Abstract
Meiotic recombination gives rise to crossovers, which are required in most organisms for the faithful segregation of homologous chromosomes during meiotic cell division. Characterization of crossover-defective mutants has contributed much to our understanding of the molecular mechanism of crossover formation. We report here a molecular analysis of recombination in a Drosophila melanogaster crossover-defective mutant, mei-9. In the absence of mei-9 activity, postmeiotic segregation associated with noncrossovers occurs at the expense of crossover products, suggesting that the underlying meiotic function for MEI-9 is in crossover formation rather than mismatch repair. In support of this, analysis of the arrangement of heteroduplex DNA in the postmeiotic segregation products reveals different patterns from those observed in Drosophila Msh6 mutants, which are mismatch-repair defective. This analysis also provides evidence that the double-strand break repair model applies to meiotic recombination in Drosophila. Our results support a model in which MEI-9 nicks Holliday junctions to generate crossovers during meiotic recombination, and, in the absence of MEI-9 activity, the double Holliday junction intermediate instead undergoes dissolution to generate noncrossover products in which heteroduplex is unrepaired.
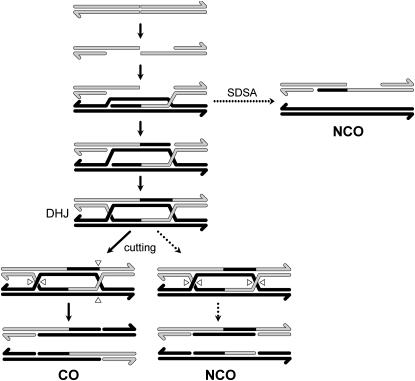
ACCURATE chromosome segregation during meiosis requires crossovers (COs) between homologous chromosomes, which are generated through meiotic recombination. A number of CO-defective mutants have been identified in model organisms (reviewed in Villeneuve and Hillers 2001; McKim et al. 2002). Much of our understanding of the molecular mechanism of meiotic recombination comes from genetic studies of the meiotic phenotypes of these mutants, molecular cloning and identification of the genes affected, and biochemical studies of the properties of the protein products of these genes. These analyses have led to the establishment of the double-strand break repair (DSBR) model for meiotic recombination (Figure 1) (Szostak et al. 1983).
Figure 1.—
DSBR model for meiotic recombination According to this model, recombination initiates with the introduction of a double-strand break (DSB) on one chromatid (shaded lines; arrows indicate 3′ ends), followed by 5′–3′ resection of the ends to leave 3′ single-stranded overhangs. One 3′ end invades the duplex of a chromatid of the homologous chromosome (solid lines), base pairing with the complementary strand and displacing the other strand as a D-loop. Synthesis follows, primed by the 3′ end of the broken chromosome and using the invaded chromosome as a template. This strand either dissociates, reannealing to the second broken end to generate an NCO by SDSA, or, alternatively, the D-loop anneals to the second free 3′ end and additional synthesis and ligation produce the double Holliday junction (DHJ) intermediate. The DHJ is resolved by cutting to generate CO or NCO products.
COs are an important product of meiotic recombination because they direct the segregation of homologous chromosomes from one another; however, meiotic recombination also gives rise to noncrossover (NCO) products. COs are easily recognized by the exchange of flanking markers, but NCOs can be distinguished from nonrecombinant chromosomes only when accompanied by gene conversion (GC). According to the DSBR model, GC results from the repair of mismatches in heteroduplex DNA (hDNA), DNA in which each strand of the duplex is derived from a different parental chromosome.
In the canonical DSBR model, COs and NCOs are alternate outcomes of resolution of a common recombination intermediate, the double Holliday junction (DHJ) structure (Figure 1). The existence of a class of mutations that reduce the number of COs but not the number of NCOs argues against this feature of the model. This class includes mutations in MUS81, MMS4, MSH4, MSH5, and MLH1 in Saccharomyces cerevisiae (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; de los Santos et al. 2001, 2003) and in mei-218, rec, and mei-9 in Drosophila (Carpenter 1982; Blanton et al. 2005). Analysis of these mutants suggests that there is a split in the recombination pathway with one branch leading to COs and the other to NCOs; these mutants are defective in the CO-specific branch. In support of this interpretation, most NCOs in S. cerevisiae are now thought to be produced by synthesis-dependent strand annealing (SDSA), with the DHJ being resolved primarily into COs (Figure 1) (Allers and Lichten 2001a).
Although the number of NCOs is not decreased in CO-defective mutants, in some cases these mutants produce NCOs with properties that distinguish them from normal NCOs, such as differences in GC tract length or repair of hDNA (Carpenter 1982; Hunter and Borts 1997; Blanton et al. 2005). One possible explanation is that these genes encode proteins that function in the CO branch and in the NCO branch, perhaps with different roles in each. It is also possible that these proteins may function solely in the CO branch, and the effect on NCOs is a consequence of an inability to complete the CO pathway (i.e., recombination events that were fated to become COs instead become NCOs). Detailed studies of the properties of NCOs produced by these mutants can provide insights into the molecular mechanism of meiotic recombination as well as specific functions of these proteins.
We recently reported analysis of NCOs in Drosophila rec mutants (Blanton et al. 2005). The average length of GC tracts among NCOs is lower in rec mutants than in wild type, suggesting that REC facilitates repair synthesis during meiotic recombination and that, as is thought to be the case in S. cerevisiae, most NCOs in Drosophila arise through SDSA. Mutations in mei-9 have a different effect on NCOs: they frequently exhibit postmeiotic segregation (PMS) (Romans 1980b; Hilliker and Chovnick 1981; Carpenter 1982, 1984; Bhagat et al. 2004). PMS arises from a failure to repair heterologies in hDNA, resulting in sister chromatids containing different sequence information after the first round of postmeiotic replication. With the possible exception of mei-9, all mutations that cause PMS in S. cerevisiae or Drosophila are in genes encoding proteins known to be involved in mismatch repair (MMR) (reviewed in Borts et al. 2000; Radford et al. 2007, this issue).
