Abstract
F1 hybrid male sterility is thought to result from interactions between loci on the X chromosome and dominant-acting loci on the autosomes. While X-linked loci that contribute to hybrid male sterility have been precisely localized in many animal taxa, their dominant autosomal interactors have been more difficult to localize precisely and/or have been shown to be of relatively smaller effect. Here, we identified and mapped at least four dominant autosomal factors contributing to hybrid male sterility in the allopatric species pair Drosophila persimilis and D. pseudoobscura bogotana. Using these results, we tested predictions of reduced recombination models of speciation. Consistent with these models, three of the four QTL associated with hybrid male sterility occur in collinear (uninverted) regions of these genomes. Furthermore, these QTL do not contribute significantly to hybrid male sterility in crosses between the sympatric species D. persimilis and D. pseudoobscura pseudoobscura. The autosomal loci identified in this study provide the basis for introgression mapping and, ultimately, for molecular cloning of interacting genes that contribute to F1 hybrid sterility.
HALDANE's (1922) rule observes that, in general, when one sex of hybrids between species is sterile or inviable, it is more frequently the heterogametic sex. The causes of this rule have been studied extensively (see reviews by Wu et al. 1996; Laurie 1997; Orr 1997), and much of the pattern seems to be explained by the “dominance theory” (Muller 1942; Orr 1993; Turelli and Orr 1995), which posits that sterility or inviability in the heterogametic sex often results from deleterious interactions between loci on a hemi-zygous X chromosome and dominant-acting loci on the autosomes. While several X-linked loci contributing to hybrid male sterility have been precisely localized (e.g., Guenet et al. 1990; Oka et al. 2004; Storchova et al. 2004), especially within Drosophila (e.g., Cabot et al. 1994; Perez and Wu 1995; MacDonald and Goldstein 1999; Orr and Irving 2001), dominant autosomal effects have typically been crudely localized or are of comparatively small effect (e.g., Moehring et al. 2006; but see Slotman et al. 2004).
The genetics of hybrid male sterility has been studied in the Drosophila pseudoobscura–D. persimilis species pair for >70 years. The U. S. subspecies of D. pseudoobscura (D. p. pseudoobscura) co-occurs with D. persimilis on the west coast of North America. The two species have been shown to hybridize in natural populations at very low levels (Dobzhansky 1973; Powell 1983), and variable amounts of introgression have been detected across regions of their genomes (Machado et al. 2002; Machado and Hey 2003; Hey and Nielsen 2004). The Bogota subspecies of D. pseudoobscura, D. p. bogotana, occurs allopatrically in South America. D. pseudoobscura and D. persimilis diverged between 0.5 and 1.0 MYA (Aquadro et al. 1991; Wang et al. 1997; Leman et al. 2005) while D. pseudoobscura and D. p. bogotana diverged between 150,000 and 200,000 years ago (Schaeffer and Miller 1991; Wang et al. 1997). Hybrid males between D. persimilis and either D. pseudoobscura subspecies are viable but sterile, while hybrid females are fertile, consistent with Haldane's (1922) rule. Previous studies have mapped the underlying genetic factors that contribute to hybrid sterility between the sympatric species pair (Dobzhansky 1936; Orr 1987, 1989; Noor et al. 2001). These factors are strongly associated with three inversions (two on the X and one on the second chromosome) that are fixed or nearly fixed, differentiating D. pseudoobscura and D. persimilis: Because these sterility-conferring loci are associated with inversions, they have not been precisely localized.
Additionally, because D. pseudoobscura and D. persimilis differ by these inversions and show multiple forms of pre- and postzygotic isolation, they have been a suitable system in which to study the effect of recombination on the evolution and maintenance of reproductive isolation. Recombination between genomes can potentially prevent the evolution or persistence of co-adapted gene complexes that confer species-specific adaptations and/or reproductive isolation between species. Thus, suppressing such recombination can allow the persistence of species despite occasional gene flow. One means for suppressing recombination and for facilitating species persistence is through chromosomal rearrangements: crossover products are not recovered from heterozygotes (hybrids) for such rearrangements.
