Abstract
In a series of seminal articles in 1974, 1975, and 1977, J. H. Gillespie challenged the notion that the “fittest” individuals are those that produce on average the highest number of offspring. He showed that in small populations, the variance in fecundity can determine fitness as much as mean fecundity. One likely reason why Gillespie's concept of within-generation bet hedging has been largely ignored is the general consensus that natural populations are of large size. As a consequence, essentially no work has investigated the role of the fecundity variance on the evolutionary stable state of life-history strategies. While typically large, natural populations also tend to be subdivided in local demes connected by migration. Here, we integrate Gillespie's measure of selection for within-generation bet hedging into the inclusive fitness and game theoretic measure of selection for structured populations. The resulting framework demonstrates that selection against high variance in offspring number is a potent force in large, but structured populations. More generally, the results highlight that variance in offspring number will directly affect various life-history strategies, especially those involving kin interaction. The selective pressures on three key traits are directly investigated here, namely within-generation bet hedging, helping behaviors, and the evolutionary stable dispersal rate. The evolutionary dynamics of all three traits are markedly affected by variance in offspring number, although to a different extent and under different demographic conditions.
PREDICTING the fate of an allele newly arisen through mutation is possibly the oldest and most recurrent problem in population genetics. It has been known for long that when the fecundity of individuals follows a Poisson distribution, the survival probability of a mutant gene lineage increases with an increase in the mean fecundity of individuals (Haldane 1927; Fisher 1930). It was later recognized that when the variance in offspring number differs from the mean, the survival probability of a mutant gene lineage decreases with an increase in the variance in offspring number (Bartlett 1955; Ewens 1969). This can be understood by realizing that a random line of genes that failed to transmit itself at any generation in the past is lost forever, and this independently of the mean fecundity of individuals. Intuitively, we thus expect that between two competing strategies with equal mean fecundity, the one with the lower variance will outpropagate its alternative.
In a series of insightful articles, Gillespie (1974, 1975, 1977) elucidated the conditions under which natural selection will decrease the variance in offspring number in panmictic populations of constant size. His key result is that the intensity of selection against the variance is proportional to the inverse of population size. The selective pressure against the variance thus decreases with increasing population size and vanishes for very large populations. This result can be understood by noting that in a population subject to regulation, the expected number of recruited offspring of an individual (i.e., its fitness) depends on its total random number of offspring produced relative to the total random number of offspring produced by its competitors. When population size becomes large, the law of large numbers informs us that the total number of offspring produced by competitors converges to a fixed value, which is given by the mean fecundity and is devoid of any variance. In this situation, a mutant allele has only to produce a mean number of offspring exceeding the mean of the resident allele to invade the population. Only when the population is small enough can the variance in offspring number play a role as important as the mean in determining the fitness of individuals. While most natural populations are arguably large, they also tend to be discontinuously distributed and typically consist of local demes connected by migration. Such a spatial structure implies that demes can be of very small size while the population as a whole remains large. This feature is renowned to affect the evolutionary process because it alters the forces shaping the changes in gene frequencies such as selection or random genetic drift (Cherry and Wakeley 2003; Roze and Rousset 2003; Whitlock 2003). We thus expect that the effect of the variance in fecundity on the average number of recruited offspring of an individual will also depend on population structure.
The nature of selection on the variance in offspring number in subdivided populations has been studied by Shpak (2005), who concluded that the conditions under which high-variance strategies are outperformed by low-variance strategies depend on the timing of the regulation of the size of the population. When density-dependent competition occurs before dispersal (i.e., soft selection), the variance in the number of offspring competing in a deme is determined by the number of individuals reproducing locally. In this case, the intensity of selection against the variance is proportional to the inverse of the size of the pool of individuals contributing to this variance, that is, deme size. However, in natural populations, density-dependent competition and competition between individuals can also occur after dispersal. Under this process (called hard selection), individuals in each deme send juveniles that will compete in other demes. Then, the individuals contributing to the variance in the number of juveniles competing in a focal deme can be separated into two pools: one small “resident” pool of individuals breeding in the focal deme and one large “foreign” pool, consisting of all those individuals breeding in different demes and that affect the demography of the focal deme through the dispersal of their progeny. This suggests that the contribution to the variance in offspring number in a focal deme should come mainly from the resident pool because the foreign pool, being very large, will contribute an essentially fixed number of individuals to each deme. Thereby, demes affect each other in a deterministic way when deme number is large Chesson (1981), a result that follows again from the law of large numbers. The nature of selection on the variance in offspring number under hard selection was recently investigated by Shpak and Proulx (2006) and their formalization suggests that selection on the variance in fecundity decreases as migration rate increases (i.e., the foreign pool of individuals becomes larger).