Molecular cloning of mei-9 revealed that it encodes the Drosophila ortholog of mammalian XPF and S. cerevisiae Rad1p (Sekelsky et al. 1995), the catalytic subunits of DNA structure-specific endonucleases required for nucleotide excision repair (Bardwell et al. 1994; Park et al. 1995). This led to the hypothesis that the function of MEI-9 in generating COs is to nick Holliday junctions in DHJ intermediates and that in the absence of MEI-9 these DHJs undergo some process that generates NCOs that are refractory to MMR (Sekelsky et al. 1995, 1998). This hypothesis predicts that most NCOs from mei-9 mutants will be identical to NCOs from wild-type flies, but the subset of NCOs that arise through MEI-9-independent processing of DHJs will exhibit PMS. An alternative hypothesis is that MEI-9 functions both in generating COs and in meiotic MMR and that PMS in mei-9 mutants is a consequence of defects in MMR. In support of this hypothesis, extracts from embryos mutant for mei-9 have defects in nick-dependent MMR (Bhui-Kaur et al. 1998); however, it is not known how this function relates to MMR during meiosis. If MEI-9 is essential for meiotic MMR, then most or all recombinants from mei-9 mutants should have PMS.
To distinguish between these two hypotheses, we conducted a molecular analysis of recombination products from mei-9 mutants. We report here that most NCOs from mei-9 mutants are indistinguishable from NCOs from wild type in that PMS is absent and GC tracts are continuous and similar in length. The subset of NCOs that did exhibit PMS often had two regions of PMS in the trans orientation. Our findings, coupled with findings from previous studies of recombination in mei-9 mutants and in an MMR-defective mutant, indicate that MEI-9 is not essential for meiotic MMR, although it may function in some specialized repair pathways, and suggest that NCOs with PMS arise from recombination events that were unable to become COs in the absence of MEI-9 activity.
MATERIALS AND METHODS
For mei-9 mutants, 120 virgin females of genotype y mei-9a f; kar ry606/ry531 cv-c were crossed to 30 males of genotype y/Dp(1;Y)y+; kar ry506 cv-c. For wild type, 20–30 females and 10 males were used. Crosses were set up in bottles containing 25 ml of standard food medium and placed at 25°. After 3 days, flies were transferred to fresh media to establish a second brood, and purine was added to the first brood bottles in the amount of 0.75 ml of 0.15, 0.18, or 0.20% (w/v) in water. This amount corresponds to 1.1 mg (9.4 μmol), 1.35 mg (11.2 μmol), and 1.5 mg (12.5 μmol) of purine per bottle. One of every 25 bottles was left untreated and adult progeny were counted to estimate the number of larvae screened. Purine dosage did not grossly affect the recovery of PMS events: we recovered 1 PMS event of 275,000 screened at 0.15%, 2 of 525,000 at 0.18%, and 2 of 550,000 at 0.20%. In contrast, the number of ry mutants that escaped killing by purine was strongly affected by purine dosage: escapers increased ∼10-fold between 0.18 and 0.15% (data not shown).
To determine whether a recombinant chromosome is CO or NCO, visible markers flanking ry (kar, which is 0.3 map units proximal to ry on the ry606 chromosome, and cv-c, which is 2.1 map units distal to ry on the ry531 chromosome) were scored in progeny surviving purine selection. Recombinant progeny were mated to kar ry506 cv-c flies of the opposite sex to detect mosaicism via germline transmission. After mating, each recombinant fly was homogenized in buffer containing proteinase K, as described (Gloor et al. 1993), and hDNA and GC tracts were determined by PCR amplification and sequencing.
To detect PMS, allele-specific PCR primers for several polymorphisms were used, as in Radford et al. (2007). Amplification with both allele-specific primers indicates PMS at that polymorphism. Each set of allele-specific PCR reactions included positive and negative controls, as well as 1:4 mixtures of DNA from both alleles to simulate a mosaic in which one allele is present in only 20% of the DNA molecules. Allele-specific PCR products were purified and sequenced to determine the length and arrangement of hDNA tracts. Additional non-allele-specific primers were also used to detect PMS. PCR products were sequenced in bulk and the chromatogram was examined for double peaks. Sequences of allele-specific primers, PCR conditions, and an example of mapping of an hDNA tract are given in Radford et al. (2007).
For one event, it was not possible to design allele-specific primers to confirm the PMS and map the hDNA tracts because of low sequence complexity in the region. Instead, non-allele-specific primers were used to amplify the region, and the product was cloned into a convenient vector for amplification in Escherichia coli. At least one clone representing each strand of the hDNA region was sequenced.
Mean GC tract lengths and statistical comparisons were calculated as described in Blanton et al. (2005). Frequency comparisons were made using Fisher's exact test with two-tailed P-values, computed by Instat 3.05 (GraphPad software).
RESULTS
To distinguish between models for the role(s) of MEI-9 in meiotic recombination, we recovered recombination events within the rosy (ry) locus, using a procedure developed by Chovnick and colleagues (Chovnick et al. 1970, 1971). The ry gene encodes xanthine dehydrogenase (XDH), which is involved in purine metabolism and is required for normal eye pigmentation. Females trans-heterozygous for ry606 and ry531, point mutations separated by 3.8 kb, were crossed to males homozygous for ry506, which deletes much of the gene. Rare rosy+ recombinants were selected by adding purine to the food during larval development. Among recombinants, COs and NCOs are distinguished from one another using markers flanking ry (see materials and methods for details). In female meiosis, only one of the two chromatids involved in any recombination event enters the oocyte, so it is not possible to determine whether a CO has an associated GC tract. In contrast, NCOs are recovered only when accompanied by a GC tract that spans one ry mutation. Hence, the COs described here may or may not be associated with GC (or PMS), but the NCOs must be associated with GC (or PMS).