Several empirical studies (e.g., Rieseberg et al. 1999; Feder et al. 2003; Panithanarak et al. 2004; Stump et al. 2005), including studies in the D. pseudoobscura group (Noor et al. 2001), support the role for chromosomal rearrangements and other regions of suppressed recombination (e.g., centromeric regions; Stump et al. 2005) in hybrids in preserving gene complexes that confer reproductive isolation (see reviews in Ortiz-Barrientos et al. 2002; Butlin 2005). Furthermore, theoretical models show that chromosomal rearrangements can facilitate the accumulation of hybrid incompatibilities between parapatric populations (e.g., Navarro and Barton 2003; Kirkpatrick and Barton 2006). In the D. pseudoobscura group, Brown et al. (2004) show that genetic factors contributing to pre- and postzygotic isolation are associated with inverted regions of the genome in the sympatric species D. pseudoobscura and D. persimilis but are associated with both inverted and probably uninverted (i.e., collinear) regions in the allopatric species D. p. bogotana and D. persimilis. The reduced recombination model of speciation directly predicts this association. However, these putative collinear-region effects were not mapped and may have been complicated by an additional fixed inversion difference between D. p. bogotana and D. persimilis on the third chromosome.
Here, we build on the results of previous studies in two significant ways. First, we use 26 microsatellite markers to demonstrate that hybrid male sterility between the allopatric species D. p. bogotana and D. persimilis maps to dominant-acting autosomal regions of the genome outside of the inversions that distinguish these species. We show that at least some of these regions do not confer hybrid sterility between the sympatric species D. pseudoobscura and D. persimilis, as predicted by the reduced recombination models of speciation. Second, we localize these dominant autosomal genetic factors to regions of the second, third, and fourth chromosomes using a large-scale backcross analysis. These factors are among the first dominant autosomal factors contributing to hybrid male sterility to be precisely mapped, and this study thus provides the basis for introgression mapping and, ultimately, molecular cloning of interacting genes that contribute to F1 hybrid sterility.
MATERIALS AND METHODS
Fly stocks and crosses:
D. pseudoobscura bogotana females carrying a white eye mutation (hereafter “bogw”) were collected as virgins and maintained for 7 days. On day 8, bogw were crossed to D. persimilis MSH 1993 (hereafter “per”) males. F1 females were backcrossed to bogw males to generate backcross males (hereafter “BCbogw males”) for fertility assays. Only male progeny bearing the white mutation were scored. The bogw strain is a subculture of the D. p. bogotana El Recreo line collected in 1978 (provided by H. A. Orr). The per line was derived from females collected at Mount Saint Helena, California, in 1993 (Noor 1995). All crosses were performed on standard sugar/yeast/agar medium at 20° ± 1° and 85% relative humidity.
Fertility assays of BCbogw males:
BCbogw males were collected as virgins and maintained for 7 days in vials containing 10–20 males. On day 8, the fertility of each backcross male was assessed by dissection of the testes in Ringer's solution following the method of Coyne (1984). A male was scored “fertile” if at least one motile sperm was observed and “sterile” if no motile sperm were observed. Treating fertility as a binary trait has been shown to be conservative (Campbell and Noor 2001), although other methods of scoring fertility exist (e.g., White-Cooper 2004). All dissected BCbogw males were labeled and stored at −20°.
Microsatellite genotyping of BCbogw males:
DNA was extracted from all dissected BCbogw males following the protocol of Gloor and Engels (1992). Microsatellite genotyping was performed in two steps. First, all 4853 BCbogw males were genotyped for markers associated with each inversion that distinguishes D. pseudoobscura (and D. p. bogotana) and D. persimilis. The markers used for this initial screen were DPSX002 (chromosome arm XL), DPSX030 (XR), and bicoid (bcd; 2) (Ortiz-Barrientos et al. 2006). These markers identified the inversion arrangement on these chromosome arms. Second, because we were interested in localizing sterility-conferring alleles that map outside the inverted regions between these species, only those 1102 BCbogw males that were hemi- or homozygous for the D. p. bogotana allele at the three inversion markers (hereafter “BCbogwLim males”) were further genotyped. This procedure thus would identify dominant D. persimilis alleles that interact with a predominantly D. p. bogotana genetic background. Surveys of other markers along the X chromosome showed that this procedure also essentially selected for an almost complete D. p. bogotana X chromosome as well as for much of the second chromosome.
BCbogwLim males were genotyped for 23 microsatellite markers distributed evenly on the second, third, and fourth chromosomes. Primer sequences for all markers used in this study are available in the supplemental data at http://www.genetics.org/supplemental/. PCR amplification followed a touchdown protocol: 95° for 1 min; 3 cycles of 94° for 30 sec, 56° for 30 sec, 72° for 30 sec; 3 cycles of 94° for 30 sec, 53° for 30 sec, 72° for 30 sec; and 30 cycles of 94° for 30 sec, 50° for 30 sec, 72° for 30 sec. PCRs were visualized on acrylamide gels on LiCor 4200 DNA sequencer/analyzers.
