Abstract
The species Arabidopsis halleri, an emerging model for the study of heavy metal tolerance and accumulation in plants, has evolved a high level of constitutive zinc tolerance. Mapping of quantitative trait loci (QTL) was used to investigate the genetic architecture of zinc tolerance in this species. A first-generation backcross progeny of A. halleri ssp. halleri from a highly contaminated industrial site and its nontolerant relative A. lyrata ssp. petraea was produced and used for QTL mapping of zinc tolerance. A genetic map covering most of the A. halleri genome was constructed using 85 markers. Among these markers, 65 were anchored in A. thaliana and revealed high synteny with other Arabidopsis genomes. Three QTL of comparable magnitude on three different linkage groups were identified. At all QTL positions zinc tolerance was enhanced by A. halleri alleles, indicating directional selection for higher zinc tolerance in this species. The two-LOD support intervals associated with these QTL cover 24, 4, and 13 cM. The importance of each of these three regions is emphasized by their colocalization with HMA4, MTP1-A, and MTP1-B, respectively, three genes well known to be involved in metal homeostasis and tolerance in plants.
METAL tolerance in plants has been considered “an example of more powerful evolution in action than industrial melanism in moths” (Antonovics et al. 1971). Therefore it has been the focus of many evolutionary studies, in which it was argued that metal tolerance could evolve rapidly following exposure to heavy metal stress (Wu et al. 1975; Al-Hiyali et al. 1988). Some heavy metals, like zinc and copper, are oligo-nutrients and thus essential in small quantities for normal plant development. To avoid metal toxicity, all plants have evolved basic tolerance mechanisms. Binding by proteins or nonprotein thiol peptides in the cytoplasm and subsequent sequestration in the vacuole are the major component processes in the cellular heavy metal detoxification (Clemens 2001; Krämer 2005). However, at so-called metalliferous sites, heavy metals can occur at highly elevated concentrations in the soil, either through ancient natural processes, as in nickel-rich serpentine soils, or through recent human activities, as in zinc- and cadmium-rich calamine soils surrounding smelters. The total metal content of contaminated sites, depending on the metal, can be up to 10- to 1000-fold higher than that of uncontaminated sites (Bert et al. 2002). At these extreme concentrations, both essential and nonessential heavy metals become toxic (Clemens 2001; Hall 2002) and only a small number of plant species have evolved tolerance to such concentrations. These species have been classified as either absolute (strict or eu-) or facultative (pseudo-) metallophytes, according to their occurrence either on contaminated sites only or on both metalliferous and nonmetalliferous soils (Lambinon and Auquier 1964).
The genetic basis of adaptive quantitative traits is still the matter of strong debates among evolutionists. Current questions concern species differences in the genetic architecture of traits related to adaptation and focus mainly on the following points: How many genes are involved? How large are their phenotypic effects? Are they involved in a pleiotropic effect (e.g., tolerance to different metals)? What are the dynamics of the alleles present at these genes? This emphasis on number of genes and sizes of effects reflects one of the oldest problems in evolutionary biology: the complexity of genetic changes underlying phenotypic evolution. Recent theoretical developments of the Fisher–Orr model suggest that fewer genes than expected according to the Fisher infinitesimal model could be involved in adaptation and that the dynamics of allele substitution at selected loci follow an approximate geometric sequence (Orr 1998a, 1999, 2002): large-effect mutations typically substitute early, whereas smaller-effect ones substitute later during adaptive walk. Heavy metal tolerance is a trait of particular interest for documenting genetic changes during adaptive walk, as high heavy metal concentration can easily be measured in soils and represents a strong directional selective pressure, resulting in the substitution of tolerance alleles at some loci.
The resolution of quantitative traits into discrete Mendelian loci analysis has made substantial progress since the development of genetic linkage maps. Quantitative trait loci (QTL) mapping has proved to be very powerful in examining complex adaptive traits (Doebley et al. 1997; Alonso-Blanco et al. 1998; Ungerer et al. 2002; Weinig et al. 2003); it provides an efficient means for determining the number of genes implicated in a trait as well as their effects and interactions, which are important for understanding the evolutionary history of a trait (Mackay 2001; Barton and Keightley 2002; Erickson et al. 2004). By identifying specific chromosomal regions where genetic variation can be associated with measurable phenotypic variation (Tanksley 1993; Doerge 2002), QTL mapping can help to detect or validate candidate genes underlying complex traits (Flint and Mott 2001; Yano 2001; Glazier et al. 2002).
In the case of heavy metal tolerance in plants, two pseudometallophyte species, Arabidopsis halleri (L.) (O'Kane & Al-Shehbaz) [syn. Cardaminopsis halleri (L.) Hayek] and Thlaspi caerulescens J. & C. Presl. recently emerged as model species (Assunção et al. 2003). These species belong to the Brassicaceae and are able to tolerate and hyperaccumulate zinc (Zn) and cadmium (Cd). A recent study performed on 33 metallicolous (M) and nonmetallicous (NM) A. halleri populations clearly established the occurrence of constitutive, or fixed, Zn tolerance in A. halleri and of quantitative variation of the degree of Zn tolerance among populations (Pauwels et al. 2006). Parsimony suggests that fixed Zn tolerance has occurred only once, probably early in the species history. Consequently, the genes underlying this tolerance are expected to be shared by all populations, irrespective of whether there is metal contamination at the site of population origin. Taking advantage of a wide range of resources that are available for its wild nontolerant close relative A. thaliana (Mitchell-Olds 2001), A. halleri can also be considered the most promising model for identifying the genetic basis of adaptation to metalliferous soil.
A global comparison of A. halleri and A. thaliana through transcription profiling was recently performed to identify genes differentially expressed and/or regulated in A. halleri compared to A. thaliana under various Zn concentrations. These studies identified A. halleri homologs of A. thaliana genes potentially involved in Zn tolerance (Becher et al. 2004; Weber et al. 2004). However, differential expression and/or regulation may just as well be the primary cause of heavy metal tolerance as its consequence or might simply be the result of the divergence time separating A. halleri and A. thaliana. Thus, definitive evidence for their implication in heavy metal tolerance is still missing.
In this study, we have applied a QTL approach to investigate the genetic basis underlying Zn tolerance in A. halleri and to define genomic regions containing the genes underlying the constitutive Zn tolerance in A. halleri as well as those involved in the recent adaptation to industrial polluted sites. The application of a QTL approach in A. halleri was highly promoted by the recent publication of the genetic linkage maps of its close relatives A. l. petraea and A. l. lyrata (Kuittinen et al. 2004; Yogeeswaran et al. 2005). The extensive conservation of marker order reported between A. lyrata subspecies and the model A. thaliana (Kuittinen et al. 2004; Yogeeswaran et al. 2005) made the prospect of transferring these resources to A. halleri even more attractive for gaining insights into adaptive evolution of heavy metal tolerance and hyperaccumulation (Clauss and Koch 2006). We performed an interspecific cross between A. halleri from a highly contaminated industrial site and A. l. petraea to generate a first-generation backcross (BC1). These progeny, segregating for Zn tolerance, were used to construct a molecular linkage map (the first reported for a cross between these two species) and to identify QTL regions for Zn tolerance in A. halleri, making full use of the previous mapping experiments conducted on A. lyrata subspecies (Kuittinen et al. 2004; Yogeeswaran et al. 2005) and of recent and current functional analyses of metal homeostasis genes in A. halleri (Becher et al. 2004; Weber et al. 2004; Filatov et al. 2006).
MATERIALS AND METHODS
Plant material:
A single cross was performed between one individual from the Zn-tolerant species A. halleri ssp. halleri (henceforth called A. halleri) (pollen donor) and one from the nontolerant species A. lyrata ssp. petraea (A. l. petraea 1) (pollen recipient). The A. halleri individual (2n = 16) originated from a site highly contaminated with Zn, Cd, and lead (Pb) (Auby, France) (Van Rossum et al. 2004). The A. l. petraea 1 individual (2n = 16) originated from an uncontaminated site in the Czech Republic (Unhost, Central Bohemia) (Macnair et al. 1999). Both species are self-incompatible and usually outcrossing. Therefore, to avoid any inbreeding depression effect, one randomly selected F1 individual was used as the male parent to fertilize a second A. l. petraea genotype (A. l. petraea 2), generating the interspecific backcross progeny (BC1). The BC1 population used for linkage map construction and QTL mapping consisted of 199 individuals. Both parental and progeny genotypes are easily maintained in time and multiplied for replication by cuttings.
Evaluation of Zn tolerance:
Twelve replicates of the four parental genotypes (A. halleri, A. l. petraea 1, A. l. petraea 2, and the F1 individual) and three replicates of each of the BC1 individuals were obtained by vegetative propagation. They were grown in the greenhouse on sand, and 8 weeks after cloning were transferred to 10-liter polycarbonate trays containing a nutrient solution in a controlled growth chamber (temperature, 20° day:15° night; light, 14 hr day:10 hr night). The plants were randomly assigned in the trays (48 plants/vessel, including one copy of each parental genotype), which in turn were rotated in the growth chamber twice a week. The nutrient solution consisted of 0.5 mm Ca(NO3), 0.2 mm MgSO4, 0.5 mm KNO3, 0.1 mm K2HPO4, 0.2 μm CuSO4, 2 μm MnCl2, 10 μm H3BO3, 0.1 μm MoO3, 10 μm FeEDDHA, and 10–3000 μm Zn added as ZnSO4. The pH of the solution was set at 6.5.