Most NCOs from mei-9 mutants do not exhibit PMS:
PMS results when mismatches in hDNA are not repaired during meiosis. When unrepaired hDNA is present in a gamete, DNA replication in the first zygotic S phase produces sister chromatids that differ in sequence. Segregation of these sisters at the first mitosis results in daughter cells that have different sequences where the hDNA was. In a metazoan such as Drosophila, PMS manifests as a mosaic individual, in which some cells have the sequence from one strand of the recombinant chromatid and other cells have the sequence from the other strand. Most of the polymorphisms used in this study do not cause a visible mutant phenotype, so these mosaics can be detected only by molecular methods. Although the two ry point mutations do cause a visible mutant phenotype (rosy-colored eyes), XDH is secreted and diffuses throughout the developing larva, allowing ry+//ry− mosaics that survive purine treatment to develop into adults that are rosy+ in eye color (Romans 1980a); therefore, mosaicism for the mutant sites also cannot be detected visibly in the recombinant fly. We used three assays to screen rosy+ adults for mosaicism: germline sampling, allele-specific PCR, and examination of chromatograms for double peaks after sequencing non-allele-specific PCR products in bulk (Radford et al. 2007). Germline sampling can detect PMS only at the ry mutant sites and does not detect all ry+//ry− mosaics (Carpenter 1982; Radford et al. 2007), but PCR-based assays provide a more sensitive method to detect PMS at the mutant sites and also at silent polymorphisms.
We previously reported analysis of 81 COs and 31 NCOs from wild-type females (Blanton et al. 2005). We have now recovered an additional 31 COs and 22 NCOs (Table 1). We screened 1.4 million larvae from mei-9 mutant females and recovered 5 COs and 32 NCOs. This represents a 90% decrease in COs and a 60% increase in NCOs compared to wild type, similar to previously published findings (Romans 1980b; Carpenter 1982).
TABLE 1.
Intragenic recombination in wild-type, Msh6, and mei-9 mutants
| Progeny screened | Crossovers
|
Noncrossovers
|
|||||
|---|---|---|---|---|---|---|---|
| Genotype | n | Frequency | PMS (%) | n | Frequency | PMS (%) | |
| Wild typea | 3,710,000 | 112 | 3.0 × 10−5 | 0 | 53 | 1.4 × 10−5 | 0 |
| Msh6b | 1,775,000 | 67 | 3.8 × 10−5 | 21 | 42 | 2.4 × 10−5 | 58 |
| mei-9 | 1,405,000 | 5 | 0.36 × 10−5 | 0 | 32 | 2.3 × 10−5 | 16 |
Includes data from Blanton et al. (2005).
Data are from Radford et al. (2007).
Using genetic approaches, Chovnick et al. (1971) found PMS to be exceedingly rare among NCOs from wild-type females. Our molecular analyses gave similar results: we did not detect PMS in any of the 112 COs or 53 NCOs from wild-type females (Table 1) (Blanton et al. 2005). We did not detect PMS in any of the five COs from mei-9 mutant females; however, 5 of 32 NCOs exhibited PMS. This frequency of PMS in NCOs from mei-9 mutants (16%) is significantly higher than that in NCOs from wild type (P = 0.0061). We previously found that meiotic recombination in an MMR mutant results in frequent PMS: we detected PMS in 14 of 66 COs (21%) and 23 of 40 NCOs (58%) derived from females mutant for Msh6 (Radford et al. 2007). The rate of PMS among NCOs from mei-9 mutants is significantly lower than that in NCOs from Msh6 mutants (P = 0.0005). These results do not support the hypothesis that MEI-9 is essential for meiotic MMR.
Most non-PMS NCOs from mei-9 mutants are indistinguishable from NCOs from wild type:
The relatively low frequency of PMS among NCOs from mei-9 mutants indicates that MEI-9 is not absolutely required for meiotic MMR. The five NCOs with PMS may represent recombination events that were fated to become COs, but in the absence of MEI-9 became NCOs through a pathway that does not involve MMR. This hypothesis predicts that the NCOs without PMS will be identical to NCOs from wild-type females. An alternative hypothesis is that MEI-9 has a function in generating crossovers and a separate, limited function in meiotic MMR. This hypothesis predicts that some recombinants from mei-9 mutants will be similar to those from an MMR mutant. To test these hypotheses, we used the multiple polymorphisms between the two ry alleles to map GC and hDNA tracts.
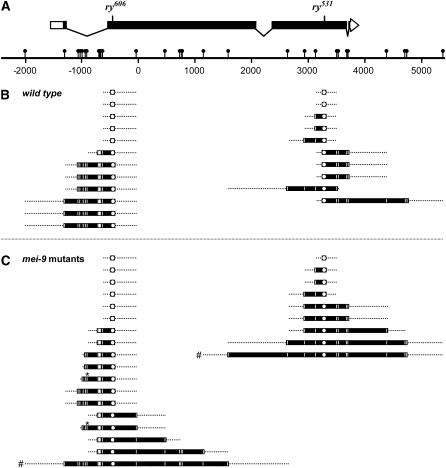
We previously described GC tracts from 29 NCOs from wild-type flies and found that, with one exception, all tracts were continuous (Blanton et al. 2005). We mapped an additional 22 GC tracts from wild-type females and found that all were continuous (Figure 2). We also mapped the 27 non-PMS GC tracts from mei-9 mutants. Two discontinuities were found, both at the −937 polymorphism (Figure 2, asterisks). The frequency of discontinuity is not significantly different between wild type and mei-9 mutants (P = 0.6). In contrast, 70% of NCOs from Msh6 mutants show evidence of discontinuities (Radford et al. 2007), which is significantly different from both wild type and mei-9 (P < 0.0001). Thus, with respect to continuity, GC tracts from mei-9 mutants are indistinguishable from those from wild-type females, but are unlike those from an MMR mutant.