More generally, we also expect that the variance in fecundity will affect not only the evolution of bet-hedging strategies but also other life-history traits. In particular, traits involving kin interactions such as sex ratio, dispersal, or altruism depend on the local relatedness arising through population structure, which is itself a dynamical variable shaped by the variance in fecundity. The effect of the variance in offspring number on the evolution of such behaviors has been overlooked so far. Apart from sex-ratio homeostasis (Verner 1965; Taylor and Sauer 1980) in panmicitic populations and sex allocation (Proulx 2000, 2004), we are aware of no model investigating the role of the fecundity variance on the evolutionary stable state of various life-history strategies.
The objective of this article is twofold. First, we integrate the classical measure of selection for within-generation bet hedging (Gillespie 1974, 1975, 1977) into the inclusive fitness and game theoretic measure of selection for structured populations (e.g., Taylor 1990; Taylor and Frank 1996; Frank 1998; Rousset and Billiard 2000; Rousset 2004, 2006). This allows us to set up a framework for studying the effect of the mean and the variance in fecundity for selection on various life-history traits and under different life-cycle assumptions (e.g., timing of regulation, population structure). Second, we apply the resulting framework to investigate the selective pressure on three life-history traits in subdivided populations under soft and hard selection regimes. These traits are within-generation bet hedging, helping between deme mates, and dispersal. Our results for selection on the fecundity variance are qualitatively consistent with those of Shpak (2005) and Shpak and Proulx (2006) but differ quantitatively because the approach endorsed in this article allows us to take the variance in gene frequency between demes explicitly into account. Our results also indicate that the magnitude of selection on both helping behaviors and dispersal rates is markedly affected by the fecundity variance and can even lead to outcomes qualitatively different from the classical predictions considering mean fecundity only.
MODEL
Life cycle and change in gene frequency:
Consider that the population consists of haploid organisms occupying an infinite number of demes of size N that are connected by migration (finite deme number is considered in the appendix). The life cycle is punctuated by the following events:
Each adult produces a large number of independently distributed juveniles. The mean and the variance of the fecundity distribution of an adult depend on its own phenotype and on the phenotype of all other deme mates.
Each juvenile disperses independently from each other with probability m to another deme.
Density-dependent ceiling regulation occurs so that only N juveniles reach adulthood in each deme. The size of each deme is thus assumed to be constant over time.
The evolution of a phenotypic trait z under selection in this population can be described by a gradual, step-by-step transformation caused by the successive invasion of mutant alleles resulting in different phenotypic effects from those of resident alleles fixed in the population. The invasion of a mutant allele can in turn be ascertained by the change in its frequency p over one generation, which for a mutant with small phenotypic effect δ (weak selection) can be written as
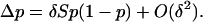 |
(1) |
The term O(δ2) is a remainder and S is Hamilton's (1964) inclusive fitness effect, which measures the direction of selection on the mutant and also provides the first-order phenotypic effect of the mutant on its probability of fixation when the population is of finite size [Roze and Rousset 2003, 2004; Rousset 2004 (pp. 99, 108–109, and 206–207), 2006]. The inclusive fitness effect (S) can be computed by the direct fitness method (Taylor and Frank 1996; Rousset and Billiard 2000), as
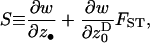 |
(2) |
where w ≡ w(z•, 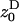 ) is a fitness function giving the expected number of adult offspring of a focal individual (FI) and FST is the relatedness between the FI and another individual randomly sampled from its deme (here Wright's measure of population structure). The partial derivatives of w are the effects of actors (i.e., individuals bearing the mutant allele) on the fitness of the FI, the actors being the FI itself with phenotype denoted z•, and the other individuals from the focal deme with average phenotype denoted
) is a fitness function giving the expected number of adult offspring of a focal individual (FI) and FST is the relatedness between the FI and another individual randomly sampled from its deme (here Wright's measure of population structure). The partial derivatives of w are the effects of actors (i.e., individuals bearing the mutant allele) on the fitness of the FI, the actors being the FI itself with phenotype denoted z•, and the other individuals from the focal deme with average phenotype denoted 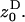 The partial derivatives can be evaluated at a candidate evolutionary stable strategy z★. This phenotype toward which selection leads through small mutational steps is obtained by solving S = 0 at
The partial derivatives can be evaluated at a candidate evolutionary stable strategy z★. This phenotype toward which selection leads through small mutational steps is obtained by solving S = 0 at 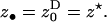 In the next section we provide expressions for both the fitness function w and the relatedness FST in terms of the means and the variances of the fecundity distributions of the individuals in the population.