Mapping hybrid male sterility:
QTL mapping was first performed with composite interval mapping (CIM) (Zeng 1994) using Windows QTL Cartographer V. 2.5 (Wang et al. 2006). We focus on our CIM results rather than our other analysis (see below) because of the longer history of confirmation of effects initially mapped using CIM. Fertility was treated as a binary trait (the presence or absence of sperm; see above). Although this violates the assumption of normality in CIM, a previous study (Moehring et al. 2004) has shown that this treatment gives essentially the same result as when a trait is continuous if CIM is based on logistic regression (e.g., Xu and Atchley 1996). Thresholds for significance were set by permutations (experiment-wise P = 0.05 and N = 1000).
Nonetheless, because our data set does violate an assumption of the CIM procedure, the QTL detected using CIM were further confirmed using the new binary multiple interval mapping (bMIM) (Li et al. 2006) procedure in Windows QTL Cartographer V.2.5 (Wang et al. 2006). Results from both forward and backward regression methods on markers are reported.
F1 hybrid male sterility is thought to result from epistatic interactions between recessive X-linked and dominant autosomal loci (Muller 1942; Orr 1993; Turelli and Orr 1995). We do not explicitly test for epistasis in this study because we have limited the data set to only those males bearing an X chromosome from D. p. bogotana, thereby identifying dominant autosomal loci contributing to sterility derived from D. persimilis. While there may be epistasis among autosomal loci, bMIM currently does not include a test for epistasis (S. Wang, personal communication), and such a test is beyond the scope of the hypotheses that we examine in this study.
We evaluated whether the same QTL are associated with hybrid male sterility in backcross hybrids between per and D. pseudoobscura (hereafter, ps) vs. between per and D. p. bogotana (hereafter, bog) using a three-way contingency test based on a log-linear model (Sokal and Rohlf 1995). For the per–ps hybridization, we used the raw data from our previous mapping study (Noor et al. 2001), limited the data set to those backcross hybrid males bearing the three inversion-associated markers from D. pseudoobscura, and examined the effects of markers closest to the ones surveyed in our per–bog backcross. Markers DPS2003 and DPS3001 were surveyed in both crosses and their associations with hybrid male sterility were compared in this manner. On the fourth chromosome, we did not have data for markers immediately adjacent to the per–bog sterility QTL in the per–ps backcross. Thus, for the fourth chromosome QTL, we performed a more conservative three-way contingency test using a marker even farther from the sterility QTL in per–bog (DPS4G1e) than the nearest marker surveyed in per–ps (Adh).
RESULTS
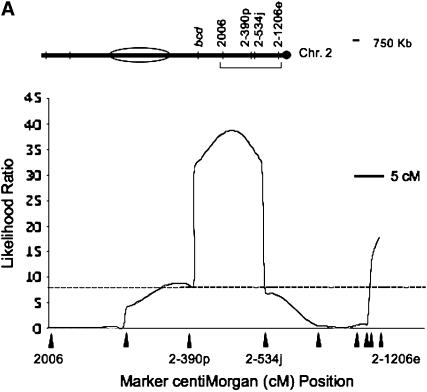
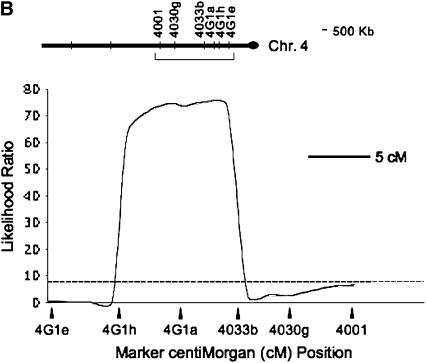
Controlling for the effects of three microsatellite markers (DPSX002, DPSX030, and bcd; see materials and methods) associated with the inversion differences between D. persimilis and D. p. bogotana allowed us to detect QTL that confer hybrid male sterility occurring outside the chromosomal rearrangements that distinguish these species. Using 26 microsatellite markers, we mapped using CIM at least four autosomal dominant QTL with large effects on hybrid male sterility that interact with a predominantly D. p. bogotana genetic background. Furthermore, we were able to localize two of these four QTL to relatively small regions: On the second chromosome, a QTL was localized to an interval of ∼840 kb between markers DPS2-390p and DPS2-534j (Figure 1A). The annotated part of this region contained 104 genes on the basis of sequence homology to D. melanogaster (Gilbert 2005; see supplemental Table 1 at http://www.genetics.org/supplemental/). The second QTL on this chromosome is associated with marker DPS2-1206e; we could not localize the size of this genomic region due to a lack of markers beyond DPS2-1206e, which lies ∼17 kb from the centromeric end of the second chromosome sequence assembly. On the fourth chromosome, a QTL was localized to an interval of ∼1.2 Mb between markers DPS4G1h and DPS4033b (Figure 1B) and is closely associated with marker DPS4G1a. The annotated part of this region contained 136 genes (Gilbert 2005; see supplemental Table 1 at http://www.genetics.org/supplemental/). We were unable to refine the location of the third chromosome QTL further because of an inversion difference between D. persimilis and D. p. bogotana. The biological and molecular functions of the 239 genes contained in the two smaller QTL intervals were evaluated using PANTHER (Mi et al. 2005) software and are included in supplemental Table 2 at http://www.genetics.org/supplemental/. Figure 2 shows the relative effects of these QTL individually and in combination on hybrid male sterility.
