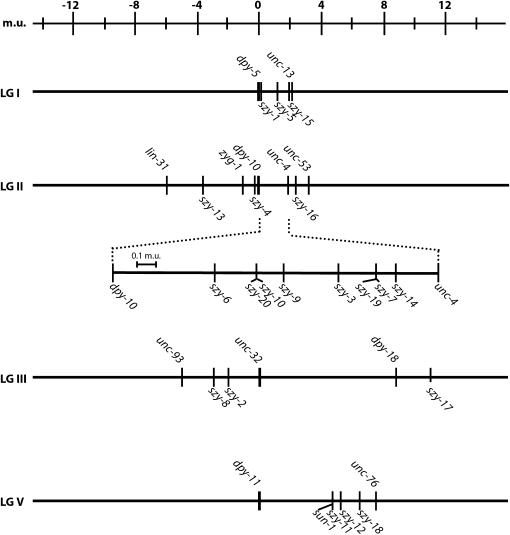
To determine how many genes were represented by this set of suppressor mutations and to position these genes for further study, we determined the genetic map position of each of the 40 mutations. We found that the suppressor mutations are distributed on four of six C. elegans chromosomes (Figure 2). Interestingly, we mapped 26 suppressors to chromosome II within the vicinity of zyg-1, initially leading us to believe that most of the mutations that we had identified were intragenic suppressors. However, this was not the case. Additional mapping placed 16 of these suppressors to the right of dpy-10 and therefore outside of the interval containing the zyg-1 locus. As most of our suppressor mutations do not map near loci known to be required for centrosome replication (spd-2, sas-4, etc.), these mutations appear to define genes not previously implicated in this process.
Figure 2.—
Genetic map showing location of szy genes. Note that the relative ordering of some szy genes within the dpy-10-unc-4 interval is not known with certainty.
Through genetic mapping and genomic sequencing, we determined that 6 of the 40 suppressors are intragenic suppressors. Each of these mutations—bs8, bs18, bs30, bs34, bs44, and bs48—were mapped to within <1 map unit of the zyg-1 locus and all were found to exhibit some degree of dominance as expected for intragenic mutations. We sequenced the entire zyg-1 locus in each of these mutants and in every case identified a unique single-base-pair substitution (Figure 3A). The mutation bs30 results in a G-to-A transition 3 bp upstream of the initiator methionine codon, suggesting that it affects expression of zyg-1. The mutations bs8, bs18, bs34, bs44, and bs48 each result in a predicted single-amino-acid substitution within the C-terminal half of zyg-1 where all previously studied mutations, including it25 and or409, are known to reside. Interestingly, only one of these suppressors, bs44, affects an amino acid residue conserved between C. elegans and the related species C. briggsae (Figure 3B). This is in stark contrast to the four loss-of-function mutations all of which affect conserved residues. This result is not unexpected given that conserved residues are likely critical for function and that most mutagenic changes in this group of residues would render the protein nonfunctional. The positions of suppressor mutations, however, appear to be less constrained, with both conserved and nonconserved residues affected.
To obtain an estimate of the number of genes defined by the 34 extragenic suppressors, we performed complementation tests on all closely linked recessive and semidominant mutations (see materials and methods). Because the bs49 mutation is dominant, we were unable to subject it to complementation analysis with closely linked suppressor mutations. As this mutation is unique in that it was the only fully dominant extragenic suppressor identified, we assigned it as an allele of a distinct gene. Interestingly, we found that our set of zyg-1 suppressor mutations defines a surprisingly large number of genes. In total, our data indicate that this set of mutations comprises 21 distinct genes, which we refer to as szy (suppressor of zyg-1) genes. However, as described below, the bs12 mutation was found to be an allele of the sun-1 gene and thus we utilize the established nomenclature. Sixteen of these genes are defined by a single allele. Of the remaining 5 szy genes, 2 are defined by two alleles, 1 by three alleles, 1 by five alleles, and 1 by six alleles. Given that so many of the szy genes are defined by one allele, we believe the screen is not yet saturated and that additional genes can be mutated to produce a suppressor phenotype.
Having established map positions, we next addressed whether our collection contained allele-specific suppressors. Allele-specific suppressors suppress only one mutant allele of the target gene and are more likely to define genes whose products physically interact with the targeted gene's product. Due to the tight linkage between zyg-1 and 11 of the szy genes, we were unable to test allele specificity in these cases—we were able to separate most of the linked suppressors from the original zyg-1 mutation but we were not able to design an effective strategy to reintroduce a zyg-1 mutation. We did, however, test at least one allele of each of the 10 unlinked szy genes. Suppressor mutations, once separated from the original zyg-1(it25) mutation, were crossed back into a zyg-1(it25) background, as well as into a zyg-1(or409) background. The zyg-1(or409) allele confers a temperature-sensitive phenotype similar in severity to that of the zyg-1(it25) allele (our unpublished data), yet, at the molecular level, zyg-1(or409) is distinct from zyg-1(it25), resulting in a ZYG-1 protein with an amino acid substitution that differs from zyg-1(it25) (Figure 3). Although we did not quantify the level of suppression in this test, all suppressor mutations tested—szy-1(bs3), szy2-(bs4), szy-5(bs7), sun-1(bs12), szy-8(bs15), szy-12(bs16), szy-11(bs22), szy-15(bs35), szy-(bs39), and szy-18(bs53)—allowed both the zyg-1(it25) and zyg-1(or409) mutants to grow at the restrictive temperature. Thus, alleles of all 10 genes tested do not exhibit allele specificity, suggesting that the majority of mutations identified in this screen are capable of suppressing more than one allele of zyg-1.