In the next section we provide expressions for both the fitness function w and the relatedness FST in terms of the means and the variances of the fecundity distributions of the individuals in the population.
Fitness function:
The direct fitness function of a FI can be decomposed into two terms:
 |
(3) |
This formulation emphasizes that fitness depends on the expected number of FI's offspring recruited in the focal deme (wp) and on the ones reaching adulthood in a foreign deme after dispersal (wd). In the appendix we show that when deme size is large, each component of fitness can be expressed as a function of only the mean and the variance of the fecundity distribution of the different individuals in the population affecting the fitness of the FI. Neglecting terms of order 1/N2 and higher order, the philopatric component of fitness can be written as
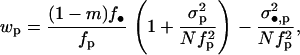 |
(4) |
where f• is the mean fecundity of the FI and fp = (1 − m)f0 + mf1 is the average of the mean number of juveniles produced by individuals in the population and coming into competition in the focal deme (Equation A18 of the appendix). This number depends on the average mean fecundity f0 of individuals breeding in the focal deme and on the average mean fecundity f1 of individuals breeding in different demes. The fitness function wp also involves the variance in the number of offspring of the FI coming into competition in the focal deme,
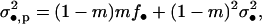 |
(5) |
where 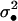 is the variance of the fecundity distribution of the FI (here, the variance of the number of offspring produced before the dispersal stage). The term (1 − m)mf• in this equation can be interpreted as the variance in offspring number entering in competition in the focal deme and caused by dispersal being a random event. The second term in Equation 5 is the additional variance in offspring number entering in competition in the focal deme and resulting from fecundity being a random variable. Accordingly, when fecundity is a fixed number and migration is random, we are left only with the first term while if fecundity is a random number but a fixed proportion of juvenile migrate, we are left only with the second term. The fitness function also depends on the average variance of the number of juveniles entering into competition in the focal deme, which reads
is the variance of the fecundity distribution of the FI (here, the variance of the number of offspring produced before the dispersal stage). The term (1 − m)mf• in this equation can be interpreted as the variance in offspring number entering in competition in the focal deme and caused by dispersal being a random event. The second term in Equation 5 is the additional variance in offspring number entering in competition in the focal deme and resulting from fecundity being a random variable. Accordingly, when fecundity is a fixed number and migration is random, we are left only with the first term while if fecundity is a random number but a fixed proportion of juvenile migrate, we are left only with the second term. The fitness function also depends on the average variance of the number of juveniles entering into competition in the focal deme, which reads
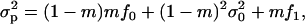 |
(6) |
where 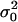 is the average variance in fecundity of individuals breeding in the focal deme.
is the average variance in fecundity of individuals breeding in the focal deme.
The component of the fitness of the FI resulting from dispersing progeny reads
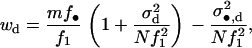 |
(7) |
where
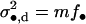 |
(8) |
is the variance in the number of offspring of the FI entering into competition in other demes (Equation A24 of the appendix). This variance depends only on dispersal being a random event and not on the variance in fecundity. This is so because the contribution of the variance in fecundity to the variance in the number of competing offspring in a given deme depends on the square of the migration rate to that deme (Equation A16 of the appendix). Since in the infinite island model two offspring descending from the same parent have a probability of zero of emigrating to the same deme, the effect of the fecundity variance on the variance in the number of competing offspring in a nonfocal deme vanishes. Finally, we need the average variance of the number of juveniles entering into competition in different demes; this is
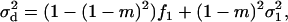 |
(9) |
where 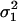 is the average variance in fecundity of individuals breeding in different demes.
is the average variance in fecundity of individuals breeding in different demes.