Zn tolerance was measured by a sequential test established by Schat and Ten Bookum (1992). This test provides a measure for tolerance by sequentially transferring plants into increasing concentrations of Zn and determining for each individual the lowest concentration at which no new root growth is produced (the EC100). Roots of plants were blackened with activated charcoal to observe new root growth more easily. The plants were grown on 10 μm of Zn for the first 3 weeks. After verification of their root growth at 10 μm of Zn, 6–12 replicates of each parental genotype and 1–3 replicates of each BC1 individual were transferred in successive weeks to 25, 50, 75, 100, 150, 250, 500, 1000, 2000, and 3000 μm of Zn. Root growth of the plants was evaluated at the end of each week. Plants were observed for at least 2 weeks after reaching their EC100, to ensure that no new root growth occurred.
Marker analysis:
Genomic DNA of the four parental genotypes and of the 199 individuals of the BC1 was extracted for marker analysis using a slightly modified Dellaporta method (Saumitou-Laprade et al. 1999). The BC1 progeny were genotyped using 65 sequence-based markers anchored in A. thaliana and 18 AFLP markers (Table 1).
TABLE 1.
Markers used in linkage map construction
| Locus name | Identification/BAC location in A. thaliana | Type of polymorphism | Annealing temperature (°) | Forward primer | Reverse primer |
|---|---|---|---|---|---|
| Linkage group 1 | |||||
| AthACS | F22L4 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| AAC/CG-88 | AFLP | ||||
| AXR1 | At1g05180 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| ACA/CG-105 | AFLP | ||||
| PhyA | At1g09570 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| ICE10 | F12F1 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| ICE13 | At1g13220 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| GI | At1g22770 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| AGG/CT-198 | AFLP | ||||
| ATTS0392 | At1g30630 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| ACA/CG-228 | AFLP | ||||
| VIP1 | At1g43700 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| At1g46768 | At1g46768 | Indel | 50 | AGGTTGATCATTTTCTAAAAGTTCTTG | TTCCTCCTCCGTATCCCTCT |
| MTP1-C | RFLP/SSCP | Dräger et al. (2004) | Dräger et al. (2004) | ||
| Linkage group 2 | |||||
| F19K23 | At1g62050 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| ACA/CG-87 | AFLP | ||||
| ACA/CG-160 | AFLP | ||||
| ACA/CG-320 | AFLP | ||||
| AAC/CG-84 | AFLP | ||||
| SLL2 | At1g66680 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| lyr132a | T23K23 | Microsat | 50 | GCCGTGAGATTAAAGAAGACG | GCAAGAGCTGATCTCCATCC |
| ACA/CG-71 | AFLP | ||||
| nga111 | F28P22 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| At1g75830b | At1g75830 | 55 | CATATCTATGCAAATTGTGTTTAATATA | ACATGGGAAGTAACAGATACACTTATGA | |
| ADH1 | At1g77120 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| Linkage group 3 | |||||
| At3g08040c | At3g08040 | Indel | 60 | TACCAACCAGCCACAGCAACC | CGCTTTGTTTCCACTATTTGACTTTG |
| MDC16 | MDC16 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| nga162 | MDC16 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| DMC1 | At3g22880 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| At2-TCA1 | T9F8 | Microsat | 50 | CAACCACACCCCTTTAGCTT | GAGAGCCCATGGAGATGAAG |
| HMA4 | At2g19110 | Indel | 62 | TGACCTGAAAATGAAAGGTGGTC | TGCATAACTCCTGCAACAGCT |
| ICE14 | F11A3 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| ACA/CG-310 | AFLP | ||||
| Linkage group 4 | |||||
| con | At2g21320 | MseI | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| At2g22430d | At2g22430 | Indel | 55 | AGTTCAGATTCAGTGGGTGG | GTAGATCTGTGAAACTCCGG |
| Ck2-alpha2 | At2g23080 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| NRAMP3e | At2g23150 | PsiI | 55 | TTGGATGTTTGGTCAAGCTAAGCCAAGTG | TGCCACGAGCAATGAGGTAGAGGATGAAT |
| At2-TA5 | T19L18 | Microsat | 50 | TCATCGGATCCATATTTGTTTG | CATTGTTGGTCGTGGCTATG |
| ELF3 | At2g25930 | Microsat | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| At2g28700d | At2g28700 | Indel | 55 | TGGGAACATGGGAGATTTTGTTATG | TGTCTTGCCCTTCACTGAAAAAGAG |
| nga361 | T16B12 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| At2g33010d | At2g33010 | Indel | 55 | TGGAGCATATCGAGAAGAAGCACTC | GGACTTCTTGTGAAGGCAAATCAGA |
| ACT3 | At2g37620 | MspI | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| ICE12 | At2g39010 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| At2g40140 | At2g40140 | Indel | 50 | TCAAAAACCCCAACCACTTC | TCCACCGATGGATTCTCTTC |
| At2g43010 | At2g43010 | Indel | 50 | CGGACTCATGGACTTGCTTT | TTCTGGGTTTGGGTTTGTTC |
| ACA/CG-130 | AFLP | ||||
| ICE11 | F11C10/F13A10 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| MTP1-A | At2g46800 | RFLP/SSCP | Dräger et al. (2004) | Dräger et al. (2004) | |
| Linkage group 5 | |||||
| nga1145 | T16F16 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| At3g28220 | At3g28220 | Indel | 50 | TGGGTCCATTTCTTGTTGTT | CCAAGCCAATTGCTCCATAG |
| At3g33530 | At3g33530 | MfeI | 50 | TGGTGGGATGTAACAACAGG | TTTAGACTGGGGCACAAAGC |
| AAC/CG-179 | AFLP | ||||
| At3g45810 | At3g45810 | DraI | 50 | TTTGCTGGTTATTGCCTACG | ACCTCTCGCTCTTGTTTCCA |
| F3H | At3g51240 | HindIII | Kuittinen et al. (2002) | Kuittinen et al. (2002) | |
| MTP3c | At3g58060 | Indel | 55 | CCATGGTCACGGTCATAGTCAT | CGCTCTGTATCGAATCTCCCAGCA |
| nga112 | At3g62650 | Microsat | Bell and Ecker (1994) | Bell and Ecker (1994) | |
| Linkage group 6 | |||||
| At4-GA2 | T18A10 | Microsat | 50 | CCTCGGGTAAAGACAGAGCA | TGGTAACACCGGAAGTTTCA |
| LD | At4g02560 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| MTP1-B | RFLP/SSCP | Dräger et al. (2004) | Dräger et al. (2004) | ||
| AthDET1 | At4g10180 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| ICE2 | K18P6 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| AthCDPK9 | MQM1 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| AAC/CG-336 | AFLP | ||||
| ACA/CG-359 | AFLP | ||||
| At5g08160 | At5g08160 | Indel | 50 | TTGTTGGTACGACTTTTTCTCG | GCCGGATCCTTGACTTTCTT |
| MT2b | RFLP | Zhou and Goldsbourgh (1995) | Zhou and Goldsbourgh (1995) | ||
| Linkage group 7 | |||||
| At4g38220 | At4g38220 | Indel | 50 | GTGGTGGTGGGAGAAGAGAG | ACCGTGTTGATTCGGAGGTA |
| At4g33160 | At4g33160 | Indel | 50 | TTGGCCTAAG TTTTTCTTTTTG | GTCATCCATGGGGAATTCAG |
| cha15f | F11C18 | Microsat | 50 | AATTCAAACAGGCGAAACCA | CTGCGAATCTCACGACTTCA |
| TSB2 | At4g27070 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| At4-TC1 | T19F6 | Microsat | 50 | CAAGGTCGAATGTTGGAGACT | GCGCACTACAAAAATGAGAGG |
| SRKg | At4g21366 | SNP | TCAAGATTGAAGCTGAGTGA | TACACAACCCGTCCCGCCAA | |
| FCA | At4g16280 | HinfI | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| ACA/CG-266 | AFLP | ||||
| ACA/CG-312 | AFLP | ||||
| PhyC | At5g35840 | Indel | Kuittinen et al. (2002) | Kuittinen et al. (2004) | |
| ATTSO191 | At5g37780 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| ICE9 | At5g40340 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| Linkage group 8 | |||||
| ATCLH2 | At5g43860 | Indel | Kuittinen et al. (2004) | Kuittinen et al. (2004) | |
| ACA/CT-86 | AFLP | ||||
| EMF2 | At5g51230 | XhoI | 54 | GTTGCAGTTTGCAAAAACGA | CATGGAATGTGACCATCTGC |
| ACA/CG-387 | AFLP | ||||
| MHJ24 | MHJ24 | Microsat | Clauss et al. (2002) | Clauss et al. (2002) | |
| NRAMP4e | At5g67330 | Indel | 55 | TTGGATGTTTGGTCAGACGAAACCCAGTG | ATAAACTGTCCGGCGTACGTACCTGTGAT |
The names and identification of the loci in A. thaliana, the type of polymorphism scored, restriction enzyme if a CAPS marker, annealing temperature, and primer sequences if newly defined markers are shown.