szy mutations suppress the centrosome duplication defect of zyg-1(it25) mutants:
We next investigated possible mechanisms whereby the extragenic suppressors restored viability to zyg-1 mutants. One obvious possibility is that the normal process of centrosome duplication is restored. However, it is also possible that other mechanisms are at work. For instance, some szy mutations might allow centrioles to form de novo rather than through the normal templated pathway (Marshall et al. 2001). As mentioned above, some suppressors could also function by activating a centrosome-independent spindle assembly mechanism. To address these issues, we analyzed spindle assembly in wild-type, zyg-1(it25), and zyg-1(it25); szy double-mutant embryos. Using immunofluorescence microscopy, we examined microtubule organization and ZYG-1 distribution at all stages between first anaphase and second telophase. In wild-type embryos, ZYG-1 can be detected as a single dot at the poles of the first mitotic spindle until late anaphase when the two centrioles of a pair separate giving rise to two dots (Figure 4A). At each spindle pole, one of the dots represents a sperm-derived centriole while the other dot marks a centriole synthesized during the initial round of centrosome duplication following fertilization. During the second cell cycle of wild-type embryos, ZYG-1 can be detected at the center of each centrosome/spindle pole as either one dot representing a centriole pair prior to anaphase or later two dots representing the separated centrioles (Figure 4B).
In zyg-1(it25) mutants, we found that ZYG-1 could still localize to centrioles (n = 41 embryos). At most cell cycle stages, we found that there were no discernable differences between wild-type and zyg-1(it25) mutants in the centriole levels of ZYG-1. In anaphase, however, most wild-type embryos appeared to possess more centriole-associated ZYG-1 than did the zyg-1(it25) mutants (Figure 4A). This difference appeared transient as the wild type and the mutant invariably possessed similar centriole levels at telophase. zyg-1(it25) embryos also never contained more than a single dot of ZYG-1 at each pole of the first mitotic spindle, indicative of a failure to duplicate the sperm-derived centrioles. In two-cell stage zyg-1(it25) embryos, we continued to detect a single dot of ZYG-1 at the center of each centrosome, but we never observed more than one microtubule-organizing center per blastomere (Figure 4B).
We next analyzed spindle assembly and ZYG-1 distribution in embryos from seven zyg-1(it25); szy double-mutant lines: szy-1(bs3); zyg-1(it25) (n = 28), szy-5(bs7); zyg-1(it25) (n = 26), zyg-1(it25) szy-6(bs9) (n = 22), zyg-1(it25) szy-4(bs17) (n = 30), zyg-1(it25) szy-9(bs32) (n = 19), zyg-1(it25); szy-17(bs39) (n = 19), and zyg-1(it25); szy-18(bs53) (n = 28). For all seven strains, we observed that the first bipolar spindle often contained poles with two ZYG-1 dots (Figure 4A), indicating that the first round of centrosome duplication had been executed properly. As in the wild type, the two ZYG-1 dots first became apparent during late anaphase. At the two-cell stage, bipolar spindles were assembled at a high frequency in all seven lines (Figure 4C). As in the wild type, ZYG-1 staining indicated that all spindles in the double mutants contained centrioles at the poles (Figure 4B and our unpublished observations). We did not observe any evidence of acentriolar spindle formation, nor did we observe any indication that centrioles were arising de novo. The number and position of ZYG-1 positive dots in the zyg-1(it25); szy double mutants were similar to those in wild type during the first (Figure 4A) and second (Figure 4B) cell cycles, suggesting that the normal pathway of centriole replication was being executed. However, in the absence of ultrastructural analysis, we cannot rule out the possibility of de novo centriole formation.
To determine if any of the szy mutations affect the localization of ZYG-1, we compared ZYG-1 staining in the double-mutant lines with that in the wild-type and the zyg-1(it25) line. We found that at all stages examined, all of the zyg-1(it25); szy double mutants possessed centrosome-associated levels of ZYG-1 that were similar to that of the parental zyg-1(it25) line. Thus, for this set of szy mutants, we do not find evidence that suppression occurs as a result of an increase in the centrosome-associated levels of ZYG-1.
szy genes function in a variety of cellular processes:
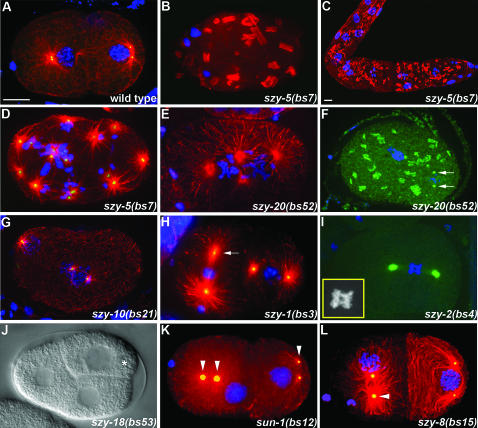
Some suppressor mutations confer phenotypes of their own. This is particularly true in cases where the mutation is in an essential gene. To determine if any of our suppressor mutations confer phenotypes on their own, we constructed zyg-1(+) derivatives of 30 of the 34 extragenic suppressors—the szy-4 and szy-13 alleles were too tightly linked to zyg-1 to remove the zyg-1(it25) mutation. For each suppressor, multiple independent zyg-1(+) derivatives were analyzed for growth defects at 25°, 20°, and where possible 16°. Interestingly, 12 of the 30 suppressors exhibit an observable phenotype (Table 2). Eight of the suppressors—szy-1(bs3), szy-2(bs4), szy-5(bs7), szy-8(bs15), szy-10(bs21), szy-15(bs35), szy-20(bs52), and szy-18(bs53)—were found to exhibit an embryonic lethal phenotype. In addition, the szy-1(bs3) mutant exhibits a partially penetrant larval arrest phenotype and a Him phenotype. Likewise the szy-5(bs7), szy-11(bs22), szy-17(bs51), and szy-20(bs52) mutants each possess a Him phenotype. The Him phenotype arises due to meiotic chromosome segregation defects that result in loss of the sex (X) chromosome and the production of X/O male progeny. Finally, we found that both alleles of the szy-14 gene—bs31 and bs38—confer a slow growth phenotype that appears to be due to smaller than normal brood sizes. Surprisingly, the phenotypes exhibited by most of these mutants are temperature sensitive; for szy-2(bs4), szy-5(bs7), szy-8(bs15), szy-15(bs35), szy-20(bs52), and szy-18(bs53), significantly higher levels of embryonic lethality were observed at 25° than at lower temperature. Likewise, the Him phenotypes of szy-11(bs22) and szy-17(bs51) were observed only at 25° and the slow growth phenotypes of the szy-14 alleles were found to be most severe at 25°. Conditional alleles are relatively rare and thus it was surprising to identify so many temperature-sensitive mutations.