Figure 2.—
GC tracts from wild-type and mei-9 mutants. (A) Schematic of the rosy locus. Intron/exon structure is shown, with coding sequences as solid areas. The positions of the selected sites corresponding to the ry606 and ry531 chromosomes are indicated. Additional polymorphisms are indicated as lollipops on the scale bar. These are all single-nucleotide polymorphisms, except for −1029 and −685, which are insertions of 1 and 4 bp, respectively, in ry531 relative to ry606. The scale is in base pairs, using the coordinate system of Bender et al. (1983). (B and C) Tract lengths observed in NCOs recovered from wild-type (B) and mei-9 mutants (C). Each bar represents an independent event, with the circle denoting the selected marker (ry606 or ry531 mutant sites). Solid bars represent the minimum tract length for each event, with coconverted sites marked by the vertical lines within the bars. Dotted lines represent the maximum tract length possible based on the next unconverted polymorphism. The asterisks mark two instances in which conversion at the −937 polymorphism was discontinuous (see materials and methods). Pound signs mark the two unusually long GC tracts (see results).
We also determined mean GC tract length among non-PMS NCOs. Recombination at ry is thought to initiate throughout the gene, rather than at one end (Clark et al. 1988); therefore, our experimental design selects for longer GC tracts, since longer tracts are more likely to cross a ry mutant site. To estimate the mean tract length in the absence of such selection (unselected tract length), we employed a statistical analysis developed previously (Hilliker et al. 1994; Blanton et al. 2005). The unselected mean GC tract length in NCOs is 425 bp for wild-type females and 509 bp for mei-9 females. The difference is statistically significant (P = 0.03). Closer inspection of the GC tracts from mei-9 mutants reveals two tracts that are much longer than the others (Figure 2, pound signs). Analysis of actual (selected) tract lengths, taking the end of each tract to be the midpoint between the last polymorphism contained within the tract and the nearest polymorphism excluded from the tract, shows that these two long tracts are more than two standard deviations greater than the mean (mean = 1076 bp, standard deviation = 1026 bp, lengths > 3700 bp). Excluding these two aberrant tracts, the mean unselected length of GCs from mei-9 mutants is 417 bp, which is not significantly different from the wild-type length (P = 0.8). This suggests that most non-PMS NCOs from mei-9 mutants are indistinguishable from NCOs from wild type in terms of GC tract length. These results also raise the possibility that loss of MEI-9 activity may lead to very long GC tracts in some instances.
Taken together, our results demonstrate that most NCOs from mei-9 mutants (25 of 32; 78%) are indistinguishable from NCOs from wild type. This finding suggests that most or all NCOs in wild-type flies arise through a MEI-9-independent pathway, providing further support for a model for meiotic recombination in which COs and NCOs arise from separate branches of the meiotic recombination pathway.
PMS NCOs from mei-9 mutants exhibit trans hDNA:
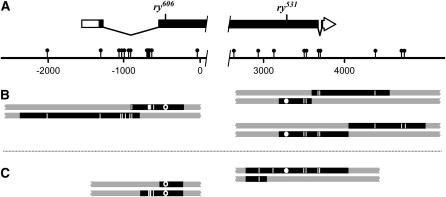
We sequenced PCR products from the mosaic flies and their progeny (see materials and methods) to determine the arrangement of hDNA in the PMS NCOs from mei-9 mutants. We classify the five PMS events from mei-9 mutants into two types (Figure 3). In the first type (three events, Figure 3B), there are two adjacent hDNA tracts in the trans configuration. In one of these, a single site between the two hDNA tracts is fully converted. In the second type (two events, Figure 3C), there is a single hDNA tract adjacent to a single site that is fully converted.
Figure 3.—
hDNA tracts from meiotic recombination in mei-9 mutants. (A) Schematic of the rosy locus. The symbols are the same as in Figure 2, but only the left and right ends of the gene are shown. (B) Three events with trans hDNA. Each pair of bars represents the two strands of a recombinant chromosome with unrepaired hDNA. Solid segments are sequences derived from the homologous chromosome, with vertical hatches showing positions of polymorphisms within hDNA and GC tracts. The position of the ry606 point mutation is denoted by an open circle and that of ry531 by a solid circle. The ends of the solid segments are drawn at the midpoints between the last included polymorphism and next excluded polymorphism. In all three cases, hDNA on each side was extensive (245–1530 bp) and included at least three polymorphisms. The event that includes ry606 showed full conversion of a single polymorphism between regions of hDNA. (C) Two events with a single hDNA tract. In both cases, a single, fully converted polymorphism is present at one end of the tract.
A prediction of models in which MEI-9 participates in meiotic MMR is that the PMS NCOs from mei-9 mutants will resemble those from Msh6 mutants. A striking feature of PMS NCOs from Msh6 mutants is that tracts were frequently discontinuous. We classify an event as discontinuous if there are three or more regions of GC, restoration, and hDNA. Of 18 PMS NCOs from Msh6 mutants that spanned three or more polymorphisms, 17 (94%) were discontinuous (Radford et al. 2007). In contrast, of the 5 PMS NCOs from mei-9 mutants (all of which spanned at least six polymorphisms), only one was discontinuous (Figure 3B, event on the left). Thus, PMS NCOs from mei-9 mutants are structurally dissimilar from PMS NCOs from an MMR mutant, strengthening our conclusion that MEI-9 is not an essential component of the canonical MMR pathway that operates during meiotic recombination.