Primers provided by T. Mitchell-Olds (personal communication).
Primers and genotyping results provided by M. Mirouze (personal communication).
Primers and genotyping results provided by M. Hanikenne (personal communication).
Primers and genotyping results provided by F. Varoquaux (personal communication).
Primers and genotyping results provided by R. Oomen (personal communication).
Primers provided by C. Schlötterer (personal communication).
A. halleri allele-specific primers are given, and genotypes have been controlled by amplification with A. l. petraea 1 allele-specific primers (V. Castric, personal communication).
The anchored markers consisted of microsatellites, indels, and polymorphisms revealed by cleaved amplified polymorphisms (CAPS), RFLP, and SSCP analysis. Some markers were selected because of their potential implication in metal homeostasis (FRD3, HMA4, MTP1, MTP3, NRAMP3, NRAMP4, and MT2B). Others had been identified by transcription profiling analyses conducted on A. halleri (At1g46768, HMA4, NRAMP3, At2g40140, At2g43010, MTP1, At3g28220, At4g33160, At4g38220, and At5g08160) (Becher et al. 2004; Weber et al. 2004; Craciun et al. 2006; S. Mari, unpublished data). The remainders were selected using their position in the A. thaliana genome to improve coverage of the A. halleri genome. Of the 65 anchored markers, 40 were previously reported for A. thaliana and/or A. l. petraea (Bell and Ecker 1994; Clauss et al. 2002; Kuittinen et al. 2002, 2004); 31 of the 40 markers had already been mapped in A. l. petraea (Kuittinen et al. 2004). In addition, 25 newly defined markers were introduced in this study. For 11 of the 25 markers, primer design and/or genotyping of the BC1 progeny were kindly performed by colleagues (Table 1). For the other 14 markers, we designed primers on the basis of the A. thaliana sequence (The Arabidopsis Information Resource database at http://www.arabidopsis.org/) using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For 4 of these 14 markers (At2-TCA1, At2-TA5, At4-GA2, and At4-TC1), primer pairs were designed to amplify small fragments of 150–400 bp. For the other 10 markers (At1g46768, At2g43010, At3g28220, At3g33530, At3g45810, At5g08160, At4g38220, At2g40140, At4g33160, and HMA4), one primer was designed in an exonic region of the gene and the second primer was placed either in the following exon or in the 5′-upstream region of the start codon. In the latter case, the possible promoter region of the gene, supposed to be highly variable between species, was targeted. For amplification of EMF2, already mapped in A. l. petraea, new primer sequences were designed as described above and used instead of the ones defined by Kuittinen et al. (2004). To obtain labeled PCR products detectable on the automated genotyper Li-Cor 4200 (Li-Cor-ScienceTec), either the forward or the reverse primer contained a 5′-tail of 19 bp (forward primer) or 20 bp (reverse primer) homologous to the universal consensus M13 primer sequence, followed by the locus-specific sequence (Oetting et al. 1995).
The following polymerase chain reaction (PCR) conditions were applied for all 65 markers, except for those that were provided through collaborations (Table 1). PCR reactions were carried out in a total volume of 15 μl containing 20 ng of template DNA, 2 mm MgCl2, 0.2 mg/ml BSA, 0.2 mm dNTP, 0.2 μm of each unlabeled primer and 0.15 μm of the M13 fluorescently labeled primer (either IRD-700 or IRD-800), 20 mm Tris–HCl (pH 8.3), 50 mm KCl, and 0.4 unit of AmpliTaq DNA Polymerase (Applied Biosytems, Foster City, CA). PCR was performed on a Perkin-Elmer (Norwalk, CT) Gene-Amp system 9700 under the following conditions: 94° for 5 min, followed by locus-specific amplification; 94° for 30 sec, annealing temperature for 45 sec, 72° for 40 sec, for eight cycles, followed by M13 labeling amplification; and 94° for 30 sec, 50° for 20 sec, 72° for 40 sec, for 30 cycles, and a final extension 72° for 7 min. The annealing temperature of the locus-specific cycles varied between 50° and 60°, depending on the locus. For CAPS markers, restriction was carried out in a total volume of 20 μl containing 10 μl of PCR product, 0.2 mm spermidine, 1× specific enzyme buffer provided by the supplier and 1 unit of restriction enzyme. Restriction was carried out by incubation for at least 4 hr at the appropriate temperature in a Perkin-Elmer Gene-Amp system 9700. Polymorphisms were revealed on agarose or polyacrylamide gels using a Li-Cor genotyper.
Amplified fragment length polymorphism (AFLP) marker analysis was performed as described by Vos et al. (1995), using EcoRI/MseI restriction enzymes. For preamplification, one nucleotide was added to EcoRI and MseI. For selective amplification, three and two nucleotides were added to EcoRI and MseI, respectively. The EcoRI selective primer was fluorescently labeled with either IRD-700 or IRD-800 for visualization of the AFLP bands on a Li-Cor genotyper 4200 (Li-Cor-ScienceTec). Polymorphic and segregating bands were scored using the program RFLPSCAN 3.0 (Scanalytics). Their sizes were determined by comparison with an appropriately labeled molecular weight marker (50–700 bp, Li-Cor-ScienceTec). AFLP markers were named according to the selective nucleotides used in selective amplification and their size.
Linkage map construction:
The A. halleri × A. l. petraea (Ah × Alp) linkage map was constructed with the Joinmap 3.0 program (Van Ooijen and Voorrips 2001). Individuals lacking information for >25% of all markers were excluded from the analysis. Linkage groups were obtained at a logarithm-of-odds (LOD) score threshold of 4. Markers along each linkage group were ordered using the sequential method implemented in Joinmap. In this method, the best order was determined by comparing the goodness of fit of the resulting map for each tested order using a threshold of 0.5 and 1.0 for the linkage groups and the loci, respectively. Kosambi's mapping function was used to translate recombination frequencies into map distances (Kosambi 1944).
Linkage map analysis:
We performed a t-test for correlated samples (Minitab, State College, PA) to test for a significant difference in marker intervals between the Ah × Alp and the A. l. petraea maps using the markers common to both mapping experiments. A t-test for correlated samples (Minitab) was also performed to compare the linkage group lengths in the Ah × Alp and either the A. l. petraea or the A. l. lyrata maps.
Marker segregation:
According to Mendelian inheritance, the A. halleri alleles are expected to segregate in a 1:1 ratio in the BC1. When dealing with an interspecific cross, segregation distortion frequently occurs. Deviations from the Mendelian ratios were tested using a chi-square test implemented by Joinmap 3.0 at a locus-by-locus significance level of α = 0.05 (Van Ooijen and Voorrips 2001).
Statistical analysis and QTL mapping:
We performed a Kruskal–Wallis test, based on Wilcoxon rank scores of the data, to test for significant differences of Zn tolerance among the four parental genotypes using the NPAR1WAY procedure in SAS (1999). A one-way analysis of variance (ANOVA) using the GLM procedure in SAS (1999) was performed on the EC100 values obtained for the replicates of the 199 BC1 individuals to determine the genotype effect. The main factor, i.e., the BC1 progeny, was considered a random effect, because the BC1 individuals tested for Zn tolerance represent a random sampling of the total BC1 population. The broad-sense heritability of Zn tolerance was calculated by dividing the genetic variance by the total phenotypic variance, using the mean square values (MS) from the ANOVA (h2 = MSgenot/(MSgenot + MSerror). Type III sums of squares were used because the data set was unbalanced, due to an unequal number of clones of each BC1 individual.
We performed a Kolmogorov–Smirnov test (Minitab) on the EC100mean values (the arithmetic mean of the EC100 values of the clones) of the BC1 genotypes as well as on the EC100mean values after logarithmic (log) transformation to test for deviation from a normal distribution. The QTL analysis for Zn tolerance was performed using the MapQTL 4.0 program (Van Ooijen et al. 2002). The LOD score threshold for QTL detection was set at 2.3 (α = 0.05) and obtained by a permutation test on the quantitative data in MapQTL. A first QTL analysis was performed using interval mapping (IM) as implemented in MapQTL. The LOD score representing the likelihood of a QTL being present has been calculated every centimorgan within the intervals along the linkage groups. Markers for which the LOD score exceeded the significance threshold were identified in each linkage group. Automatic cofactor selection was performed by MapQTL on these markers for their use as cofactors in multiple QTL models (MQM) analysis. We performed MQM mapping twice while adjusting the selection of the cofactors to obtain the best possible set of QTL, i.e., showing maximal LOD scores. One- and two-LOD support intervals were obtained using Mapchart 2.1 (Voorrips 2002). The estimated additive genetic effect (a) and the percentage of variance explained by each QTL (R2) were calculated in IM. We tested for significant interactions between QTL using the GLM procedure in SAS (1999) on both raw and log-transformed EC100mean values. The model involved three factors corresponding to the genotypes of the BC1 individuals at the markers (HMA4, MTP1-A, and MTP1-B) closest to or at the three QTL. These factors were considered either random or fixed. A box plot analysis was performed on the markers HMA4, MTP1-A, and MTP1-B. We performed a Kruskal–Wallis test, based on Wilcoxon rank scores of the data, to test for significant differences of Zn tolerance among the eight genotypic groups using the NPAR1WAY procedure in SAS (1999).