TABLE 2.
Phenotypes of szy alleles
| Plate phenotypea
|
||||||
|---|---|---|---|---|---|---|
| Strain | Temperature | Defect | % Embb | SDc | Nd | Cytological defects |
| sun-1(bs12)e | 25° | None | 1.2 | 1.1 | 772 | Detached centrosomes |
| szy-1(bs3) | 20° | Emb, Him, Lva | 45.1 | 12.4 | 1099 | Detached centrosomes, elongated centrosomes |
| szy-2(bs4) | 25° | Emb ts | 61.8 | 14.9 | 632 | Anaphase bridging of chromatin |
| szy-5(bs7) | 25° | Emb ts, Him | 74.1 | 14.7 | 174 | Complex phenotype: abnormal microtubule cytoskeleton, supernumerary centrosomes and chromosomes, detached centrosomes |
| szy-8(bs15) | 20° | Emb ts | 10.8 | 10.8 | 512 | Detached centrosomes |
| szy-10(bs21)f | 25° | Emb ts | 81.3 | 11.7 | 104 | Supernumerary centrosomes and chromosomes |
| szy-11(bs22) | 25° | Him | NDg | ND | ND | ND |
| szy-14(bs31)f | 25° | Slow growth | ND | ND | ND | ND |
| szy-14(bs38)f | 25° | Slow growth | ND | ND | ND | ND |
| szy-15(bs35) | 25° | Emb ts | 54.9 | 14.2 | 280 | Consistent defect not detected |
| szy-18(bs53)h | 25° | Emb ts | 93.8 | 7.9 | 536 | Abnormal cell cycle timing |
| szy-17(bs51) | 25° | Him | ND | ND | ND | ND |
| szy-20(bs52)f | 25° | Emb ts, Him | 54.3 | ND | 186 | Complex phenotype: abnormal microtubule cytoskeleton, abnormal cell cycle, detached centrosomes, defective cytokinesis |
Phenotypes scored on plates with a dissecting microscope include embryonic lethality (Emb), high incidence of males (Him), larval arrest (Lva), and slow growth. Temperature-sensitive (ts) designation indicates the phenotype was significantly more severe or more fully penetrant at 25°.
Average percentage of embryos that fail to hatch.
Standard deviation.
Number of embryos tested.
Complete strain genotype: zyg-1(it25)/+; sun-1(bs12) + unc-76(e911)/sun-1(bs12) szy-18(bs53) +.
All contained the dpy-10(e128) marker.
Not done.
Complete strain genotype: zyg-1(it25)/+; sma-1(e30) + szy-18(bs53)/+ sun-1(bs12) szy-18(bs53).
It is possible that zyg-1(it25) and one or more of the szy mutations exhibit mutual suppression. That is, not only would a szy mutation suppress zyg-1(it25) defects, but also the zyg-1(it25) mutation would suppress szy defects. A comparison of the numbers reported in Tables 1 and 2 would seem to suggest that animals carrying szy-5(bs7), szy-10(bs21), szy-15(bs35), or szy-18(bs53) all grow better if the zyg-1(it25) mutation is also present. However, one should be careful to note that these zyg-1(it25); szy double mutants were assayed for growth at 23.5° and 24° (Table 1), while the corresponding szy single mutants were assayed at 25° (Table 2). To assess the ability of the zyg-1(it25) mutation to suppress a szy mutation the single and double mutants need to be assayed at the same temperature. Indeed when the szy-5(bs7) mutant is assayed at 24°, rather than at 25°, we find that the level of embryonic lethality drops to 31.4 ± 18.2% (n = 262). Thus at this temperature, 68.6% of szy-5(bs7) embryos survive. In comparison, only 51.5% of szy-5(bs7); zyg-1(it25) embryos survive at 24° (Table 1). Therefore at 24° we find no evidence of mutual suppression. Nonetheless, it is still possible that zyg-1(it25) suppresses one or more of the szy mutations including szy-5(bs7), but such a determination will require growth comparisons over a range of temperatures.