Figure 1.—
QTL associated with hybrid male sterility on the (A) second and (B) fourth chromosomes. Plots are the likelihood-ratio (LR) test statistic as determined by composite interval mapping on the y-axis. The significance thresholds were determined by permutation testing to be an LR of ∼7.8 and are indicated by dashed horizontal lines. Marker locations are represented by solid elongated triangles on the x-axis of the QTL plot. The markers flanking the significant QTL are indicated by name. Above the QTL plots are diagrams of the chromosomes with the physical location of relevant markers indicated by hatch marks. An inversion is represented by the presence of an open oval. A bracket indicates the area magnified in the QTL plot below. Scale bars (representing either recombinational or physical distance) are given.
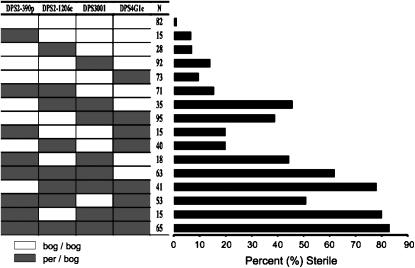
Figure 2.—
Relative effects of each QTL on hybrid male sterility. Hybrid male genotypes are given on the y-axis and sterility (as determined by sperm motility) is given on the x-axis. Heterozygous QTL (hybrid males carry both the D. persimilis and D. p. bogotana alleles) are indicated by shaded rectangles, and homozygous QTL (hybrid males carry only the D. p. bogotana allele) are indicated by open rectangles. A marker associated with each QTL is used to determine the average sterility of males hetero- or homozygous for that QTL. “N” is the number of males of a given genotype.
We further confirmed the presence and location of these QTL using bMIM. Table 1 shows the additive effects associated with the three major QTL, as well as the microsatellite markers flanking each QTL, for both regression forward and backward selection on markers. The results from bMIM (Table 1) and those from CIM (Figure 1) were highly comparable, aside from minor movements of the exact peak location. Using a backward regression method in bMIM also detected the fourth QTL near the centromeric end of chromosome 2 (data not shown), as did CIM.
TABLE 1.
Location and additive effect of the three major QTL detected using bMIM
| QTL | Chromosome | Additivity | Flanking markers |
|---|---|---|---|
| Forward regression | |||
| 1 | 2 | 2.634 | DPS2-390p; DPS2-534j |
| 2 | 3 | 2.049 | DPS3001; DPS3026 |
| 3 | 4 | 1.784 | DPS4G1a; DPS4033b |
| Backward regression | |||
| 1 | 2 | 2.047 | DPS2-390p; DPS2-534j |
| 2 | 3 | 2.008 | DPS3001; DPS3026 |
| 3 | 4 | 1.756 | DPS4G1h; DPS4G1a |
Previously, Brown et al. (2004) showed that genes likely associated with collinear regions significantly decreased fertility in hybrids between D. persimilis and D. p. bogotana but do not have detectable effects on fertility in hybrids between D. persimilis and D. pseudoobscura. This analysis was based on a comparison between the proportion of sterile males from a D. p. bogotana backcross and the proportion of sterile males from a D. pseudoobscura backcross. Here, we directly evaluate whether the same collinear-region QTL are associated with hybrid male sterility in hybrids between the two hybridizations by using a three-way contingency test based on a log-linear model (Sokal and Rohlf 1995). In backcross hybrids, the major QTL on the second, third, and fourth chromosomes all contribute significantly to hybrid male sterility in hybrids between D. persimilis and D. p. bogotana but do not contribute significantly to sterility between D. persimilis and D. pseudoobscura, and the differences in effect are all statistically significant (G2 = 94.05, P < 0.001; G2 = 159.97, P < 0.001; and G2 = 61.72, P < 0.001, respectively; supplemental Table 1 at http://www.genetics.org/supplemental/). Thus, these genomic regions contribute significantly more to hybrid male sterility in hybridizations of the allopatric species than in hybridizations of the sympatric species, as predicted by the reduced recombination model of speciation.