RESULTS
Linkage map construction:
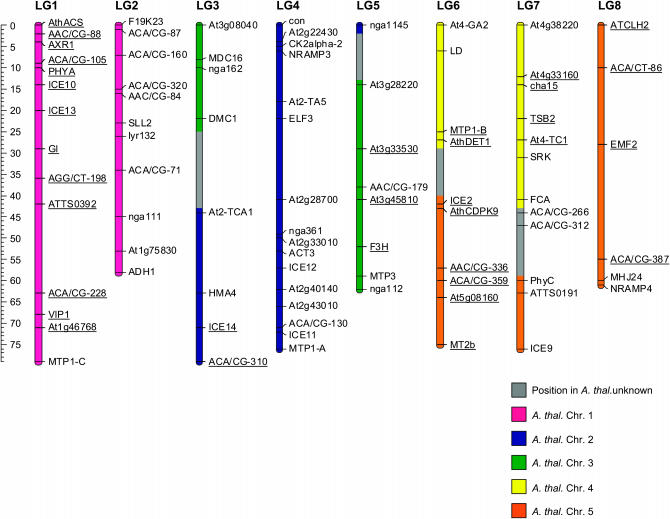
Of the 199 BC1 individuals, 196 could be genotyped successfully at >75% of all markers and were used for the map construction. A total of 85 markers were assigned to eight linkage groups (LG1–LG8) by using a LOD score threshold of 4 (Table 1). The lengths of the linkage groups varied from 57 to 80 cM and summed to a total of 567 cM. The average distance between two adjacent markers was 6.6 cM, ranging from 1 to 27 cM (Figure 1).
Figure 1.—
Linkage map of the A. halleri × A. l. petraea BC1 progeny constructed with Joinmap 3.0. The homology with the A. thaliana chromosomes is indicated by the colors. The regions where translocation occurred are in gray. The position in A. thaliana of the anonymous markers was inferred by integration of the A. l. petraea and A. l. lyrata mapping experiments. Markers in segregation distortion are underlined.
Comparative analysis of the A. halleri × A. l. petraea map with the maps of A. l. petraea and A. l. lyrata:
The transition from eight chromosomes in the Ah × Alp map to five chromosomes in the A. thaliana genome can be explained by five main chromosomal rearrangements as described for A. l. petraea (Kuittinen et al. 2004; Koch and Kiefer 2005) and A. l. lyrata (Yogeeswaran et al. 2005). These consist of three fusions between the linkage groups LG1/LG2, LG3/LG4, and LG7/LG8 and two reciprocal translocations between LG3/LG5 and LG6/LG7 (Figure 1). Marker order in the Ah × Alp and A. thaliana maps was generally similar. The order of the 31 marker loci shared by the Ah × Alp and A. l. petraea linkage maps was identical, with one exception on LG1. The positions in the interspecific map of the loci (PhyA and AXR1) did not agree with those reported for A. l. petraea, but rather with those expected from A. thaliana.
The marker distances obtained on the marker intervals between the markers common to the Ah × Alp and A. l. petraea maps were not significantly different in both mapping experiments (P = 0.27). The linkage group lengths differed significantly between the Ah × Alp map and either the A. l. petraea (P = 0.03) or the A. l. lyrata map (P = 0.02) (Table 2).
TABLE 2.
Comparison of mean linkage group length in the A. halleri × A. l. petraea, the A. l. petraea, and the A. l. lyrata maps
| Linkage group | A. halleri × A. l. petraeaa | A. l. petraeaa | A. l. lyrataa |
|---|---|---|---|
| LG1 | 78 | 74 | 39 |
| LG2 | 57 | 58 | 6 |
| LG3 | 80 | 64 | 69 |
| LG4 | 76 | 67 | 47 |
| LG5 | 63 | 54 | 60 |
| LG6 | 75 | 61 | 76 |
| LG7 | 76 | 78 | 49 |
| LG8 | 62 | 59 | 61 |
Lengths of the linkage groups are given in centimorgans.
Two duplication events of the MTP1 gene (a single-copy gene on A. thaliana chromosome 2 and A. lyrata LG4) were detected in A. halleri. We mapped the three copies identified in A. halleri (MTP1-A, MTP1-B, and MTP1-C) (Dräger et al. 2004) on three different linkage groups (LG4, LG6, and LG1, respectively). The A. halleri MTP1-A copy mapped to the lower arm of LG4 beyond the marker ICE11 (the expected position from A. thaliana) and can therefore be considered as the ortholog of the A. lyrata and A. thaliana MTP1 gene.
Markers in segregation distortion:
At a locus-by-locus significance level of 0.05, 34 markers (40%) showed distorted segregation and were found on six of the eight linkage groups (Figure 1). The segregation ratio bias was highly directional. Of the 34 distorted markers, 31 showed an excess of the A. l. petraea 1/A. l. petraea 2 homospecific (i.e., originating from the same species) allelic combination compared to the A. halleri/A. l. petraea 2 heterospecific (i.e., originating from different species) genotype. Only three markers, all located on linkage group LG5, showed the opposite pattern, i.e., an excess of the heterospecific combination. With a single exception (on LG5), distorted markers were always linked to markers distorted in the same direction, indicating that the segregation bias was due to meiotic events rather than genotyping errors.
Evaluation of Zn tolerance:
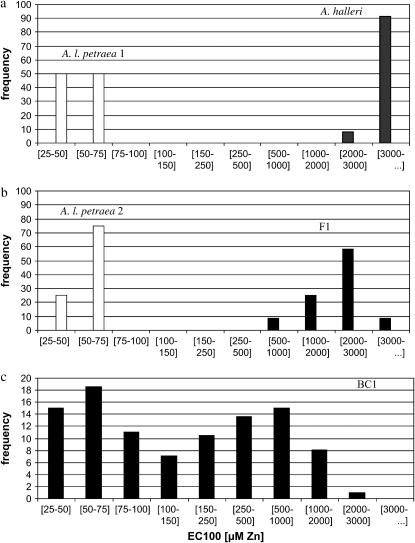
A Kruskal–Wallis test showed a highly significant difference among the tolerance levels of the four parental lines of the BC1 (P < 0.0001) (Figure 2, a and b). Pairwise comparisons revealed a significant difference in Zn tolerance between the A. halleri parental clones (n = 12; EC100mean = 2917 μm Zn) and the A. l. petraea 1 parental clones (n = 6; EC100mean = 38 μm Zn) (P < 0.0001). The Zn tolerance of the A. halleri parental clones was probably underestimated because, even at the highest concentration applied in the test (3000 μm Zn), all clones except one still showed new root growth. The Zn tolerance of the F1 parental clones (n = 12; EC100mean = 1708 μm Zn) differed significantly from the tolerance of the A. l. petraea 2 (n = 8; EC100mean = 44 μm Zn) (P < 0.0001) and A. halleri parental clones (P < 0.0001). This indicates partial dominance of Zn tolerance in A. halleri, even though the underestimation of Zn tolerance for the A. halleri parental clones precludes any estimation of the dominance coefficient. No significant difference was identified between Zn tolerance of A. l. petraea 1 and A. l. petraea 2 parental clones (P = 0.3519). The genotype effect of Zn tolerance of the BC1 individuals was highly significant (F = 2.22; P < 0.0001). Broad-sense heritability of Zn tolerance in the BC1 was high (h2 = 0.69). No transgressive segregation of Zn tolerance was observed (Figure 2c).
Figure 2.—
Frequency distribution of Zn tolerance of the parental clones A. halleri, A. l. petraea 1, A. l. petraea 2, and F1, and the BC1 derived from A. halleri × A. l. petraea. Zn tolerance was measured by a sequential Zn exposure test in hydroponic solution. (a) Frequency distribution of Zn tolerance of the A. halleri and A. l. petraea 1 parental genotypes. (b) Frequency distribution of Zn tolerance of the A. l. petraea 2 and F1 parental genotypes. (c) Frequency distribution of Zn tolerance of the BC1 progeny; EC100mean values were used.