The embryonic lethal phenotypes associated with some of the szy mutations suggest that the corresponding genes are important for normal embryonic development. To determine what roles these genes play, we cytologically examined lines carrying the embryonic lethal mutations by immunostaining gonads and embryos for tubulin, DNA, and centrosomes. Most of these suppressor mutants were found to exhibit clearly observable phenotypes. The most striking phenotype is that associated with the szy-5(bs7) mutant. In 52% of these embryos (n = 27), tubulin staining revealed the presence of large cytoplasmic structures. In some of the most severely affected szy-5(bs7) embryos, the robust arrays of microtubules observed in wild-type embryos are completely absent, with all tubulin appearing in an aggregated form (Figure 5B). These aggregates were also found throughout the hermaphrodite germ line (Figure 5C). We also observed that 56% (n = 27) of szy-5(bs7) embryos possess cells with an excess of centrosomes and DNA. It is likely that in many cases this phenotype is due to cytokinesis failure as a consequence of the tubulin aggregation defect. However, we found several examples of szy-5(bs7) embryos carrying extra centrosomes and DNA that lacked the tubulin aggregates (Figure 5D). This suggests that szy-5(bs7) mutants possess two independent defects: tubulin aggregation and supernumerary centrosomes/chromosomes. The origin of the extra centrosomes in these szy-5(bs7) embryos is not currently clear. Given that all embryos with an excess number of centrosomes also contained an excess number of chromosomes, the most plausible explanation is a defect in cytokinesis. Live imaging of szy-5(bs7) embryos will be needed to address this issue.
Figure 5.—
szy genes are required for proper cell division. Confocal (A–I, K, and L) or DIC (J) images of szy mutants are shown. Confocal images are maximal-intensity projections of multiple optical sections of embryos/gonads stained for tubulin (red) and/or DNA (blue). Some embryos are also stained for centrosomes (green) with antibodies to SPD-2 (A, D, F–I, K, and L). (A) A wild-type embryo at the beginning of the two-cell stage. (B) A szy-5(bs7) one-cell embryo and (C) proximal gonad showing the tubulin aggregation phenotype. (D) A one-cell szy-5(bs7) embryo and (E) a one-cell szy-20(bs52) embryo each with excess centrosomes and DNA. (F) A one-cell szy-20(bs52) embryo with cytoplasmic aggregates of SPD-2. Arrows indicate centrosomes. (G) A multinucleated szy-10(bs21) embryo with multiple centrosomes. (H) A two-cell szy-1(bs3) embryo with an elongated centrosome (arrow). (I) A szy-2(bs4) embryo with anaphase bridging of chromatin. Inset shows a magnified view of chromatin. (J) A szy-18(bs53) embryo with an unusual three-cell configuration resulting from delayed division of the P1 blastomere (asterisk). (K) Two-cell sun-1(bs12) and (L) szy-8(bs15) embryos displaying detached centrosomes (arrowheads). Bars in A and C, 10 μm.
A variety of severe defects were observed in szy-20(bs52) embryos. Multinucleate cells with supernumerary centrosomes were observed in 32% (n = 22) of szy-20(bs52) embryos (Figure 5E). Live-imaging analysis revealed that this defect arises from cytokinesis failure (our unpublished observations). szy-20(bs52) embryos also display defects in other processes (Table 2). A tubulin aggregation phenotype similar to but less severe than that of szy-5(bs7) embryos was observed in 10% (n = 40) of szy-20(bs52) embryos (Figure 5F). Interestingly, in both szy-5(bs7) and szy-20(bs52) animals, centriole proteins, such as SPD-2 (Figure 5F) and ZYG-1 (our unpublished observations), also appear in aggregate form. In some instances these aggregates colocalize with the tubulin aggregates but in other instances they do not. There are a number of possible explanations for this phenotype. For example, centriole proteins might be aggregating in response to an inappropriately activated centrosome replication pathway or centrosomes might be undergoing fragmentation. Additional molecular, cytological, and genetic analysis should be helpful in distinguishing between the various possibilities.
Like the szy-5(bs7) and szy-20(bs52) mutants, embryos carrying the szy-10(bs21) mutation were found to possess multinucleate cells with supernumerary centrosomes (17%, n = 12 embryos) (Figure 5G). These embryos, however, lack the protein aggregation phenotype seen in the szy-5(bs7) and szy-20(bs52) mutants. As is the case with szy-5(bs7) and szy-20(bs52)mutants, the excess centrosomes of szy-10(bs21) embryos are always accompanied by an excess of chromosomes, suggesting a defect in cytokinesis.
The szy-1(bs3), szy-2(bs4), and szy-18(bs53) mutants each possess a defect that is unique among the szy mutants. In 66% (n = 32) of szy-1(bs3) embryos, some centrosomes were found to exhibit an odd morphological defect. Affected centrosomes have an elongated appearance (Figure 5H). Many spindles were observed that possessed one elongated pole and one normal-looking pole although some spindles with two elongated poles were observed. The szy-2(bs4) mutation affects the separation of chromosomes. In 50% of the mutant embryos (n = 14), we observed blastomeres containing chromatin bridges between the separating sets of anaphase chromosomes (Figure 5I). There is an interesting developmental aspect to this phenotype, as the later divisions seem to be affected more than the earlier divisions. In contrast, the szy-18(bs53) mutation affects cell cycle timing. Wild-type embryos exhibit asynchronous divisions beginning at the two-cell stage where the anterior blastomere AB divides ∼2 min ahead of its posterior sister P1 (Brauchle et al. 2003). We noted that in szy-18(bs53) embryos this asynchrony is exaggerated with P1 dividing on average 10.5 min (n = 6 embryos) after AB. This timing defect results in embryos with an unusual three-cell configuration (Figure 5J). Interestingly, this phenotype has been noted to occur when DNA synthesis is inhibited (Encalada et al. 2000; Brauchle et al. 2003). In such cases, AB and P1 are delayed in S phase, but P1 is delayed to a much greater extent than AB. Similarly, we found that the underlying cause of this phenotype is an S-phase delay (our unpublished data). Further analysis will be needed to determine if the szy-18(bs53) mutation identifies a pathway regulating S phase and its associated events such as DNA synthesis and centriole duplication.