Equations 4 and 7 highlight that both the means (f) and the variances (σ2) of the fecundity distributions of individuals in the population affect the fitness of a FI. This result is an approximation that follows from the assumption that demes are of large size. Conversely, if demes are small, the fitness function w will also depend on the kurtosis, skewness, and other measures of the dispersion of the fecundity distributions (see Equation A6 of the appendix). By contrast, when fecundity follows a Poisson distribution, fitness depends only on mean fecundity. This is an exact result (see Poisson-distributed fecundity in the appendix), holding whatever the sizes of demes, which is usually invoked in applications of inclusive fitness theory (e.g., Comins et al. 1980; Rousset and Ronce 2004) or population genetics (e.g., Karlin and McGregor 1968; Ewens 1969; Gillespie 1975; Ewens 2004), because it greatly simplifies the calculations. To investigate how a distribution of fecundity with a variance in excess of a Poisson-distributed fecundity might affect selection, we consider here the fate of mutant alleles affecting three different life-history traits:
The mutant results in a reduction in both the mean (by C) and the variance (by D) of its random number of juveniles produced before dispersal.
The mutant causes an action that reduces its mean fecundity by some cost C but that increases the mean fecundity of each individual in the deme (excluding the actor) by a benefit B/(N − 1).
The mutant expresses a decrease (or increase) in the dispersal rate set by the resident allele.
Assumptions for the variance in fecundity:
According to our assumption that the deme sizes remain constant over time, the mean fecundity f of individuals in a monomorphic population has to be large to cancel out the fluctuations of deme sizes induced by demographic stochasticity. If the variance in fecundity is of the same order of magnitude as the mean, all terms involving a variance in the fitness functions (Equations 4 and 7) will vanish because they are divided by the mean squared (i.e., σ2/f2 → 0 when f becomes large). We are then left with a situation where the fitness of a FI depends only on its mean fecundity of that of the other actors in the population. However, when the variance in fecundity is of higher order of magnitude than the mean, the coefficient of variation defined as
 |
(10) |
(e.g., Lynch and Walsh 1998, p. 23) remains positive under large mean fecundity (i.e., σv > 0 for f large). For the rest of this article, we assume that the fecundity distribution has a variance of larger order of magnitude than the mean and that the mean fecundity is finite but large enough to prevent demographic stochasticity.
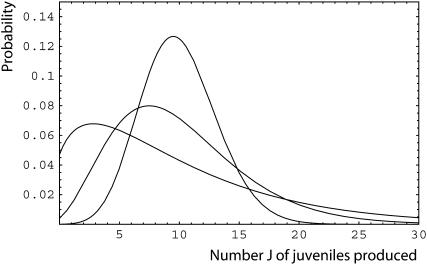
Several distributions closely related to the Poisson distribution have a variance of larger order of magnitude than the mean. Such distributions can be conveniently obtained by mixing the rate of occurrence of birth events f of the Poisson distribution with other distribution (i.e., mixed Poisson distributions; Willmot 1986). For instance, the negative-binomial distribution, which is advocated to be the basis of the fertility in humans and other species (Caswell 2000), can be obtained by mixing the birth rate f by a gamma distribution. This distribution has mean f and variance f + f2/α, where the parameter α allows tuning this variance away from that of a Poisson distribution (Figure 1). When the mean becomes large, the coefficient of variation of this distribution remains positive (σv → 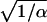 ). Similarly, when clutch size follows a Poisson distribution and the whole clutch has a probability (1 − s) of complete failure, the resulting distribution has mean sf and variance fs + f2s(1 − s).
). Similarly, when clutch size follows a Poisson distribution and the whole clutch has a probability (1 − s) of complete failure, the resulting distribution has mean sf and variance fs + f2s(1 − s).
Figure 1.—
Negative-binomial fecundity probability distribution 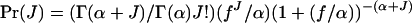 for the number J of juveniles produced by an individual. The mean is given by f and the variance by f + f2/α. For all curves we have a mean of f = 10, while α = ∞ for the curve with the highest peak (i.e., Poisson distribution), α = 5 for the curve with the intermediate peak, and α = 1.5 for the curve with the lowest peak.
for the number J of juveniles produced by an individual. The mean is given by f and the variance by f + f2/α. For all curves we have a mean of f = 10, while α = ∞ for the curve with the highest peak (i.e., Poisson distribution), α = 5 for the curve with the intermediate peak, and α = 1.5 for the curve with the lowest peak.