QTL mapping of Zn tolerance:
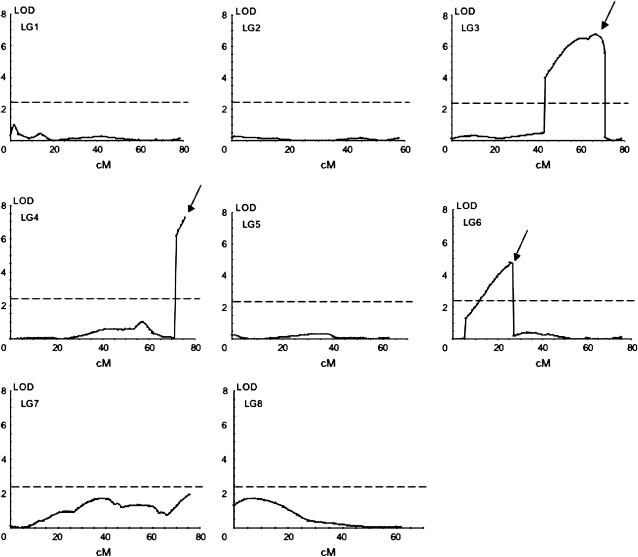
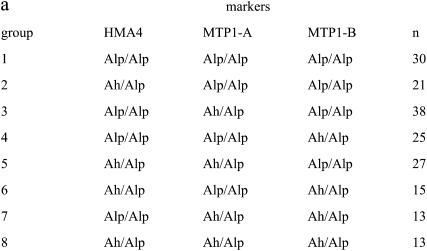
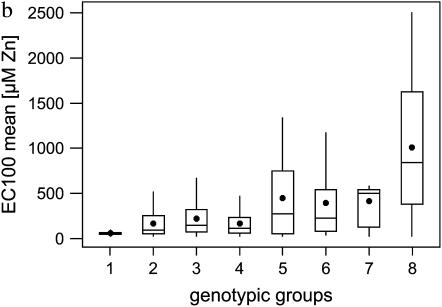
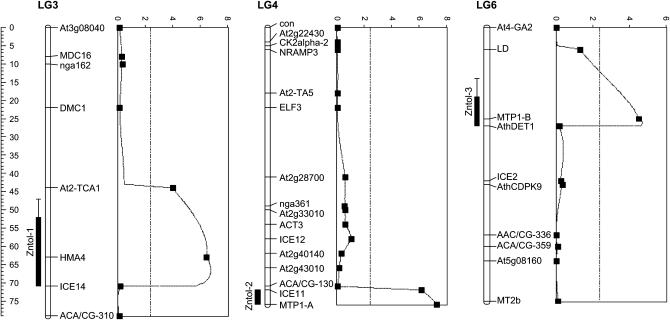
Three QTL located on linkage groups LG3, LG4, and LG6 were identified by IM and subsequent MQM mapping (Figure 3). The QTL were named Zntol-1, -2, and -3, respectively, for linkage group LG3, LG4, and LG6. LOD scores of Zntol-1, -2, and -3, calculated by the MQM module of MapQTL, were 6.46, 7.28, and 4.52, respectively (Table 3). In the five other linkage groups, the LOD scores did not exceed the significance threshold value of 2.4 (Figure 3). The individual contribution of each QTL to the phenotypic variance was 12.2, 11.2, and 5.6%, respectively, for the QTL Zntol-1, -2, and -3 (Table 3). They accounted together for 29% of the total phenotypic variance, which represents 42% of the genetic variance. For each of the QTL, the A. halleri allele increased Zn tolerance (Table 3). Pairwise interactions between the QTL were significant at α = 0.05 using the raw EC100mean data. However, loss of significance was observed using the log-transformed EC100mean values, indicating a statistical artifact (Table 4). Identical results were obtained independently of considering the main factors HMA4, MTP1-A, and MTP1-B fixed or random. As showed by box plots, eight genotypic groups were obtained by considering the markers HMA4, MTP1-A, and MTP1-B, closest to or at the three QTL positions, characterized by the presence or absence of the A. halleri allele at one, two, or three markers (Figure 4). A Kruskal–Wallis test revealed significant differences of Zn tolerance among the eight groups (P < 0.0001). For Zntol-1, -2, and -3, one-LOD support intervals of 19, 4, and 8 cM were reported, respectively. Two-LOD support intervals were 24, 4, and 13 cM for the three QTL, respectively (Figure 5). Six markers colocalized with the LOD support intervals. Markers ICE14 and HMA4 mapped in the LOD support interval of Zntol-1, markers MTP1-A and ICE11 colocalized with Zntol-2, and markers AthDET1 and MTP1-B colocalized with Zntol-3.
Figure 3.—
LOD score profiles for Zn tolerance. Names of linkage groups are given at the top left corner of each graph. Map positions are plotted along the abscissa. LOD scores are plotted along the y-axis. Dashed lines correspond to the LOD score threshold (2.3) for QTL detection at an error level of α = 0.05. QTL are indicated by arrows above the LOD score profile.
TABLE 3.
QTL for Zn tolerance
| QTLa | LG/Markerb | Positionc | LOD Scored | R2e | af |
|---|---|---|---|---|---|
| Zntol-1 | LG3/HMA4 | 64.6 | 6.46 | 12.2 | −280.861 |
| Zntol-2 | LG4/MTP1-A | 75.5 | 7.28 | 11.2 | −266.055 |
| Zntol-3 | LG6/MTP1-B | 24.7 | 4.52 | 5.6 | −197.275 |
QTL are named by the trait and ordered from 1 to 3.
Linkage group on which QTL were mapped and marker at QTL position or closest to QTL position.
Position of marker at or closest to QTL (in centimorgans).
LOD score of marker at or closest to QTL.
Percentage of explained variance of the marker at or closest to QTL.
Additive effects are given as the difference between the means of the two genotypic groups of BC1 individuals (negative value implies that the A. halleri allele increases Zn tolerance compared to the A. l. petraea allele).
TABLE 4.
Epistatic interactions between QTL for Zn tolerance
| On raw EC100mean data
|
On log-transformed EC100mean data
|
|||
|---|---|---|---|---|
| Source | Fa | Pb | Fa | Pb |
| Zntol-1 | 31.53 | <0.0001 | 16.83 | <0.0001 |
| Zntol-2 | 41.85 | <0.0001 | 30.43 | <0.0001 |
| Zntol-3 | 30.23 | <0.0001 | 25.56 | <0.0001 |
| Zntol-1 × Zntol-2 | 4.75 | 0.0306 | 0.03 | 0.8664 |
| Zntol-1 × Zntol-3 | 5.51 | 0.0200 | 0.38 | 0.5592 |
| Zntol-2 × Zntol-3 | 3.96 | 0.0482 | 0.11 | 0.7428 |
Values of F-statistic, considering the markers fixed or random factors.
Significance levels, considering the markers fixed or random factors.
Figure 4.—
Box plot of Zn tolerance. (a) Genotypic groups of BC1 individuals at the markers HMA4, MTP1-A, and MTP1-B are numbered from 1 to 8. The number of BC1 individuals in each group is given by “n.” (b) Box plot of Zn tolerance by genotypic groups. The boxes represent the interquartile range, with arithmetic means indicated by solid circles and medians indicated by horizontal lines. Whiskers connect the nearest observations within 1.5 times the interquartile ranges of the lower and upper quartiles.
Figure 5.—
LOD score profiles and LOD support intervals for the QTL for Zn tolerance in the BC1 progeny. The linkage groups LG3, LG4, and LG6 on which a QTL for Zn tolerance was detected are represented. The LOD score profiles along the linkage groups are given at the right of each linkage group. Map positions are plotted along the vertical axis. LOD scores are plotted along the horizontal axis. Dashed lines correspond to the LOD score threshold (2.3) for QTL detection at an error level of α = 0.05. LOD support intervals are given at the left of each linkage group. The position of a QTL is shown as the interval over which the LOD score is within 1 or 2 log units of its maximum value, i.e., at the most likely position of the QTL. Bars indicate 1-LOD (10-fold) support intervals and whiskers (lines extending beyond bars) indicate 2-LOD (100-fold) support intervals.
DISCUSSION
Previous combinations of classical functional and transcriptional analyses have identified several genes potentially involved in the adaptation of A. halleri to high heavy metal concentrations. However, definitive evidence for their implication in heavy metal tolerance has not yet been provided, and the genetic mechanisms underlying this trait are still largely unknown. This article contributes to a better understanding of the genetic architecture of Zn tolerance by applying a QTL mapping approach to the Zn-tolerant species A. halleri. The linkage map, constructed on A. halleri × A. l. petraea BC1 progeny, revealed three QTL regions determining Zn tolerance, in which we would expect genes involved in the adaptation of A. halleri to high heavy metal concentrations to be located.
Genetic architecture of Zn tolerance in A. halleri:
Until recently, the constitutive nature of Zn tolerance in A. halleri (Pauwels et al. 2006) rendered its genetic analysis inaccessible. Macnair et al. (1999) was the first to circumvent this major handicap by analyzing the segregation of Zn tolerance in interspecific crosses performed between A. halleri and its closest nontolerant relative A. l. petraea. On the basis of segregation analysis of one F2 progeny, the authors hypothesized a single major gene determining Zn tolerance in A. halleri, as already described for other metals and species (Schat et al. 1993; Smith and Macnair 1998). Considering the two modes observed in the frequency distribution of the EC100mean values of the BC1 individuals, one might expect this distribution to indicate also the presence of a single major gene in the determinism of Zn tolerance. Our results based on a QTL analysis firmly establish that three additive QTL located on three different linkage groups are involved in the evolution of Zn tolerance in A. halleri. We believe that the results of the QTL analysis and the distribution of the phenotypic data are not contradictory for two main reasons. First, a significant deviation from the distribution expected under the hypothesis of a single major gene governing Zn tolerance was observed (P < 0.001). Second, the QTL analysis performed on log-transformed EC100 values, which is expected to improve the normal distribution of the phenotypic data, did not modify the results (data not shown). With the exception of a minor QTL on LG8 slightly exceeding the LOD score threshold, no other QTL and no major variation of the explained variance for the identified QTL were detected.