DISCUSSION
In this study, we fine map at least three D. persimilis autosomal dominant QTL that confer hybrid male sterility in a D. p. bogotana genetic background. These QTL are located outside the chromosomal inversions that differentiate the two arms of the X chromosome and on the second chromosome in D. persimilis and D. p. bogotana. Furthermore, we demonstrate that the effects of these QTL on hybrid male sterility are greater in crosses between D. persimilis and D. p. bogotana than between D. persimilis and D. pseudoobscura.
These results confirm a prediction of the reduced recombination model of speciation: In allopatric species where the potential for gene flow does not exist, genetic factors associated with reproductive isolation should reside both within and outside genomic regions experiencing reduced recombination (in this case, within and outside chromosomal inversions); in sympatric species, factors associated with reproductive isolation should reside preferentially within genomic regions experiencing reduced recombination (within inversions). Thus, in the D. pseudoobscura species group, chromosomal rearrangements appear to contribute to the maintenance of species persistence by restricting recombination between genomic regions that contain genetic factors underlying reproductive isolation, while gene flow has homogenized collinear regions.
Chromosomal rearrangements have also been proposed to contribute directly to reproductive isolation via strong underdominance (White 1969) resulting from meiotic difficulties. This type of chromosomal speciation model is distinct from the recombination–reduction models in that hybrid sterility results from the chromosomal rearrangements directly, not from effects of loci captured within the rearrangement. Although bearing some recent support (Delneri et al. 2003), this model remains controversial (Coyne and Orr 2004). In contrast, our results directly support a prediction of the reduced recombination chromosomal speciation models.
A previous study by Brown et al. (2004) showed that almost all hybrid males between D. persimilis and D. p. pseudoobscura homozygous or hemi-zygous for the three inverted regions were fertile. In contrast, only about one-third of the male hybrids between D. persimilis and D. p. bogotana homozygous or hemi-zygous for the three inverted regions were sterile, suggesting that other factors conferring sterility likely occur in collinear regions. By selecting only those D. p. bogotana backcross hybrid males that were homozygous or heterozygous for the three inversions, we built on this work by directly localizing these additional hybrid male sterility loci to small regions on the second and fourth chromosomes and a region of the third chromosome. Our findings are also consistent with DNA sequence surveys of these species, which indicate extensive introgression in collinear regions between D. pseudoobscura and D. persimilis but probably not between D. persimilis and D. p. bogotana (Machado and Hey 2003).
The sterility effects of these three autosomal regions are dominant, as hybrid males heterozygous (i.e., carrying both the D. persimilis and the D. p. bogotana allele) for these loci are more likely to be sterile. The dominance theory proposed to explain Haldane's rule suggests that F1 hybrid male sterility often results from a deleterious interaction between a hemi-zygous (recessive) sex chromosome effect and dominant autosomal effects. While many studies have precisely localized the recessive X chromosomal effects (Guenet et al. 1990; Cabot et al. 1994; Perez and Wu 1995; MacDonald and Goldstein 1999; Orr and Irving 2001; Oka et al. 2004; Storchova et al. 2004; Moehring et al. 2006), most have failed to pinpoint the locations of individually significant dominant autosomal effects with high confidence (but see Slotman et al. 2004).
Although we identify at least three such dominant autosomal regions, the effect of each individual allele appears to be weak as well (Figure 2): No single factor caused complete or nearly complete sterility. Figure 2 shows that the effect on the sterility phenotype increases with the addition of each QTL, although the QTL must interact with D. p. bogotana factors that were not identified in this study (such as the X chromosome). As expected, the highest proportion of hybrid male sterility (83%) occurred in hybrid males that were heterozygous for all four of the QTL.