Relatedness:
The variance in fecundity will also affect the dynamics of relatedness between two individuals sampled in the same deme after dispersal. At equilibrium, relatedness satisfies the recursion
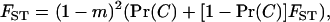 |
(11) |
where (1 − m)2 is the probability that the two individuals are of philopatric origin and Pr(C) is the probability that they descend from the same parent.
Using Equation 10 and Equation A31 from the appendix, assuming that both deme size and mean fecundity are large, and keeping only terms of leading order we find that the coalescence probability is given by
 |
(12) |
where 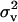 is called the effective variance by Gillespie (1975, p. 407).
is called the effective variance by Gillespie (1975, p. 407).
Using Equations 4, 7, and 12 we investigate in the next section the selective pressure on the three life-history traits discussed above.
EXAMPLES
Within-generation bet hedging:
Here we investigate the conditions under which an organism is selected to decrease its mean fecundity to reduce its variance in fecundity. The setting is the same as the one investigated by Shpak and Proulx (2006). The fecundity means and variances of the various classes of actors affecting the fitness of a FI bearing a mutant bet-hedging allele are evaluated as is usually done by the direct fitness method (e.g., Frank 1998; Rousset and Billiard 2000; Rousset 2004) and are presented in Table 1. Substituting these functionals into the direct fitness function (Equation 3) allows us to calculate explicitly the selective pressure on the mutant allele (Equation 2). Evaluating this selective pressure at the phenotypic value of the resident allele ( ) and taking into account only terms of leading order [i.e., neglecting terms of order 1/N2, 1/f2, 1/(Nf) and beyond], we find that the inclusive fitness effect becomes
) and taking into account only terms of leading order [i.e., neglecting terms of order 1/N2, 1/f2, 1/(Nf) and beyond], we find that the inclusive fitness effect becomes
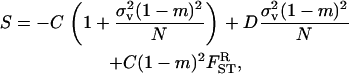 |
(13) |
where
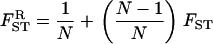 |
(14) |
is the relatedness between the FI and an individual randomly sampled with replacement from its deme (thus including the FI with probability 1/N) and
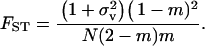 |
(15) |
TABLE 1.
| Symbol | Value for within-generation bet hedging | Value for helping |
|---|---|---|
| f• | f(1 − Cz•) |  |
| f0 | 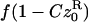 |
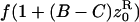 |
| f1 | f(1 − Cz1) | f(1 + (B − C)z1) |
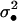 |
σ2(1 − Dz•) | σ2 |
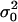 |
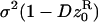 |
σ2 |
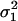 |
σ2(1 − Dz1) | σ2 |
f, baseline mean fecundity of each individual in a monomorphic population; σ2, baseline fecundity variance of each individual in a monomorphic population; z•, phenotype of a focal individual;  average phenotype of individuals from the focal deme (
average phenotype of individuals from the focal deme (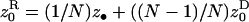 );
); 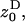 average phenotype of individuals from the focal deme but excluding the FI; z1, average phenotype of individuals from different demes.
average phenotype of individuals from the focal deme but excluding the FI; z1, average phenotype of individuals from different demes.
The inclusive fitness effect S is composed of three terms. First, there is a direct cost to the FI stemming from the reduction in its (mean) fecundity. Second, there is a direct benefit to the FI stemming from the reduction in its variance in fecundity, which is proportional to the probability (1 − m)2/N that two randomly sampled offspring in the focal deme descend from the FI and on the coefficient of variation squared (i.e., effective fecundity). Third, we have to account for an indirect benefit resulting from the reduction in competition in the focal deme faced by the FI's offspring, which results from its own and its relatives1 reduced fecundity. This decrease in kin competition is proportional to the probability (1 − m)2 that two offspring produced in the focal deme compete against each other. Since we assumed large deme size, there are no effects on fitness resulting from relatives lowering their fecundity variance as such effects are of order 1/N2. However, the variance in fecundity increases relatedness (Equation 15) and thus has an impact on the intensity of kin competition. Rearranging the inclusive fitness effect, the low-variance mutant spreads when the threshold cost to benefit ratio satisfies
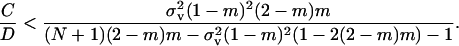 |
(16) |
This threshold decreases with an increase in migration rate and deme size and with a decrease in the variance in fecundity. The inequality cannot be satisfied when the coefficient of variation is vanishingly small (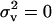 ) or when migration is complete (m = 1).