DISCUSSION
In this study, we set out to conduct a careful analysis of meiotic gene conversion and postmeiotic segregation in mei-9 mutants. Insights into meiotic functions of MEI-9 are obtained from consideration of the frequency of PMS, the structure of hDNA in recombination products that exhibited PMS, and comparisons with PMS events from a mutant defective in MMR.
The frequency of PMS in mei-9 mutants:
We found PMS in only 5 of 32 (16%) NCOs from mei-9 mutants (Table 1). This is a lower frequency than that in previous studies, in which the frequency of PMS in NCOs from mei-9 mutants was 60–100% (Romans 1980b; Carpenter 1982; Bhagat et al. 2004). We considered several possible explanations for the different results. First, we would observe a lower frequency of PMS if mosaic flies did not survive purine selection as efficiently in our experiments as in previous studies. The degree to which ry+//ry− mosaic larvae survive purine selection is dependent on the dose of purine used (Romans 1980a). It is not possible to compare purine doses used in experiments in different laboratories, because the final concentration of purine depends on the volume of food in each bottle, which is usually not specified. We used three different amounts of purine: 1.1 mg, 1.35 mg, and 1.5 mg per bottle (see materials and methods). There was no difference in frequency of PMS events within this range. There was, however, a 10-fold increase in survival of ry− larvae between the two lowest doses, suggesting that the purine selection we employed was not a substantial barrier to recovery of ry+//ry− mosaics.
A second possible source of the difference in PMS frequency in our experiments vs. previous studies is that the methods we employed (germline sampling and molecular assays) gave false negatives or the methods used previously (germline sampling and staining of adult tissues for XDH activity) gave false positives. In previous studies (Romans 1980b; Carpenter 1982; Bhagat et al. 2004), PMS was not detected in recombination events from wild-type females, suggesting that the high frequencies of PMS reported for recombination events from mei-9 mutants did not result from false positives. Conversely, several considerations suggest that the low frequency of PMS we found is not due to false negatives. First, controls run alongside every allele-specific PCR reaction (see materials and methods) gave the expected results, and we routinely detected the mutant allele when it comprised only 20% of the available template molecules (the lowest concentration tested). Second, results from each of the three methods we used were internally consistent (e.g., the single rosy+ recombinant that transmitted a ry− chromosome through the germline was also classified as PMS in both molecular assays we employed). Third, using identical methodologies, we found high levels of PMS in recombination events from Msh6 mutants (Radford et al. 2007), indicating that these methods are efficient in detecting mosaicism.
On the basis of the arguments presented above, we conclude that the low frequency of PMS in our experiments is not an artifact resulting from inability to recover mosaic flies or inability to detect mosaicism. Rather, our results appear to reflect a real difference in PMS frequency, presumably due to other differences in experimental design. Although the different studies all used the genetically null mutation mei-9a, which is a point mutation predicted to destroy nuclease activity (Yildiz et al. 2004), we used different ry alleles than did Romans (1980b) and Carpenter (1982). The alleles they used (ry5 and ry41) are deletions of 19 and 3 bp, which generate small insertion–deletion loops in hDNA. In contrast, the alleles we used (ry606 and ry531) are missense mutations that generate base–base mismatches in hDNA. In S. cerevisiae, loops are repaired by different proteins than base–base mismatches (Marsischky et al. 1996). Importantly, the S. cerevisiae ortholog of MEI-9 (Rad1) has been directly implicated in the repair of loops in meiotic hDNA, but no role in repair of base–base mismatches has been identified (Kirkpatrick and Petes 1997; Kearney et al. 2001; Stone and Petes 2006). If this aspect of meiotic MMR is conserved in Drosophila, then the relatively high rates of PMS reported by Romans and by Carpenter may be due to defects in loop repair. Our alleles did include two insertion–deletion heterologies: ry531 has an insertion of 1 bp at −1029 and an insertion of 4 bp at −685 relative to ry606. We did not find an increased frequency of PMS at these sites relative to single-nucleotide polymorphisms; however, nearby base–base mismatches (6 and 14 bp flanking the 4-bp insertion and 33 and 32 bp flanking the 1-bp insertion) are expected to stimulate efficient repair of the loops by canonical long-tract MMR (Detloff and Petes 1992).
The ry alleles we used also differed from those used by Romans and by Carpenter in the level of sequence heterology. There are no sequence differences between ry5 and ry41 other than the small deletions at the mutant sites (S. J. Radford, unpublished data). In contrast, we chose alleles that differed from one another at many sites (∼0.4% heterology, Figure 2A) so that we could map conversion and PMS tracts with high resolution. In S. cerevisiae, the presence of even a few mismatches decreases the rate of meiotic recombination dramatically (Borts and Haber 1987), but mutations in MMR genes restore normal rates of recombination (Chambers et al. 1996; Hunter et al. 1996). Drosophila differs in that high levels of heterology do not affect the frequency of meiotic recombination (Hilliker et al. 1991). Likewise, loss of MMR does not increase the frequency of meiotic recombination between highly polymorphic chromosomes, although it does allow recovery of recombination events that would have been lost in our selection scheme due to repair of hDNA restoring a mutant allele (Radford et al. 2007). In the experiments of Romans and Carpenter, the only heterology in hDNA crosses would be the small unpaired loop caused by the deletion mutation, whereas recombination between the alleles we used generates hDNA with multiple base–base mismatches. It is possible that these numerous mismatches provide greater opportunity for recruitment of MMR machinery in mei-9 mutants, leading to the lower frequency of PMS that we observed.
The relatively low frequency of PMS in our experiments compared to previous experiments may be due to one or more of the reasons discussed above; however, the source of the difference does not affect the data and interpretations described below.