To attempt to examine whether genetic introgression has occurred between D. persimilis and D. p. bogotana in the major QTL intervals on the second and fourth chromosomes, two 900-bp regions (one within each interval) were randomly selected for amplification and sequencing. The region on the second chromosome contains coding DNA while the region on the fourth chromosome contains only noncoding DNA. These sequences were obtained for two D. persimilis strains, two D. p. bogotana strains, two D. pseudoobscura strains, and one D. miranda (outgroup) strain. Sequences obtained are available in GenBank (accession nos. EF392818–EF392831). No fixed differences were detected between D. persimilis and D. p. bogotana or between D. persimilis and D. pseudoobscura in the ∼1800 bp amplified so it appears that the differentiation associated with sterility between these species may be rather localized. We refrained for further analysis of our sequence data because of the presently unrefined scale of our QTL mapping.
Because the QTL identified in this study were mapped only in a single line of D. persimilis (MSH 1993) and in a single line of D. p. bogotana (El Recreo), it is also possible that the alleles detected in these hybrids could reflect intraspecific polymorphism for alleles associated with sterility. However, Brown et al. (2004) did not observe dramatic differences in hybrid male sterility among three backcross lineages of D. pseudoobscura or among three backcross lineages of D. p. bogotana, suggesting that our mapping results may be representative of much of these species. Further, because precision mapping of any phenotypic trait typically involves scoring hundreds, if not thousands, of individuals for that trait, this caveat applies to almost all QTL mapping studies published to date. Because this study localizes the QTL to relatively small autosomal regions, it does provide a basis for future introgression studies of these factors that cause hybrid male sterility between D. persimilis and D. p. bogotana and for the eventual cloning of “sterility genes.”
Acknowledgments
We thank K. M. Brown for performing many of the testes dissections and fertility assays; S. Wang for help using WinQTL Cartographer; M. Lavine and C. Simpson for help on statistical procedures; N. Kandul, A. Moehring, S. Schaeffer, and anonymous reviewers for helpful comments on this manuscript; and A. Somerville for technical assistance. This work was funded by a Sigma Xi grant-in-aid of research to A.S.C. and National Science Foundation grants 0509780 and 0549893 to M.A.F.N.
References
- Aquadro, C. F., A. L. Weaver, S. W. Schaeffer and W. W. Anderson, 1991. Molecular evolution of inversions in Drosophila pseudoobscura: the amylase gene region. Proc. Natl. Acad. Sci. USA 88: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. M., L. M. Burk, L. M. Henagan and M. A. F. Noor, 2004. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution 58: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Butlin, R. K., 2005. Recombination and speciation. Mol. Ecol. 14: 2621–2635. [DOI] [PubMed] [Google Scholar]
- Cabot, E. L., A. W. Davis, N. A. Johnson and C.-I. Wu, 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. V., and M. A. F. Noor, 2001. Assessing hybrid male fertility in Drosophila species: correlation between sperm motility and production of offspring. Dros. Inf. Serv. 84: 6–9. [Google Scholar]
- Coyne, J. A., 1984. Genetic basis of male sterility in hybrids between two closely related species of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4444–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis et al., 2003. Engineering evolution to study speciation in yeasts. Nature 422: 68–72. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1936. Studies of hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1973. Is there gene exchange between Drosophila pseudoobscura and Drosophila persimilis in their natural habitats? Am. Nat. 107: 312–314. [Google Scholar]
- Feder, J. L., J. B. Roethele, K. Filchak, J. Niedbalski and J. Romero-Severson, 2003. Evidence of inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics 163: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, D. G., 2005. DroSpeGe, a public database of Drosophila species genomes (http://insects.eugenes.org/DroSpeGe). [DOI] [PMC free article] [PubMed]
- Gloor, G. B., and W. R. Engels, 1992. Single-fly DNA preps for PCR. Dros. Inf. Serv. 71: 148–149. [Google Scholar]
- Guenet, J., C. Nagamine, D. Simon-Chazottes, X. Montagutelli and F. Bonhomme, 1990. Hst-3: an X-linked hybrid sterility gene. Genet. Res. 53: 163–165. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109. [Google Scholar]