Interestingly, six of the suppressor mutations confer a common phenotype: loss of close association between the centrosome and nuclear envelope. This “detached centrosome” defect was observed in 54% (n = 28) of sun-1(bs12) embryos (Figure 5K), in 64% (n = 11) of szy-8(bs15) embryos (Figure 5L), and in 23% (n = 22) of szy-5(bs7) embryos (our unpublished observations). Embryos carrying the szy-1(bs3), szy-10(bs21), or szy-20(bs52) mutations also occasionally exhibit this defect. In sun-1(bs12) mutant embryos, centrosome detachment was most often observed during the early part of the cell cycle when the microtubule asters organized by the centrosomes were relatively small. This detachment appears to be only temporary as prophase-stage blastomeres typically were found to possess centrosomes and nuclei in close association. Despite this defect, there is no significant embryonic lethality associated with the sun-1(bs12) mutation (Table 2), indicating that continuous association between the nucleus and the centrosome is not essential for viability.
The sun-1 gene is a regulator of centrosome duplication:
As a mechanistic link between centrosome duplication and nuclear association had not been previously established, the identification of genes that participate in linking the centrosome to the nucleus was an unexpected outcome of our screen. To begin to understand the mechanisms that tie centrosome–nuclear attachment to duplication, we set out to molecularly identify one of the suppressors with a detached centrosome phenotype and discovered that the bs12 mutation is an allele of the sun-1 gene. We accomplished this by mapping the bs12 mutation to the right arm of chromosome V between the morphological markers sma-1 and unc-76. Within this 3.18-Mbp region of DNA, the only gene known to have a role in attaching the centrosome to the nucleus is sun-1 (Malone et al. 2003). We therefore sequenced the sun-1 genomic region in the bs12 mutant and found a single G-to-A transition in a conserved residue within the 5′-splice site of the third intron. Translation of the improperly spliced message would be expected to produce a truncated protein lacking a putative transmembrane domain and the conserved C-terminal SUN domain. As noted, sun-1(bs12) is a weak allele with no effect on embryonic viability (Table 2), and thus this mutation likely reduces, but does not eliminate, proper splicing of the sun-1 message.
Our results indicate that SUN-1 is a negative regulator of ZYG-1. Yet, the sun-1(bs12) mutation only marginally affects the viability of the zyg-1(it25) mutant (Figure 1C and Table 1). Given that sun-1(bs12) is a weak allele, the lack of a robust effect might be due to insufficient inactivation of sun-1. To address this, we used RNAi to silence expression of sun-1 in the zyg-1(it25) strain and assayed suppression. Under these conditions we still observed only weak suppression of the embryonic lethal phenotype (3.5%, n = 367). However, under the same conditions, RNAi of sun-1 in wild-type worms resulted in a high level (87%, n = 209) of embryonic lethality. Given that almost 90% of the embryos die due to loss of sun-1 activity, the highest level of suppression one could expect to observe would be ∼10%. Thus, the high level of lethality caused by RNAi of sun-1 precluded us from utilizing embryonic viability as an accurate readout for suppression.
We therefore chose to assay the effect of sun-1(RNAi) directly on centrosome duplication using two approaches. One approach was to stain fixed embryos for tubulin, DNA, and ZYG-1. We analyzed 31 one- and two-cell zyg-1(it25) embryos that had been subjected to sun-1(RNAi). Silencing of sun-1 severely impaired the association of centrosomes and nuclei and led to chromosome segregation defects and aneuploidy. Unfortunately, this made it extremely difficult to accurately assign developmental stages to affected embryos. Nevertheless we found numerous two-cell zyg-1(it25); sun-1(RNAi) embryos with evidence of centrosome duplication (Figure 6A). In most instances, we could clearly detect ZYG-1 staining at the center of these centrosomes, indicating that centrioles were present and that suppression by sun-1(RNAi) involved restoration of centriole replication. In these experiments we also compared the level of ZYG-1 staining between the 31 zyg-1(it25); sun-1(RNAi) embryos and 26 zyg-1(it25) embryos that were mock-RNAi treated (i.e., those that were grown on bacteria carrying only the empty RNAi feeding vector L4440). If suppression is due to an increase in ZYG-1 levels at the centrosome, we might expect that centrioles in zyg-1(it25); sun-1(RNAi) embryos would stain more intensely than controls. However, we found that zyg-1(it25) embryos subject to sun-1(RNAi) did not appear to possess any more centrosome-associated ZYG-1 than controls. In fact, some embryos subject to sun-1(RNAi) appeared to have less ZYG-1 present at centrioles than control embryos (Figure 6A). Thus, we do not find evidence that loss of SUN-1 activity results in elevated levels of ZYG-1 at the centrosome.
Figure 6.—
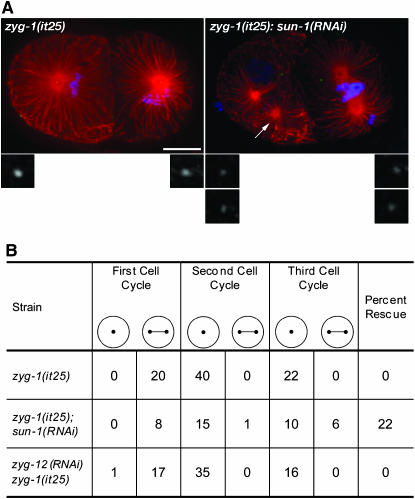
sun-1(RNAi), but not zyg-12(RNAi), suppresses the centrosome duplication defect in zyg-1(it25) mutants. (A) zyg-1(it25) and zyg-1(it25); sun-1(RNAi) embryos were stained for microtubules (red), DNA (blue), and ZYG-1 (green). The zyg-1(it25); sun-1(RNAi) embryo exhibits evidence of centrosome duplication as well as loss of contact between a nucleus and a centrosome (arrow). Insets show threefold magnified views of ZYG-1 staining at centrosomes. Bar, 10 μm. (B) Spindle assembly in zyg-1 embryos depleted of SUN-1 or ZYG-12. Embryos were isolated from mothers grown at 24° and early development was analyzed by 4D-DIC imaging at the same temperature. In columns 2–7, the number of spindle assembly events resulting in either a monopolar (·) or a bipolar (·–·) spindle is tallied for the first three cell cycles. Note that not all embryos were scored for the third cell cycle. In the last column, the percentage of bipolar spindles formed, indicating successful centrosome duplication, is shown.