) or when migration is complete (m = 1).
While the intensity of selection on the variance in fecundity is proportional to the inverse of population size in panmictic populations (Gillespie 1974, 1975, 1977; Demetrius and Gundlach 2000; Demetrius 2001), we see from Equation 13 that the intensity of selection on this variance is proportional to (1 − m)2/N in subdivided populations when regulation occurs after dispersal. Thus, selection on the variance decreases as the migration rate and/or deme size increases. This is so because the contribution of the variance in fecundity of a focal individual to the variance in the number of offspring competing in a given deme after dispersal depends on the square of the migration rate to that deme. Under infinite-island model assumptions, this contribution vanishes for all demes except for the focal deme (compare Equation 5 and Equation 8). The finding that selection on the variance is of intensity (1 − m)2/N corroborates the results of Shpak and Proulx (2006). Indeed, the two first terms of the inclusive fitness effect are consistent with Equation 20 of Shpak and Proulx (2006). However, we note that the approach endorsed here takes into account the effect of the variance in gene frequency between demes on the expected change of allele frequency at the level of the population given by Equation 1. Indeed, the third term of the inclusive fitness effect, which involves kin competition, depends on the variance in gene frequency among demes (i.e., FST) and is not considered by Shpak and Proulx (2006) but its presence here is consistent with previous population genetic derivation of selection on the mean fecundity in subdivided populations (e.g., Roze and Rousset 2003, 2004; Rousset 2004). In the appendix, we complement our analyses by presenting the condition under which an organism is selected to decrease its mean fecundity to reduce its variance in fecundity when regulation occurs before dispersal (soft selection). Selection on the variance in fecundity then scales as 1/N as was already established by Shpak (2005).
Helping behaviors:
Here, we investigate the effect of the variance in fecundity on the condition under which an organism should be prepared to sacrifice a fraction of its resources to help its deme mates. The setting is exactly the same as that in Taylor (1992a), except for the assumptions regarding the distribution of fecundity. While Taylor considers an infinite fecundity, we assume here as in the previous section that the fecundity is finite but large and that its distribution has a variance in excess of a Poisson-distributed fecundity. The means and the variances of the fecundities of the various classes of individuals affecting the fitness of a FI are given in Table 1. Evaluating the selective pressure on the helping allele at the phenotypic value of the resident allele (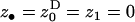 ) and taking into account only terms to leading order, the selective pressure on helping becomes
) and taking into account only terms to leading order, the selective pressure on helping becomes
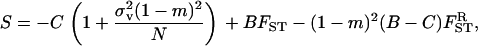 |
(17) |
where 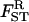 and FST are given by Equation 14 and Equation 15, respectively. This inclusive fitness effect S is also composed of three terms: first, the direct cost of helping; second, the benefit received by the FI from its neighbors; and third, the cost of the increase in competition in the focal deme faced by the FI's offspring caused by it and its neighbors expressing the helping trait. In Taylor's (1992a) original setting we have FST = (
and FST are given by Equation 14 and Equation 15, respectively. This inclusive fitness effect S is also composed of three terms: first, the direct cost of helping; second, the benefit received by the FI from its neighbors; and third, the cost of the increase in competition in the focal deme faced by the FI's offspring caused by it and its neighbors expressing the helping trait. In Taylor's (1992a) original setting we have FST = (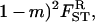 which inserted into Equation 17 cancels out all the benefit terms. However, in the present situation we have FST > (
which inserted into Equation 17 cancels out all the benefit terms. However, in the present situation we have FST > (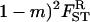 (see Equation 12) because relatedness is inflated due to the increased frequency of coalescence events stemming from the higher variance in fecundity. This results in a situation where the fecundity benefit B is no longer canceled out by the concomitant increase in kin competition. Rearranging this selective pressure, we find that helping spreads when