- Hey, J., and R. Nielsen, 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M., and N. Barton, 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics 147: 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman, S. C., Y. Chen, J. E. Stajich, M. A. F. Noor and M. K. Uyenoyama, 2005. Likelihoods from summary statistics: recent divergence between species. Genetics 171: 1419–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., S. Wang and Z. B. Zeng, 2006. Multiple-interval mapping for ordinal traits. Genetics 173: 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, S. J., and D. B. Goldstein, 1999. A quantitative genetic analysis of male sexual traits distinguishing the sibling species Drosophila simulans and D. sechellia. Genetics 153: 1683–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. A., and J. Hey, 2003. The causes of phylogenetic conflict in a classic Drosophila species group. Proc. Biol. Sci. 270: 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus sequence data: the case of Drosophila pseudoobscura and its close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Mi, H., B. Lazareva-Ulitsky, R. Loo, A. Kejariwal, J. Vandergriff et al., 2005. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33: D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring, A. J., J. Li, M. D. Schug, S. G. Smith, M. deAngelis et al., 2004. Quantitative trait loci for sexual isolation between Drosophila simulans and D. mauritiana. Genetics 167: 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring, A. J., A. Llopart, S. Elwyn, J. A. Coyne and T. F. Mackay, 2006. The genetic basis of postzygotic reproductive isolation between Drosophila santomea and D. yakuba due to hybrid male sterility. Genetics 173: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Navarro, A., and N. H. Barton, 2003. Accumulating postzygotic isolation in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Noor, M. A., 1995. Speciation driven by natural selection in Drosophila. Nature 375: 674–675. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci, Y. Almendarez, J. Reiland et al., 2001. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55: 512–521. [DOI] [PubMed] [Google Scholar]
- Oka, A., A. Mita, N. Sakurai-Yamatani, H. Yamamoto, N. Takagi et al., 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1987. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and D. persimilis. Genetics 116: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1989. Localization of genes causing postzygotic isolation in two hybridizations involving Drosophila pseudoobscura. Heredity 63: 231–237. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1993. A mathematical model of Haldane's rule. Evolution 47: 1606–1611. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1997. Haldane's rule. Annu. Rev. Ecol. Syst. 28: 195–218. [Google Scholar]
- Orr, H. A., and S. Irving, 2001. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics 158: 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos, D., J. Reiland, J. Hey and M. A. F. Noor, 2002. Recombination and the divergence of hybridizing species. Genetica 116: 167–178. [PubMed] [Google Scholar]
- Ortiz-Barrientos, D., A. S. Chang and M. A. F. Noor, 2006. A recombinational portrait of the Drosophila pseudoobscura genome. Genet. Res. 87: 23–31. [DOI] [PubMed] [Google Scholar]
- Panithanarak, T., H. C. Hauffe, J. F. Dallas, A. Glover, R. G. Ward et al., 2004. Linkage-dependent gene flow in a house mouse chromosomal hybrid zone. Evolution 58: 184–192. [DOI] [PubMed] [Google Scholar]
- Perez, D. E., and C.-I. Wu, 1995. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. R., 1983. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc. Natl. Acad. Sci. USA 80: 492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, S. W., and E. L. Miller, 1991. Nucleotide sequence analysis of Adh genes estimates the time of geographic isolation of the Bogota population of Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 88: 6097–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman, M., A. dellaTorre and J. R. Powell, 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics 167: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry. W. H. Freeman, New York.
- Storchova, R., S. Gregorova, D. Buckiova, V. Kyselova, P. Divina et al., 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Stump, A. D., M. C. Fitzpatrick, N. F. Lobo, S. Traore, N. Sagnon et al., 2005. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102: 15930–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. L., J. Wakeley and J. Hey, 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z-B. Zeng, 2006. Windows QTL Cartographer 2.5. North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm).
- White, M. J. D., 1969. Chromosomal rearrangements and speciation in animals. Annu. Rev. Genet. 3: 75–98. [Google Scholar]
- White-Cooper, H., 2004. Spermatogenesis: analysis of meiosis and morphogenesis, pp. 45–75 in Drosophila Cytogenetics Protocols, edited by D. S. Henderson. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Wu, C.-I., N. A. Johnson and M. F. Palopoli, 1996. Haldane's rule and its legacy: Why are there so many sterile males? Trends Ecol. Evol. 11: 281–284. [DOI] [PubMed] [Google Scholar]
- Xu, S., and W. R. Atchley, 1996. Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics 143: 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]