We used a second approach to more accurately measure the ability of sun-1(RNAi) to suppress the centrosome duplication defect of zyg-1(it25) mutants. Specifically, we used 4D-DIC imaging to follow bipolar spindle formation during the first several cell cycles in zyg-1(it25) and zyg-1(it25); sun-1(RNAi) embryos. In this assay, assembly of a bipolar spindle indicates a successful centriole duplication event during the previous cell cycle, while assembly of a monopolar spindle indicates failure. At 24°, control zyg-1(it25) embryos exhibit a complete block to centriole duplication (Figure 6B). In such embryos, the first spindle is always bipolar (n = 20 events) owing to the separation of the sperm-donated centrioles. However, these centrioles invariably fail to duplicate, resulting in the formation of monopolar spindles in all blastomeres during the second and third cell cycles (n = 62 events). In zyg-1(it25); sun-1(RNAi) embryos the first spindle is also invariably bipolar. However, depletion of sun-1 allowed 7 of 32 blastomeres to assemble a bipolar spindle during the second and third cell cycles. Curiously, the effect of depleting sun-1 on centrosome duplication is more apparent during the third cell cycle than during the second cell cycle (Figure 6B). This could indicate a more significant role for SUN-1 in these later cell cycles or else that less zyg-1 activity is needed later in development. In any event, this result confirms that suppression arises as a result of loss of sun-1 activity, indicating that SUN-1 antagonizes the activity of ZYG-1.
SUN-1 has distinct roles in centrosome duplication and nuclear association:
Our identification of SUN-1 at once suggests the presence of a novel regulatory mechanism governing centrosome duplication. We envision two alternative models. It is possible that SUN-1 in concert with at least some other components of the centrosome–nucleus attachment complex operates in a direct manner to regulate centrosome duplication. In such a scenario, these proteins would be bifunctional, operating to anchor the centrosome to the nucleus and independently to regulate duplication. Alternatively, these proteins might function in an indirect manner to regulate centrosome duplication simply through their ability to maintain association between the centrosome and nucleus. The nucleus might be associated with an inhibitor of duplication, and thus by tethering the centrosome to the nucleus SUN-1 might maintain contact between the centrosome and this inhibitory signal.
The indirect model predicts that any condition that detaches the centrosome from the nucleus will suppress zyg-1 mutations. To test this model we sought to break the nucleus–centrosome connection via other means. In addition to SUN-1, the hook-related protein ZYG-12 is known to be required for proper centrosome–nucleus attachment (Malone et al. 2003). Interestingly, we did not identify alleles of zyg-12 in our screen. This might be due to the lack of saturation in our screen or alternatively that inhibition of zyg-12 does not suppress zyg-1 loss-of-function mutations. To investigate a potential role for ZYG-12 in centrosome duplication, we performed zyg-12(RNAi) in a zyg-1(it25) background and assayed for suppression. Since zyg-12 is an essential gene, we decided to assay the effect of zyg-12(RNAi) on centrosome duplication directly by analyzing spindle assembly in 4D-DIC recordings of zyg-1(it25) embryos depleted of zyg-12 by RNAi. In most of the zyg-12(RNAi) zyg-1(it25) embryos examined (n = 18) we observed a dramatic loss of association between the centrosomes and the nucleus, indicating that we had significantly inhibited zyg-12 activity (our unpublished data). However, all 51 spindle-assembly events recorded during the second and third cell cycles resulted in formation of monopolar spindles (Figure 6B). Thus, despite significant inhibition of zyg-12, the zyg-1(it25) centrosome duplication phenotype is not suppressed. This result demonstrates that suppression does not simply result from freeing centrosomes from the nuclear envelope, indicating that sun-1 functions to regulate centrosome duplication independent of its role in centrosome–nuclear attachment.
DISCUSSION
The isolation and characterization of suppressors provide a powerful approach to uncover the mechanisms regulating centrosome replication. Historically, the application of suppressor genetics led to one of the most important discoveries in the centrosome field. γ-Tubulin, a central player in microtubule nucleation, was first identified in the fungus Aspergillus in a screen for mutations that suppress the lethality of a temperature-sensitive β-tubulin mutation (Weil et al. 1986; Oakley and Oakley 1989). In applying this approach to a zyg-1 mutant, we have identified 21 genes with potentially important roles in regulating centrosome duplication and thus have laid the foundation for future studies aimed at understanding regulatory inputs that provide temporal and spatial control of this process.
A number of the suppressor mutations identified in our screen appear to define essential genes as they are associated with lethal phenotypes. This demonstrates that the design of our screen successfully cleared a major hurdle, as essential genes can be particularly difficult to identify via this approach. On the one hand, strong loss-of-function mutations in essential genes might strongly suppress the zyg-1 duplication defect but cause such a debilitating growth defect of their own that they would be missed. On the other hand, weaker alleles might not cause a significant growth defect but also might not be potent enough to suppress zyg-1 lethality. On the basis of the results, our screen appears sensitive enough to identify both strong and weak alleles of essential genes (Figure 1C and Table 2). For instance, we identified the mutation szy-1(bs3), which while producing a significant level of embryonic lethality of its own still modestly suppresses zyg-1 lethality, and we identified a very weak allele of the essential gene sun-1.