MEI-9 in repair of meiotic hDNA:
We used identical conditions and methodologies to assess the occurrence and structural properties of PMS events from mei-9 mutants and Msh6 mutants (Radford et al. 2007). The frequency of PMS among NCOs from mei-9 mutants was significantly lower than the frequency among NCOs from Msh6 mutants (Table 1), leading us to conclude that MEI-9 is not essential for meiotic MMR of the base–base mismatches present in these experiments. It remains possible that MEI-9 has one or more specialized roles in repair of meiotic hDNA. As discussed above, MEI-9 may be important in repair of unpaired loops in hDNA. It is also possible that MEI-9 is required for MMR of recombination intermediates and products formed during certain NCO pathways, such as DHJ dissolution (see below), because DNA structures are formed that require the endonuclease function of MEI-9 for MMR access. A third possible role for MEI-9 is in short-patch repair. The high frequency of discontinuity among PMS NCOs from Msh6 led us to propose the existence of a short-patch repair system that operates when canonical MMR is compromised (Radford et al. 2007). Only one PMS NCO from mei-9 was discontinuous (Figure 3B, event on left). The structure of this chromatid—GC at a single polymorphic site between two tracts of hDNA in the trans orientation—suggests that the region of GC is due to “early MMR” (see below), rather than short-patch repair. The apparent absence of short-patch repair in NCOs from mei-9 mutants may indicate that MEI-9 is required for this process. In Schizosaccharomyces pombe, there is evidence for a short-patch excision repair pathway that operates on recombination intermediates and that requires the ortholog of MEI-9, Rad16 (Fleck et al. 1999). The lack of short-patch repair in mei-9 mutants could also be due to the presence of MSH6, if binding of MSH6 complexes to mismatches prevents access by short-patch repair proteins. This hypothesis does not exclude a role for MEI-9 in short-patch repair.
The origin of PMS in mei-9 mutants:
Examination of the structure of hDNA in NCOs associated with PMS from mei-9 mutants provides insights into possible functions for MEI-9 in meiotic recombination. Three of the five PMS events we recovered from mei-9 mutants had two tracts of hDNA in the trans orientation (Figure 3B), and two had a single tract of hDNA adjacent to a single site that was fully converted (Figure 3C). It is possible that these latter two events had trans hDNA that went undetected. Tracts of hDNA that do not include a polymorphism cannot be detected. On the basis of the nearest polymorphism adjacent to the site of full GC, the second region of hDNA would have to have been <304 bp in one case and <424 bp in the other. These lengths are both within two standard deviations of the mean hDNA tract length computed using the midpoint between polymorphisms contained and excluded within each of the eight hDNA tracts detected (mean = 805 bp, standard deviation = 381 bp); therefore, we cannot exclude the possibility that in these two cases there was a second hDNA tract that did not include any heterologies.
We may also fail to detect trans hDNA if one tract undergoes repair that restores the sequence originally on that chromatid. If each of the tracts in a molecule with trans hDNA can be repaired independently, and there is no bias toward restoration repair, we would predict the occurrence of events in which one of the hDNA tracts underwent conversion repair to generate an extensive region of full GC adjacent to a region of hDNA. We did not recover any events with this structure; however, given the small sample size (n = 2), failure to observe such events cannot be interpreted as evidence against the hypothesis that the two tracts might be repaired independently. Independent repair of trans hDNA tracts could also result in long GC tracts if both tracts are repaired toward conversion. This is a possible explanation for the two unusually long GC tracts we observed among NCOs from mei-9 mutants (Figure 2, pound signs).
Three of the five PMS events from mei-9 mutants (one from the first class and both from the second class) had a tract of full gene conversion (Figure 3). In all three cases, the GC tract included only a single polymorphic site. A region of full conversion would result if initiation of recombination involves formation of a double-strand gap or if there is partial repair of hDNA. In S. cerevisiae meiotic recombination, mismatches formed during the initial strand invasion are thought to undergo “early” repair (i.e., repair occurs during strand invasion or synthesis, Figure 1), whereas mismatches further away from the site of initiation are thought to be repaired later in the recombination process (Detloff et al. 1992; Alani et al. 1994; Foss et al. 1999; Hillers and Stahl 1999; Stahl and Hillers 2000). Meiotic recombination in S. cerevisiae is initiated at hotspots, which are frequently in promoter regions (Gerton et al. 2000), so early repair results in higher rates of GC for markers near the 5′ end of the gene. In Drosophila, recombination at ry is thought to initiate and terminate throughout the gene rather than at a hotspot at one end (Clark et al. 1988; Radford et al. 2007), so we cannot determine whether the sites of full GC are near the site of initiation. Nonetheless, the positions of the sites of full GC—between two tracts of trans hDNA in one event and at the end of a single hDNA tract in two events—are consistent with the hypothesis that these short conversion tracts arose from early repair.
Formation of trans hDNA is not consistent with models in which recombination is initiated by nicks, as in the models proposed by Holliday (1964) and by Meselson and Radding (1975). Rather, trans hDNA is consistent with models in which recombination is initiated by a double-strand break (DSB), because this allows both strands of the cut duplex to receive information from the homologous chromosome (Figure 1). In S. cerevisiae and S. pombe, the DSBR model is supported by physical detection of DSBs and key recombination intermediates (Collins and Newlon 1994; Schwacha and Kleckner 1994, 1995; Cervantes et al. 2000; Allers and Lichten 2001b). This type of analysis is not possible in Drosophila, since recombination occurs asynchronously in a small subset of cells in the germline; however, many of the proteins involved in the early stages of meiotic recombination in fungi are conserved in Drosophila, including an ortholog of the protein that catalyzes DSB formation in yeast (McKim and Hayashi-Hagihara 1998). Our finding of trans hDNA provides molecular support to strengthen the case made by protein conservation for the application of the DSBR model to Drosophila meiotic recombination.