Different arguments can also be proposed to explain the discrepancy between our results and Macnair's conclusions. First, the methodologies to assess tolerance were different. In Macnair's study, Zn tolerance was evaluated at a single fixed concentration (250 μm) (Macnair et al. 1999), whereas in our study a multiple concentration test was applied to measure tolerance of the BC1 individuals; the latter is assumed more appropriate for assessing quantitative variations in tolerance levels (Schat and Ten Bookum 1992). Moreover, Macnair et al. (1999) used the lack of chlorosis as a subjective measure of tolerance rather than root growth. Second, the A. halleri genotypes used in both studies did not originate from the same metalliferous site. The A. halleri individual used in the QTL analysis was collected from Auby (France), a site with a very high metal contamination of relatively recent date (the beginning of the 20th century) resulting from the proximity to a Zn smelter factory (Van Rossum et al. 2004). In contrast, the A. halleri genotype used in Macnair's study (Macnair et al. 1999) originated from a suburb of Langelsheim (Germany); this site was reported to have become contaminated because of medieval metal-mining activities (Weber et al. 2004) and is characterized today by lower metal contamination levels. However, historical and genealogical data show high genetic relationships between German and French populations and suggest that A. halleri has been introduced in France from M sites such as Langelsheim located in Germany (Pauwels et al. 2005). Consequently, the different origin of A. halleri in both studies is not expected to contribute significantly to the discrepancies observed, even though the existence of specific local adaptations to metal contamination in both A. halleri populations might not be excluded.
The three QTL identified in this study explain 42% of the genetic variance of Zn tolerance, which means that we could have missed some QTL. The so-called “Beavis effect” predicts that in experiments using progeny sizes of ∼100 individuals, fewer QTL are identified than with larger progeny sizes of ∼400 (Beavis 1994). Moreover, estimates of genetic effects were reported to be inflated in experiments using progeny sizes of 100 compared to the ones using progenies of 400 individuals (Beavis 1994; Kearsey and Farquhar 1998; Xu 2003). According to the “Beavis effect,” a progeny of intermediate size (∼200 individuals), such as the one used in this QTL analysis of Zn tolerance, still suffers from a reduction in QTL detection power and an inflation of the estimates of the QTL effects (Beavis 1994; Kearsey and Farquhar 1998; Xu 2003). Such a reduction in power leading to a failure to detect a number of QTL in this experiment could explain the difference observed between broad-sense heritability of Zn tolerance in the BC1 progeny and the variance explained by the QTL. Segregation distortion is also believed to reduce the power of QTL detection and to affect the estimates of QTL effects, because it reduces the effective size of the progeny by reducing the size of one genotypic class (Bradshaw et al. 1998). Segregation distortion was reported for 40% of all markers in the BC1 population. It is therefore possible that some QTL for Zn tolerance, probably of minor effect, have not been detected. Among the QTL that have been detected, only the QTL Zntol-3 is located in a distorted region and showed a deficit in heterospecific allelic combinations. It is highly probable that this affected the estimation of the QTL effect since the mean Zn tolerance value was calculated on less heterospecific genotypes than the one calculated on the homospecific genotypes.
Evolutionary dynamics of metal tolerance in A. halleri:
At all three QTL positions, the tolerance-enhancing allele originated from the A. halleri parent, as expected under continuous directional selection (Orr 1998b). Because we used an A. halleri individual of metallicolous origin in our study, the identified QTL might reflect the constitutive tolerance observed in M and NM populations and the enhanced tolerance occurring in M populations on recently colonized industrial sites (Pauwels et al. 2006). In the present state of our knowledge, it remains unclear whether specific alleles at all three QTL are involved in enhanced tolerance or whether one or more QTL are specific for the constitutive tolerance. Crosses involving NM accessions might provide conclusive results to distinguish between these two hypotheses.
The QTL analysis of Cd tolerance was recently addressed in a subset of the interspecific BC1 population used in this study (M. Courbot, G. Willems, P. Motte, S. Arvidsson, P. Saumitou-Laprade and N. Verbruggen, unpublished results). Among the QTL identified for Zn and Cd tolerance, one was involved in both adaptive traits as inferred from the colocalization of the QTL Zntol-1 and the major QTL (>45% of genetic variance explained) for Cd tolerance. Because Zn and Cd are very often associated in soils naturally enriched in Zn (Ernst 1974), it is most parsimonious to hypothesize that Zn constitutive tolerance initially evolved in A. halleri refuge through the fixation of a QTL conferring tolerance to both metals. Increased tolerance to Zn and Cd might have been achieved more recently on industrial sites surrounding Zn smelters. In these sites, the concentrations of Zn and Cd are much higher than ever observed in naturally metal-enriched sites and this could have driven the selection of mechanisms specific to either Zn or Cd tolerance. The results presented here are in fair agreement with the predictions of the Fisher–Orr model of adaptation that describes the entire adaptive walk taken by a population to move toward a new fitness optimum and suggests that the size of the first factor fixed is fairly large (Orr 2005). However, the validity of this hypothesis should be verified using QTL mapping of heavy metal tolerance in recombinant populations from interspecific crosses between A. l. petraea and A. halleri from NM populations, which should reveal mainly the QTL associated with first adaptation (constitutive tolerance), and from intraspecific crosses between independently founded M and NM populations, which should detect QTL associated with the more recent adaptation to industrial polluted sites (Pauwels et al. 2005).
The interspecific map covers most of the A. halleri genome and synteny with other Arabidopsis genomes is high:
The extensive conservation of marker order between the interspecific Ah × Alp map and A. thaliana was in line with previous results reported for the A. l. petraea (Kuittinen et al. 2004; Koch and Kiefer 2005) and A. l. lyrata (Yogeeswaran et al. 2005) maps. The only discrepancy in marker order between the A. l. petraea and the interspecific map was observed for the loci PhyA and AXR1 (LG1/AL1), but, as suggested by Kuittinen et al. (2004), their order in A. l. petraea “could well be consistent with the order expected from A. thaliana.” The large inversion observed on AL6 of the A. l. petraea linkage map (Kuittinen et al. 2004) and confirmed in A. l. lyrata (Yogeeswaran et al. 2005) as well as in another close relative, Capsella rubella (Boivin et al. 2004), was not detected on the corresponding linkage group LG6 on the Ah × Alp map. Nevertheless, the low marker density in this region probably precluded the detection of this inversion.
Less efficient recombination has been reported in interspecific crosses due to the genetic divergence between the parental lines belonging to different species (Williams et al. 1995; Bernacchi and Tanksley 1997). However, the nonsignificant difference of the marker interval sizes between the Ah × Alp and the A. l. petraea map suggests that the recombination between the A. halleri and A. l. petraea genomes was as efficient in the interspecific hybrid as in the intraspecific cross. Significant differences of linkage group lengths were observed between the Ah × Alp map and either the A. l. petraea map or the A. l. lyrata map: the total length was increased in our mapping experiment compared to the A. lyrata maps. Since the inverse would be rather expected in such interspecific crosses in which absence of homology could reduce recombination and subsequent observed genome size, we assume that this just indicates the less complete saturation of the A. lyrata genetic linkage maps. We believe that our interspecific map covers most of the A. halleri genome, since markers situated near the extremities of A. thaliana chromosomes were used in the interspecific cross for map construction. Moreover, the markers located on the extremity of the LG4 (AL4, AlyLG3), LG6 (AL6, AlyLG7), and LG8 (AL8, AlyLG8) lower arms were nearer to the A. thaliana chromosome extremities in the interspecific map than in the A. l. petraea and the A. l. lyrata maps (Kuittinen et al. 2004; Yogeeswaran et al. 2005).
The genome sizes reported for A. thaliana (0.16 pg) and its close relatives (∼ 0.26 pg) (Johnstone et al. 2005) indicate that one or more deletion events might have accompanied the transition from eight to five chromosomes, characterizing the genome of A. thaliana. Ideally, the comparative analyses of the Ah × Alp and the A. lyrata maps with the A. thaliana genome should take these events into account. In this regard, the sequencing project on A. l. lyrata (http://www.jgi.doe.gov/sequencing/why/CSP2006/AlyrataCrubella.html) will be very valuable, since this will provide us with an exhaustive knowledge of the genome of the Arabidopsis relatives.
Segregation distortion:
At a significance threshold of 0.05, we might expect 5% of all markers to show distorted segregation by chance; in the BC1 population, we greatly exceeded this proportion. A failure to show the expected Mendelian ratios is rather common in interspecific crosses for different reasons (Zamir and Tadmor 1986; Bernacchi and Tanksley 1997; Jenczewski et al. 1997). In the BC1 progeny, segregation bias could be explained, at least partially, by the divergence time between the parental lines, which has been estimated to be about half (X. Vekemans, personal communication) the divergence time between A. thaliana and A. l. petraea (5.8 MYA) (Koch et al. 2001). Outbreeding depression, which in our case is supported by the large majority of the distorted markers (92%) showing an excess of homospecific vs. heterospecific allelic combinations, could also be at the origin of the high segregation distortion observed in the BC1 progeny. Finally, because the F1 and all the BC1 individuals belong to maternal progenies collected on A. l. petraea, the highly directional segregation bias observed in the BC1 could also indicate a negative interaction between the A. halleri alleles at the nuclear loci (or at closely linked loci) and the maternally inherited cytoplasmic genotype corresponding to A. l. petraea (Fishman et al. 2001).