An important issue with suppressor screens is specificity: How many of the suppressors identify genes that have a functionally relevant interaction with the target gene? For instance, collagen mutations have been found to suppress temperature-sensitive glp-1 mutations in C. elegans (Maine and Kimble 1989), and indeed in the course of our work we found that some collagen mutations also provide modest suppression of zyg-1 (our unpublished data). However, on the basis of map position and phenotypic analysis it appears that our screen filtered out this nonspecific class of suppressors. A second line of evidence supporting the specificity of our approach is our cytological analysis, which has demonstrated that many of the szy mutants have centrosome or microtubule-related defects (Figure 5). Third, we have cloned one of these szy genes (sun-1) and found it to be a gene with an established centrosome-related function (Malone et al. 2003). These three lines of evidence argue that many of the genes identified here are specifically involved in the process of centrosome duplication.
Given these arguments for the specificity of our approach, why are there so many szy genes? In fact, on the basis of the large fraction of szy genes defined by a single allele (16 of 21), this screen is not yet saturated and thus more szy genes must exist. One possible explanation for this surprising result might be that there are multiple inputs that regulate zyg-1 activity. For instance, zyg-1 activity might be regulated as a means to coordinate duplication with other cell cycle events, to ensure that centrosomes are not replicated more than once per cell cycle or to prevent the de novo formation of centrioles (Delattre and Gonczy 2004). Thus the large number of szy genes might simply reflect the presence of multiple regulatory circuits that fine tune zyg-1 activity. Alternatively, or in addition, zyg-1 activity might be governed by a large multisubunit complex and loss of activity of any one constituent could compromise zyg-1 regulation.
In light of the fact that many of our szy mutants share at least one phenotype in common—detached centrosomes—it is tempting to speculate that the factors encoded by these genes assemble into a multifunctional complex that anchors the centrosome to the nuclear envelope while also regulating duplication. One of these factors is SUN-1, a member of a conserved family of inner nuclear envelope proteins that play pivotal roles in linking the nucleus to the microtubule or actin cytoskeletons (Starr and Fischer 2005). SUN-domain-containing proteins target adapter proteins such as ZYG-12 (Malone et al. 2003) or the actin-binding protein ANC-1 (Starr and Han 2002) to the outer nuclear envelope. We found, however, that SUN-1-dependent regulation of centrosome duplication does not involve ZYG-12. These results show that the role played by SUN-1 in centrosome duplication is distinct from its role in providing a physical linkage between the centrosome and the nucleus. Recently, Mps3, a homolog of SUN-1, has been shown to be required for duplication of the yeast spindle pole body, an organelle analogous to the animal centrosome (Jaspersen et al. 2006). Our finding differs, as we define a negative role for SUN-1 in centrosome duplication rather than the positive role described in this recent work. This might indicate that SUN-domain-containing proteins play a complex role in this process or else that the function of such proteins has changed over the course of evolution. Further analysis should help clarify this issue.
Nuclear proteins are emerging as important regulators of the centrosome and microtubule cytoskeleton. Spindle assembly factors such as TPX2 and NuMA are sequestered in the nucleus during interphase (Harel and Forbes 2004), and the nucleolar protein nucleophosmin associates with centrosomes and inhibits their duplication (Okuda et al. 2000). These factors are all under the control of the small GTPase Ran, a key regulator of nucleocytoplasmic transport (Harel and Forbes 2004; Wang et al. 2005). RanGTP functions in this capacity by antagonizing the activity of importins that bind to and inhibit TPX2 and NuMA. Also, RanGTP in a complex with the export receptor Crm1 promotes association of nucleophosmin with centrosomes. Intriguingly, disruption of the Ran/importin regulatory cascade in C. elegans results in some phenotypes that are similar, though not identical, to those observed in our screen (Askjaer et al. 2002). These include detached centrosomes, chromatin bridges, and P1 cell cycle delays. However, the map position of most szy genes does not coincide with that of Ran or its known effectors and thus if Ran is involved in ZYG-1 regulation, some szy genes might represent a new class of Ran effectors.
The identification of szy genes has provided us the opportunity to begin to address the mechanisms that regulate centrosome duplication. Further study of the szy genes, including molecular analysis, should allow us to quickly dissect the regulatory networks involved. In some cases we are likely to establish new ties to existing cellular processes as suggested by the phenotypic similarity of some our mutants to the defects that arise when other vital processes are perturbed. Equally as important, some of our mutants have novel phenotypes, such as the striking protein aggregation phenotype seen in szy-5 and szy-20 mutants, suggesting completely novel forms of regulation. Thus, analysis of the szy genes is likely to provide important insights into centrosome duplication and to once again validate the power of genetics in tying together seemingly unrelated cellular processes.
Acknowledgments
We thank Kevin Kopish for technical assistance; Edward Kipreos for helpful suggestions; and Andy Golden, Kathryn Stein, and Nick Miliaras for comments on the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH) and by the National Institute of Diabetes and Digestive and Kidney Diseases. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
References
- Askjaer, P., V. Galy, E. Hannak and I. W. Mattaj, 2002. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 13: 4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh, J., and M. Bornens, 2004. The centrosome in evolution, pp. 93–122 in Centrosomes in Development and Disease, edited by E. A. Nigg. Wiley-VCH GmbH, Weinheim, Germany.