(see Equation 12) because relatedness is inflated due to the increased frequency of coalescence events stemming from the higher variance in fecundity. This results in a situation where the fecundity benefit B is no longer canceled out by the concomitant increase in kin competition. Rearranging this selective pressure, we find that helping spreads when
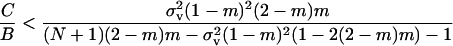 |
(18) |
is satisfied. As in the preceding section, the threshold cost to benefit ratio decreases with an increase in the migration rate and deme size and with a decrease in the variance in fecundity. When the coefficient of variation is vanishingly small (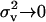 ), we have C/B < 0 so that the direction of selection on helping is determined solely by direct fecundity benefits. The behavior is then selected for if −C > 0, that is, if the number of juveniles counted before any competition stage is increased (Taylor 1992a; Rousset 2004). Here, helping can be selected for at a direct fecundity cost to the actor whenever
), we have C/B < 0 so that the direction of selection on helping is determined solely by direct fecundity benefits. The behavior is then selected for if −C > 0, that is, if the number of juveniles counted before any competition stage is increased (Taylor 1992a; Rousset 2004). Here, helping can be selected for at a direct fecundity cost to the actor whenever  We note that Taylor's seminal hard selection model and relaxations of its life-cycle assumptions have been investigated by several authors (Taylor and Irwin 2000; Irwin and Taylor 2001; Perrin and Lehmann 2001; Rousset 2004; Roze and Rousset 2004; Gardner and West 2006; Lehmann 2006; Lehmann et al. 2006).
We note that Taylor's seminal hard selection model and relaxations of its life-cycle assumptions have been investigated by several authors (Taylor and Irwin 2000; Irwin and Taylor 2001; Perrin and Lehmann 2001; Rousset 2004; Roze and Rousset 2004; Gardner and West 2006; Lehmann 2006; Lehmann et al. 2006).
Dispersal:
Finally, we examine the effect of the variance in fecundity on the evolutionary stable (ES) dispersal rate. The setting is exactly the same as in Frank (1985, 1998), with the exception that we assume again as in the previous sections that fecundity is finite but large and that its distribution has a variance in excess of a Poisson-distributed fecundity. The fitness functions w of a focal parent bearing a mutant genotype affecting the dispersal rate of its offspring are more complicated than those investigated so far because we have to take into account the survival rate s of juveniles during dispersal. Let us designate by d the evolving dispersal rate and by d• the average dispersal rate of the offspring of a FI. A proportion 1 − d• of the offspring produced by the FI remain in the focal deme and a fraction sd• of these offspring enter in competition in another deme after dispersal. The direct fitness function for the evolution of dispersal can then be obtained by setting (1 − m) ≡ 1 − d• in the numerator of the first ratio in Equation 4 and m ≡ sd• in the numerator of the first ratio in Equation 7, while all the remaining terms of these fitness functions are given in Table 2. The fitness function for dispersal can then be written as
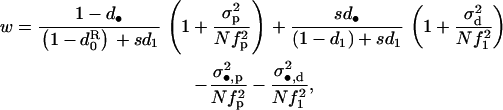 |
(19) |
which, for a large mean fecundity and vanishing coefficient of variation (σv = 0), reduces to the classical fitness function for dispersal (e.g., Frank 1998; Gandon and Rousset 1999; Perrin and Mazalov 2000). Substituting the fitness function into Equation 2, evaluating the selective pressure at the candidate ES dispersal rate, that is, at 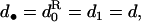 and taking into account only terms to leading order, we find that the selective pressure on dispersal becomes
and taking into account only terms to leading order, we find that the selective pressure on dispersal becomes
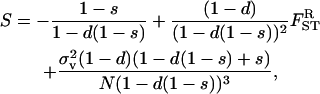 |
(20) |
where  and FST are given by Equation 14 and Equation 15 with the backward migration rate expressed as m ≡ sd/(1 − d + sd), which represents the probability that an individual sampled in a deme is an immigrant. This selective pressure on dispersal is decomposed into three terms: first, the classical direct cost of increasing the dispersal rate, which varies directly with the survival probability s during dispersal; second, the indirect benefit stemming from the reduction in competition in the focal deme faced by the FI's offspring, which results from it and its neighbors' offspring dispersing (e.g., Hamilton and May 1977; Taylor 1988; Frank 1998; Gandon and Michalakis 1999; Gandon and Rousset 1999; Perrin and Mazalov 2000); and third, a benefit resulting from the dispersing progeny of the FI reducing the variance in its number of offspring reaching adulthood in the focal deme. This term is new and is driven by Equation 5. By dispersing, juveniles minimize the variance in the number of offspring of a focal parent entering into competition in the focal deme. Since the variance in the number of offspring entering locally into competition (Equation 5) dominates the variance in the number of offspring entering into competition in different demes by dispersing (Equation 8), dispersal decreases the overall variance in the number of offspring entering into competition. As a consequence, fitness increases through dispersing and the ES dispersal rates are boosted by higher variance in fecundity. Figure 2 compares the ES dispersal rate obtained from the present model with the classical ES dispersal rate (e.g., Hamilton and May 1977; Frank 1998; Gandon and Rousset 1999; Perrin and Mazalov 2000) obtained under the vanishing coefficient of variation (