Linkage map construction and, more precisely, estimation of recombination frequencies, can be affected by segregation distortion (Fishman et al. 2001; Kuittinen et al. 2004). However, because we applied stringent goodness-of-fit thresholds to minimize the effects of segregation distortion on the linkage map construction and observed macrosynteny between the Ah × Alp map and the A. l. petraea map, as well as between the Ah × Alp map and A. thaliana, we believe that the current map is quite robust.
Colocalization of known heavy metal homeostasis genes with the QTL for Zn tolerance:
The length of the LOD support intervals associated with the QTL for Zn tolerance reported in this experiment precludes the direct identification of the underlying genes. In A. thaliana, for instance, 1 cM has been reported to correspond to an average of 250 kb or ∼40 genes (Mauricio 2001). However, the correspondence between genetic and physical distances in the close relatives of A. thaliana is not known. In this context, the sequencing of the A. l. lyrata genome (http://www.jgi.doe.gov/sequencing/why/CSP2006/AlyrataCrubella.html) also will be very useful. On the current map, we reported the colocalization of three genes, HMA4, MTP1-A, and MTP1-B, with the three QTL regions for Zn tolerance.
HMA4 is a member of the family of P-type ATPases and colocalized with the QTL Zntol-1. In A. thaliana, HMA4 is expressed mainly in the roots and was shown to be involved in the root-to-shoot transport of Zn and Cd (Mills et al. 2003; Hussain et al. 2004). Compared to the A. thaliana orthologous gene, HMA4 was shown to be highly overexpressed in the roots of the Zn/Cd-tolerant and hyperaccumulator species T. caerulescens, indicating a possible role in translocation, as well as in the shoots where HMA4 may be involved in Zn/Cd detoxification (Bernard et al. 2004; Papoyan and Kochian 2004).
The metal homeostasis genes MTP1-A and MTP1-B mapped to the QTL Zntol-2 and Zntol-3, respectively. These genes are homologous to MTP1 from A. thaliana, formerly known as ZAT (van der Zaal et al. 1999), and to ZTP1 from T. caerulescens (Assunção et al. 2001), a cation diffusion facilitator, and were clearly shown by functional analysis to interact with zinc homeostasis in A. halleri (Dräger et al. 2004; Krämer 2005). In microarrays hybridized with labeled shoot cRNA, normalized signal intensities for ZAT/AtMTP1 were between 14- and 23-fold higher in A. halleri compared to A. thaliana (Becher et al. 2004). Under control conditions (1 μm Zn), the root steady-state MTP1 transcript levels in A. halleri were approximately equivalent to those in A. thaliana. Nevertheless, after exposure to 100–300 μm Zn, root MTP1 transcript abundance increased incrementally in A. halleri, but not in A. thaliana or in A. l. petraea (Dräger et al. 2004). Using the same BC1 population as the one investigated in this study, to separately analyze the expression of the MTP1 loci, Dräger et al. (2004) performed semiquantitative RT–PCR on RNA extracted from the shoots of selected BC1 individuals containing the different copies. In three BC1 individuals harboring the copies AhMTP1-B and AhMTP1-C, the transcript level of AhMTP1-B on average was 5.7-fold higher than the one of AhMTP1-C. The expression of AhMTP1-C was more or less equivalent to the expression of the MTP1 gene in A. l. petraea. In BC1 individuals carrying the copies AhMTP1-A and AhMTP1-B, the transcript levels of both loci together on average were 11.1-fold higher than the one of A. l. petraea MTP1 (Dräger et al. 2004).
The colocalization with the QTL for Zn tolerance (this study) and the differential expression and/or regulation demonstrated for AhMTP1-A and AhMTP1-B in response to Zn (Dräger et al. 2004) provide strong arguments in favor of adaptive modifications of these specific metal homeostasis genes (or their regulatory regions) in relation with Zn tolerance in A. halleri. This may also be the case for HMA4, which was described previously in T. caerulescens (Bernard et al. 2004; Papoyan and Kochian 2004). These genes can consequently be considered as good candidates for Zn tolerance and are currently being submitted to deeper investigations.
Zn tolerance and Zn accumulation remain unlinked in A. halleri:
Recently, an interspecific crossing scheme between A. halleri and A. l. petraea was used for identifying QTL involved in Zn accumulation in A. halleri (Filatov et al. 2006). By comparing gene expression of Zn-accumulating F3 families to non-Zn-accumulating F3 families, the authors identified 237 genes that were more expressed in the accumulating progenies. Deducing the chromosomal position of these genes from the A. l. petraea linkage map reported by Kuittinen et al. (2004), the authors identified 20 and 18 adjacent genes, respectively, belonging to two regions located on chromosomes 3 and 7 (Filatov et al. 2006), corresponding to LG3 and LG7 of the Ah × Alp map. None of these regions were identified in the QTL analysis of Zn tolerance reported here. These results confirm previous studies in which Zn hyperaccumulation and tolerance were shown to segregate independently in an A. halleri × A. l. petraea F2 progeny (Macnair et al. 1999) and suggest that in A. halleri both traits are expected to be governed, at least partially, by different genes.
In conclusion, our search for QTL controlling Zn tolerance in A. halleri revealed three genomic regions in which three metal homeostasis genes colocalized. To minimize the LOD support intervals associated with the QTL, we are currently increasing the marker density of the Ah × Alp map and producing second-generation backcross progenies. Finally, the Ah × Alp map constitutes a powerful tool available to the scientific community working on metal homeostasis genes: any gene of interest can be mapped on our material and characterized for its relationships with the QTL of Zn tolerance in A. halleri.
Acknowledgments
We thank Robert Dron, Claire Feutrie, and Eric Schmitt for technical advice and support in taking care of the plants and Maxime Pauwels and Stéphane Fenart for help in phenotyping. We are very grateful to Mark Macnair for providing A. lyrata seeds and to Vincent Castric, Adrian Radu Craciun, Marc Hanikenne, Charles Langley, Michel Lebrun, Stéphanie Loubet, Marie Mirouze, Tom Mitchell-Olds, Ronald Oomen, Outi Savolainen, Christian Schlötterer, Sébastien Thomine, and Fabrice Varoquaux for providing primer pairs and/or help in genotyping. We are also very grateful to Steve Barnes, Joel Cuguen, Ute Krämer, Patrick de Laguérie, Henk Schat, Pascal Touzet, and Xavier Vekemans for scientific discussions and helpful comments on the manuscript. This research was supported by the Contrat de Plan Etat/Région Nord-Pas de Calais (Programme de Recherché Concerté), the European Fonds Européens de Développement Régional (contract no. 79/1769), the Programme National/Action Concertée Incitative du Fond National de la Science ECCO (contract no. 04 2 9 FNS), the Belgian Science Policy (Interuniversity Attraction Pole Programme V/13), and by a grant from the Fonds National de la Recherche Scientifique (FRFC 2.4565.02), in addition to a doctoral fellowship to G.W. from the European Research Training Network Metalhome (HPRN-CT-2002-00243).
References
- Al-Hiyali, S. A., T. McNeilly and A. D. Bradshaw, 1988. The effect of zinc contamination from electricity pylons: evolution in a replicated situation. New Phytol. 110: 571–580. [Google Scholar]
- Alonso-Blanco, C., S. E. El-Assal, G. Coupland and M. Koornneef, 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics, J., A. D. Bradshaw and R. G. Turner, 1971. Heavy metal tolerance in plants. Adv. Ecol. Res. 7: 1–85. [Google Scholar]
- Assunção, A. G. L., P. Da Costa Mar, S. De Folter, R. Vooijs, H. Schat et al., 2001. Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 24: 217–226. [Google Scholar]
- Assunção, A. G. L., H. Schat and M. G. M. Aarts, 2003. Thlaspi caerulescens, an attractive model species to study heavy metal hyperaccumulation in plants. New Phytol. 159: 351–360. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Beavis, W. D., 1994. The power and deceit of QTL experiments: lessons from comparative QTL studies, pp. 250–266 in Proceedings of the 49th Annual Corn and Sorghum Industry Research Conference, edited by A. S. Trade. ASTA, Chicago.