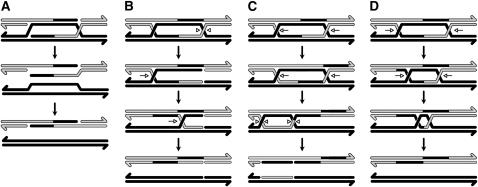
Following initiation by a DSB, four possible sources of trans hDNA can be envisioned (Figure 4, A–D): (A) two-ended SDSA; (B) cutting of the DHJ intermediate at a single HJ followed by branch migration of the second HJ through the nick; (C) branch migration of both HJs past the point of initial resection, followed by resolution by HJ nicking; and (D) branch migration of both HJs toward one another, followed by decatenation by a topoisomerase (DHJ dissolution). If the two events with a single hDNA tract did not actually have trans hDNA, they may have arisen through SDSA or through resolution of a DHJ intermediate by nicking (Figure 1).
Figure 4.—
Models for the formation of trans hDNA. (A) Two-ended SDSA. According to the canonical synthesis-dependent strand annealing (SDSA) model (Formosa and Alberts 1986), after strand invasion and synthesis the nascent strand dissociates from the template and anneals to the single-stranded overhang on the other side of the DSB. In two-ended SDSA, both nascent strands dissociate prior to ligation and reanneal to the unresected ends of the break. (B) Single-junction cutting and branch migration. One Holliday junction in the DHJ is cut (arrowheads) and the other undergoes branch migration (arrows) past the nicks. (C) Branch migration and resolution. Both Holliday junctions in the DHJ undergo branch migration in the same direction, past the region of strand invasion and synthesis. Resolution then occurs by cutting. (D) DHJ dissolution. The two Holliday junctions undergo branch migration toward one another, after which the two chromatids are decatenated. All four models predict that one chromatid will contain trans hDNA, but only DHJ dissolution results in a product that does not contain nicks.
Our results indicate that MEI-9 is not an essential component of the canonical MMR pathway during meiotic recombination. As described above, we cannot exclude the possibility that the occurrence of PMS in mei-9 mutants may result from specialized roles for MEI-9 in MMR; however, PMS might also be an indirect consequence of failure to generate COs in the absence of MEI-9, leading to the use of a pathway that generates NCOs that are refractory to MMR. In proliferating cells, MMR is stimulated by nicks (reviewed in Kunkel and Erie 2005). Similarly, it has been suggested that MMR that occurs later in meiotic recombination is directed by nicks introduced during DHJ resolution (Foss et al. 1999; Hillers and Stahl 1999; Stahl and Hillers 2000). Among the possible sources of trans hDNA enumerated above, DHJ dissolution (Figure 4D) is unique in that nicks are not present after the molecules participating in recombination are resolved into independent duplexes. The absence of nicks might make the products of dissolution resistant to MMR.
Consideration of the structures of hDNA in PMS NCOs provides support for the hypothesis that MEI-9 is required to resolve DHJs into COs and that in the absence of MEI-9 these intermediates undergo dissolution to generate NCOs with trans hDNA (Figure 4D); however, we also detected trans hDNA in PMS events from Msh6 mutants (Radford et al. 2007). As described above, there are several possible sources of trans hDNA. We propose that trans hDNA in NCOs from mei-9 mutants comes from DHJ dissolution. It is possible that dissolution is also the source of trans hDNA in NCOs from Msh6 mutants. For this to be true, DHJ dissolution would have to be a significant source of NCOs even in the presence of MEI-9. If the products of dissolution are always refractory to MMR, then we would expect to recover PMS events with trans hDNA from wild-type females. The rarity of PMS in wild-type females stands in opposition to this prediction. Alternatively, the products of dissolution might be subject to MMR, but the MEI-9 endonuclease may be required to make nicks to allow MMR, since these products do not already have nicks present. Given the similar frequencies of trans hDNA in mei-9 and Msh6 mutants (3/32 vs. 6/40, P = 0.7), we cannot exclude this possibility. Another explanation for the presence of trans hDNA among NCOs from Msh6 mutants, however, is that this trans hDNA arises from one of the other mechanisms described above (Figure 4, A–C). This hypothesis does not exclude the possibility that PMS NCOs from mei-9 mutants arise through DHJ dissolution.
In conclusion, our finding that PMS events from mei-9 mutants often have trans hDNA supports the basic features of the DSBR model for meiotic recombination and is consistent with a role for MEI-9 in generating crossovers by nicking DHJ intermediates. Our results also demonstrate that MEI-9 is not essential for meiotic MMR of base–base mismatches, but may have specialized roles in repair of loops, in repair of products that lack nicks, and in short-patch repair that occurs when the canonical MMR pathway is disrupted. Future studies with different recombination substrates and with mutants that are simultaneously defective in CO formation and canonical MMR, such as mei-9; Msh6 double mutants, can further test these hypotheses and should provide significant new insights into mechanisms of meiotic recombination.
Acknowledgments
We thank Cassie Bowen, Meaghan Martin, Sushmita Mukherjee, Mathilde Sabourin, and Preston Sloop for assistance in collecting flies and setting up crosses; Joseph Ibrahim for statistical analysis; and Bob Duronio, Dale Ramsden, Scotty Kyzer, and members of the Sekelsky lab for helpful discussions. S.J.R. was supported by a Thomas S. and Caroline H. Royster, Jr. Graduate Fellowship. This work was supported by a grant from the National Institute of General Medical Sciences (R01 GM-61252) to J.S.