- Bettencourt-Dias, M., A. Rodrigues-Martins, L. Carpenter, M. Riparbelli, L. Lehmann et al., 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15: 2199–2207. [DOI] [PubMed] [Google Scholar]
- Bobinnec, Y., A. Khodjakov, L. M. Mir, C. L. Rieder, B. Edde et al., 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143: 1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauchle, M., K. Baumer and P. Gonczy, 2003. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13: 819–827. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada, R., E. Isnenghi, M. Culotti and G. von Ehrenstein, 1981. Genetic analysis of temperature-sensitive embryogenesis mutants in Caenorhabditis elegans. Dev. Biol. 84: 193–205. [DOI] [PubMed] [Google Scholar]
- Church, D. L., K. L. Guan and E. J. Lambie, 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121: 2525–2535. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., T. Muller-Reichert, L. Pelletier, B. Habermann, A. Desai et al., 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7: 815–829. [DOI] [PubMed] [Google Scholar]
- Delattre, M., and P. Gonczy, 2004. The arithmetic of centrosome biogenesis. J. Cell Sci. 117: 1619–1630. [DOI] [PubMed] [Google Scholar]
- Delattre, M., S. Leidel, K. Wani, K. Baumer, J. Bamat et al., 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6: 656–664. [DOI] [PubMed] [Google Scholar]
- Delattre, M., C. Canard and P. Gonczy, 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16: 1844–1849. [DOI] [PubMed] [Google Scholar]
- Doxsey, S., 2001. Re-evaluating centrosome function. Nat. Rev. Mol. Cell. Biol. 2: 688–698. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., P. R. Martin, J. B. Phillips, R. Lyczak, D. R. Hamill et al., 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., S. Pichler, M. Kirkham and A. A. Hyman, 1999. a Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147: 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., H. Schnabel, T. Kaletta, A. D. Amores, T. Hyman et al., 1999. b Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144: 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck, R., Y. D. Stierhof, C. J. Wilkinson and E. A. Nigg, 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hamill, D. R., A. F. Severson, J. C. Carter and B. Bowerman, 2002. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3: 673–684. [DOI] [PubMed] [Google Scholar]
- Harel, A., and D. J. Forbes, 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16: 319–330. [DOI] [PubMed] [Google Scholar]
- Hirsh, D., and R. Vanderslice, 1976. Temperature-sensitive developmental mutants of Caenorhabditis elegans. Dev. Biol. 49: 220–235. [DOI] [PubMed] [Google Scholar]
- Jaspersen, S. L., A. E. Martin, G. Glazko, T. H. Giddings Jr., G. Morgan et al., 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kemp, C. A., K. R. Kopish, P. Zipperlen, J. Ahringer and K. F. O'Connell, 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6: 511–523. [DOI] [PubMed] [Google Scholar]
- Kemphues, K. J., M. Kusch and N. Wolf, 1988. a Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics 120: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues, K. J., J. R. Priess, D. G. Morton and N. S. Cheng, 1988. b Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., R. W. Cole, B. R. Oakley and C. L. Rieder, 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10: 59–67. [DOI] [PubMed] [Google Scholar]
- Kirkham, M., T. Muller-Reichert, K. Oegema, S. Grill and A. A. Hyman, 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587. [DOI] [PubMed] [Google Scholar]
- Leidel, S., and P. Gonczy, 2003. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4: 431–439. [DOI] [PubMed] [Google Scholar]
- Leidel, S., M. Delattre, L. Cerutti, K. Baumer and P. Gonczy, 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Maine, E. M., and J. Kimble, 1989. Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans. Development 106: 133–143. [DOI] [PubMed] [Google Scholar]
- Malone, C. J., L. Misner, N. Le Bot, M. C. Tsai, J. M. Campbell et al., 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115: 825–836. [DOI] [PubMed] [Google Scholar]
- Marshall, W. F., Y. Vucica and J. L. Rosenbaum, 2001. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 11: 308–317. [DOI] [PubMed] [Google Scholar]
- Miwa, J., E. Schierenberg, S. Miwa and G. von Ehrenstein, 1980. Genetics and mode of expression of temperature-sensitive mutations arresting embryonic development in Caenorhabditis elegans. Dev. Biol. 76: 160–174. [DOI] [PubMed] [Google Scholar]
- Oakley, C. E., and B. R. Oakley, 1989. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338: 662–664. [DOI] [PubMed] [Google Scholar]
- O'Connell, K. F., 2000. The centrosome of the early C. elegans embryo: inheritance, assembly, replication, and developmental roles. Curr. Top. Dev. Biol. 49: 365–384. [DOI] [PubMed] [Google Scholar]
- O'Connell, K. F., C. M. Leys and J. G. White, 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149: 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, K. F., K. N. Maxwell and J. G. White, 2000. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222: 55–70. [DOI] [PubMed] [Google Scholar]
- O'Connell, K. F., C. Caron, K. R. Kopish, D. D. Hurd, K. J. Kemphues et al., 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105: 547–558. [DOI] [PubMed] [Google Scholar]
- Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian et al., 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103: 127–140. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., N. Ozlu, E. Hannak, C. Cowan, B. Habermann et al., 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14: 863–873. [DOI] [PubMed] [Google Scholar]
- Sankaran, S., and J. D. Parvin, 2006. Centrosome function in normal and tumor cells. J. Cell Biochem. 99: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder, G., 2004. Centrosome duplication and it regulation in the higher animal cell, pp. 167–189 in Centrosomes in Development and Disease, edited by E. A. Nigg. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Starr, D. A., and J. A. Fischer, 2005. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27: 1136–1146. [DOI] [PubMed] [Google Scholar]
- Starr, D. A., and M. Han, 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298: 406–409. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Wang, W., A. Budhu, M. Forgues and X. W. Wang, 2005. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 7: 823–830. [DOI] [PubMed] [Google Scholar]
- Weil, C. F., C. E. Oakley and B. R. Oakley, 1986. Isolation of mip (microtubule-interacting protein) mutations of Aspergillus nidulans. Mol. Cell. Biol. 6: 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Yoder, J. H., and M. Han, 2001. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell 12: 2921–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]