- Becher, M., I. N. Talke, L. Krall and U. Krämer, 2004. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37: 251–268. [DOI] [PubMed] [Google Scholar]
- Bell, C., and J. Ecker, 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144. [DOI] [PubMed] [Google Scholar]
- Bernacchi, D., and S. D. Tanksley, 1997. An interspecific backcross of Lycopersicon esculentum × L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147: 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, C., N. Roosens, P. Czernic, M. Lebrun and N. Verbruggen, 2004. A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett. 569: 140–148. [DOI] [PubMed] [Google Scholar]
- Bert, V., I. Bonnin, P. Saumitou-Laprade, P. de Laguerie and D. Petit, 2002. Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytol. 155: 47–57. [DOI] [PubMed] [Google Scholar]
- Boivin, K., A. Acarkan, R. S. Mbulu, O. Clarenz and R. Schmidt, 2004. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae: a comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135: 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, H. D., Jr., K. G. Otto, B. E. Frewen, J. K. McKay and D. W. Schemske, 1998. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss, M. J., and M. A. Koch, 2006. Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci. 11: 449–459. [DOI] [PubMed] [Google Scholar]
- Clauss, M. J., H. Cobban and T. Mitchell-Olds, 2002. Cross-species microsatellite markers for elucidating population genetic structure in Arabidopsis and Arabis (Brassicaeae). Mol. Ecol. 11: 591–601. [DOI] [PubMed] [Google Scholar]
- Clemens, S., 2001. Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 475–486. [DOI] [PubMed] [Google Scholar]
- Craciun, A. R., M. Courbot, F. Bourgis, P. Salis, P. Saumitou-Laprade et al., 2006. Comparitive cDNA-AFLP analysis of Cd-tolerant and -sensitive genotypes derived from crosses between the Cd hyperaccumulator Arabidopsis halleri and Arabidopsis lyrata ssp. Petraea. J. Exp. Bot. 57: 2967–2983. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., 2002. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3: 43–52. [DOI] [PubMed] [Google Scholar]
- Dräger, D. B., A.-G. Desbrosses-Fonrouge, C. Krach, A. N. Chardonnens, R. C. Meyer et al., 2004. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 39: 425–439. [DOI] [PubMed] [Google Scholar]
- Erickson, D. L., C. B. Fenster, H. K. Stenoien and D. Price, 2004. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13: 2505–2522. [DOI] [PubMed] [Google Scholar]
- Ernst, W. H. O., 1974. Schwermetalvegetation der Erde. Gustav Fischer Verlag, Stuttgart.
- Filatov, V., J. Dowdle, N. Smirnoff, B. Ford-Lloyd, H. J. Newbury et al., 2006. Comparison of gene expression in segregating families identifies genes and genomic regions involved in a novel adaptation, zinc hyperaccumulation. Mol. Ecol. 15: 3045–3059. [DOI] [PubMed] [Google Scholar]
- Fishman, L., A. J. Kelly, E. Morgan and J. H. Willis, 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J., and R. Mott, 2001. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat. Rev. Genet. 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Glazier, A. M., J. H. Nadeau and T. J. Aitman, 2002. Finding genes that underlie complex traits. Science 298: 2345–2349. [DOI] [PubMed] [Google Scholar]
- Hall, J. L., 2002. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 53: 1–11. [PubMed] [Google Scholar]
- Hussain, D., M. J. Haydon, Y. Wang, E. Wong, S. M. Sherson et al., 2004. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenczewski, E., M. Gherardi, I. Bonnin, J. M. Prosperi, I. Olivieri et al., 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691. [Google Scholar]
- Johnstone, J. S., A. E. Pepper, A. E. Hall, Z. J. Chen, G. Hodnett et al., 2005. Evolution of genome size in Brassicaceae. Ann. Bot. 95: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, M. J., and A. G. Farquhar, 1998. QTL analysis in plants: Where are we now? Heredity 80: 137–142. [DOI] [PubMed] [Google Scholar]
- Koch, M. A., and M. Kiefer, 2005. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species–Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Koch, M., B. Haubold and T. Mitchell-Olds, 2001. Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88: 534–544. [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Krämer, U., 2005. MTP1 mops up excess zinc in Arabidopsis cells. Trends Plant Sci. 10: 313–315. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., M. Aguadé, D. Charlesworth, A. De Haan, B. Lauga et al., 2002. Primers for 22 candidate genes for ecological adaptations in Brassicaceae. Mol. Ecol. Notes 2: 258–262. [Google Scholar]
- Kuittinen, H., A. A. de Haan, C. Vogl, S. Oikarinen, J. Leppala et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambinon, J., and P. Auquier, 1964. La flore et la végétation des terrains calaminaires de la Wallonie septentrionale et de la Rhénanie aixoise. Types chorologiques et groupes écologiques. Natura Mosana 16: 113–130. [Google Scholar]
- Mackay, T. F., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Macnair, M. R., V. Bert, S. B. Huitson, P. Saumitou-Laprade and D. Petit, 1999. Zinc tolerance and hyperaccumulation are genetically independent characters. Proc. Biol. Sci. 266: 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio, R., 2001. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat. Rev. Genet. 2: 370–380. [DOI] [PubMed] [Google Scholar]
- Mills, R., G. Krijger, P. Baccarini, J. L. Hall and L. Williams, 2003. Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J. 35: 164–176. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds, T., 2001. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 16: 693–700. [Google Scholar]
- Oetting, W. S., H. K. Lee, D. J. Flanders, G. L. Wiesner, T. A. Sellers et al., 1995. Linkage analysis with multiplexed short tandem repeat polymorphism using infrared fluorescence and M13 tailed primers. Genomics 30: 450–458. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1998. a The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1998. b Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1999. The evolutionary genetics of adaptation: a simulation study. Genet. Res. 74: 207–214. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2002. The population genetics of adaptation: the adaptation of DNA sequences. Int. J. Org. Evol. 56: 1317–1330. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Papoyan, A., and L. V. Kochian, 2004. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 136: 3814–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, M., P. Saumitou-Laprade, A. Holl, D. Petit and I. Bonnin, 2005. Multiple origin of metallicolous populations of the pseudometallophyte Arabidopsis halleri (Brassicaceae) in central Europe: the cpDNA testimony. Mol. Ecol. 14: 4403–4414. [DOI] [PubMed] [Google Scholar]
- Pauwels, M., H. Frérot, I. Bonnin and P. Saumitou-Laprade, 2006. A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study: Arabidopsis halleri (Brassicaceae). J. Evol. Biol. 19: 1838–1850. [DOI] [PubMed] [Google Scholar]
- SAS, 1999. Statistical Analysis Systems. SAS Institute, Cary, NC.
- Saumitou-Laprade, P., Y. Piquot, O. Raspé, J. Bernard and K. Vrieling, 1999. Plant DNA fingerprinting and profiling, pp. 17–38 in DNA Profiling and DNA Fingerprinting, edited by J. T. Epplen and T. Lubjuhn. Birkhauser, Basel, Switzerland.
- Schat, H., and W. M. Ten Bookum, 1992. Genetic control of copper tolerance in Silene vulgaris. Heredity 68: 219–229. [Google Scholar]
- Schat, H., E. Kuiper, W. M. Ten Bookum and R. Vooijs, 1993. A general model for the genetic control of copper tolerance in Silene vulgaris: evidence from crosses between plants from different tolerant populations. Heredity 70: 142–147. [Google Scholar]
- Smith, S. E., and M. R. Macnair, 1998. Hypostatic modifiers cause variation in degree of copper tolerance in Mimulus guttatus. Heredity 80: 760–768. [Google Scholar]
- Tanksley, S. D., 1993. Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, J. L. Modliszewski, T. F. Mackay and M. D. Purugganan, 2002. Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160: 1133–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zaal, B. J., L. W. Neuteboom, J. E. Pinas, A. N. Chardonnens, H. Schat et al., 1999. Overexpression of a novel Arabidopsis gene related to putative zinc transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 119: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. Joinmap 3.0: Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Van Ooijen, J. W., M. P. Boer, R. C. Jansen and C. Maliepaard, 2002. MapQTL 4.0: Software for the Calculation of QTL Positions on Genetic Maps. Plant Research International, Wageningen, The Netherlands.
- Van Rossum, F., I. Bonnin, S. Fenart, M. Pauwels, D. Petit et al., 2004. Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-incompatible and heavy-metal-tolerant species. Mol. Ecol. 13: 2959–2967. [DOI] [PubMed] [Google Scholar]
- Voorrips, R. E., 2002. Mapchart: software for the graphical presentation of linkage maps and QTLs. Heredity 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans and T. van de Lee, 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M., E. Harada, C. Vess, E. Roepenack-Lahaye and S. Clemens, 2004. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J. 37: 269–281. [DOI] [PubMed] [Google Scholar]
- Weinig, C., J. R. Stinchcombe and J. Schmitt, 2003. QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Mol. Ecol. 12: 1153–1163. [DOI] [PubMed] [Google Scholar]
- Williams, C. G., M. M. Goodman and C. W. Stuber, 1995. Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics 141: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., A. D. Bradshaw and D. A. Thurman, 1975. The potential for evolution of heavy metal tolerance in plants. III. The rapid evolution of copper tolerance in Agrostis stolonifera. Heredity 34: 165–187. [Google Scholar]
- Xu, S., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., 2001. Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4: 130–135. [DOI] [PubMed] [Google Scholar]
- Yogeeswaran, K., A. Frary, T. L. York, A. Amenta, A. H. Lesser et al., 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir, D., and Y. Tadmor, 1986. Unequal segregation of nuclear genes in plants. Bot. Gaz. 147: 355–358. [Google Scholar]
- Zhou, J., and P. Goldsbourgh, 1995. Structure, organization and expression of metallothionein gene family in Arabidopsis. Mol. Gen. Genomics 248: 318–328. [DOI] [PubMed] [Google Scholar]