References
- Alani, E., R. A. G. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. a Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. b Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8: 225–231. [DOI] [PubMed] [Google Scholar]
- Bardwell, A. J., L. Bardwell, A. E. Tomkinson and E. C. Friedberg, 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Bender, W., P. Spierer and D. S. Hogness, 1983. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J. Mol. Biol. 168: 17–33. [DOI] [PubMed] [Google Scholar]
- Bhagat, R., E. A. Manheim, D. E. Sherizen and K. S. McKim, 2004. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenet. Genome Res. 107: 160–171. [DOI] [PubMed] [Google Scholar]
- Bhui-Kaur, A., M. F. Goodman and J. Tower, 1998. DNA mismatch repair catalyzed by extracts of mitotic, postmitotic, and senescent Drosophila tissues and involvement of mei-9 gene function for full activity. Mol. Cell. Biol. 18: 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton, H. L., S. J. Radford, S. McMahan, H. M. Kearney, J. G. Ibrahim et al., 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., and J. E. Haber, 1987. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science 237: 1459–1465. [DOI] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1982. Mismatch repair, gene conversion, and crossing-over in two recombination-defective mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 5961–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1984. Meiotic roles of crossing-over and of gene conversion. Cold Spring Harbor Symp. Quant. Biol. 49: 23–29. [DOI] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chambers, S. R., N. Hunter, E. J. Louis and R. H. Borts, 1996. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 16: 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick, A., G. H. Ballantyne, D. L. Baillie and D. G. Holm, 1970. Gene conversion in higher organisms: half-tetrad analysis of recombination within the rosy cistron of Drosophila melanogaster. Genetics 66: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick, A., G. H. Ballantyne and D. G. Holm, 1971. Studies on gene conversion and its relationship to linked exchange in Drosophila melanogaster. Genetics 69: 179–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. H., A. J. Hilliker and A. Chovnick, 1988. Recombination can initiate and terminate at a large number of sites within the rosy locus of Drosophila melanogaster. Genetics 118: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, I., and C. S. Newlon, 1994. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell 76: 65–75. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., J. Loidl, B. Larkin and N. M. Hollingsworth, 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., N. Hunter, C. Lee, B. Larkin, J. Loidl et al., 2003. The mus81/mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., and T. D. Petes, 1992. Measurements of excision repair tracts formed during meiotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., M. A. White and T. D. Petes, 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, O., E. Lehmann, P. Schar and J. Kohli, 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21: 314–317. [DOI] [PubMed] [Google Scholar]
- Formosa, T., and B. M. Alberts, 1986. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47: 793–806. [DOI] [PubMed] [Google Scholar]
- Foss, H. M., K. J. Hillers and F. W. Stahl, 1999. The conversion gradients at HIS4 of Saccharomyces cerevisiae. II. A role for mismatch repair directed by biased resolution of the recombinational intermediate. Genetics 153: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P-element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. I. Heteroduplex rejection and restoration of Mendelian segregation. Genetics 153: 555–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., and A. Chovnick, 1981. Further observations on intragenic recombination in Drosophila melanogaster. Genet. Res. 38: 281–296. [DOI] [PubMed] [Google Scholar]
- Hilliker, A. J., S. H. Clark and A. Chovnick, 1991. The effect of DNA sequence polymorphisms on intragenic recombination in the rosy locus of Drosophila melanogaster. Genetics 129: 779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A. J., G. Harauz, A. G. Reaume, M. Gray, S. H. Clark et al., 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, R., 1964. A mechanism for gene conversion in fungi. Genet. Res. 78: 282–304. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N. M., L. Ponte and C. Halsey, 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, N., S. R. Chambers, E. J. Louis and R. H. Borts, 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15: 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton and T. D. Petes, 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., and T. D. Petes, 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387: 929–931. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., and D. A. Erie, 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710. [DOI] [PubMed] [Google Scholar]
- Marsischky, G. T., N. Filosi, M. F. Kane and R. D. Kolodner, 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10: 407–420. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., and A. Hayashi-Hagihara, 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., J. K. Jang and E. A. Manheim, 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Meselson, M., and C. M. Radding, 1975. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 72: 358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.-H., T. Bessho, T. Matsunaga and A. Sancar, 1995. Purification and characterization of the XPF-ERCC1 complex of human DNA repair excision nuclease. J. Biol. Chem. 230: 22657–22660. [DOI] [PubMed] [Google Scholar]
- Radford, S. J., M. Sabourin, S. McMahan and J. Sekelsky, 2007. Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics 176: 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romans, P., 1980. a Effects of purine selection on survival of Drosophila mosaic for xanthine dehydrogenase (XDH) activity. Dros. Inf. Serv. 55: 132–133. [Google Scholar]
- Romans, P., 1980. b Gene conversion in mei-9a, a crossover defective mutant in D. melanogaster. Dros. Inf. Serv. 55: 130–132. [Google Scholar]
- Ross-Macdonald, P., and G. S. Roeder, 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Sekelsky, J., K. S. McKim, G. M. Chin and R. S. Hawley, 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J., K. C. Burtis and R. S. Hawley, 1998. Damage control: the pleiotropy of DNA repair genes in Drosophila melanogaster. Genetics 148: 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., and K. J. Hillers, 2000. Heteroduplex rejection in yeast? Genetics 154: 1913–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J. E., and T. D. Petes, 2006. Analysis of the proteins involved in the in vivo repair of base–base mismatches and four-base loops formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 173: 1223–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Villeneuve, A. M., and K. J. Hillers, 2001. Whence meiosis? Cell 106: 647–650. [DOI] [PubMed] [Google Scholar]
- Yildiz, Ö., H. Kearney, B. C. Kramer and J. Sekelsky, 2004. Mutational analysis of the Drosophila repair and recombination gene mei-9. Genetics 167: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]