and FST are given by Equation 14 and Equation 15 with the backward migration rate expressed as m ≡ sd/(1 − d + sd), which represents the probability that an individual sampled in a deme is an immigrant. This selective pressure on dispersal is decomposed into three terms: first, the classical direct cost of increasing the dispersal rate, which varies directly with the survival probability s during dispersal; second, the indirect benefit stemming from the reduction in competition in the focal deme faced by the FI's offspring, which results from it and its neighbors' offspring dispersing (e.g., Hamilton and May 1977; Taylor 1988; Frank 1998; Gandon and Michalakis 1999; Gandon and Rousset 1999; Perrin and Mazalov 2000); and third, a benefit resulting from the dispersing progeny of the FI reducing the variance in its number of offspring reaching adulthood in the focal deme. This term is new and is driven by Equation 5. By dispersing, juveniles minimize the variance in the number of offspring of a focal parent entering into competition in the focal deme. Since the variance in the number of offspring entering locally into competition (Equation 5) dominates the variance in the number of offspring entering into competition in different demes by dispersing (Equation 8), dispersal decreases the overall variance in the number of offspring entering into competition. As a consequence, fitness increases through dispersing and the ES dispersal rates are boosted by higher variance in fecundity. Figure 2 compares the ES dispersal rate obtained from the present model with the classical ES dispersal rate (e.g., Hamilton and May 1977; Frank 1998; Gandon and Rousset 1999; Perrin and Mazalov 2000) obtained under the vanishing coefficient of variation (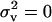 in Equations 20 and 15).
in Equations 20 and 15).
TABLE 2.
| Symbol | Value for dispersal |
|---|---|
| f• | f |
| fp | 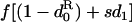 |
| f1 | 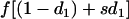 |
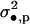 |
(1 − d•)d•f + (1 − d•)2σ2 |
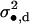 |
sd•f |
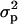 |
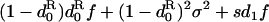 |
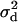 |
(1 − d1)d1f + (1 − d1)2σ2 + sd1f |
d•, dispersal rate of the offspring of the focal individual; 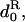 average dispersal rate of the offspring of individuals from the focal deme (
average dispersal rate of the offspring of individuals from the focal deme (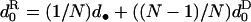 ); d1, average dispersal rate of the offspring of individuals from different demes
); d1, average dispersal rate of the offspring of individuals from different demes
Figure 2.—
Evolutionary stable dispersal rate graphed as a function of deme size N with survival rate set to s = 0.9. The solid line is the classical ES dispersal rate obtained by assuming that the progeny distribution is Poisson or has an infinite mean. The dotted line (curve between solid and dashed curves) corresponds to the ES dispersal rate when 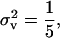 while the dashed line corresponds to the ES dispersal rate when
while the dashed line corresponds to the ES dispersal rate when 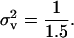 Fecundity distributions corresponding to such variances are given in Figure 1.
Fecundity distributions corresponding to such variances are given in Figure 1.
DISCUSSION
In this article, we integrated the classical measure of selection for within-generation bet hedging (Gillespie 1974, 1975, 1977) into the game theoretic and inclusive fitness measure of selection for structured populations (e.g., Taylor 1990; Rousset and Billiard 2000; Rousset 2004, 2006). The resulting framework in which fitness depends explicitly on both the means and the variances of the fecundity distribution can be applied to study the evolution of various life-history traits (i.e., sex ratio, helping, reproductive effort), under soft and hard selection regimes. The selective pressures on three traits were directly investigated here, namely within-generation bet hedging, helping behaviors, and the evolutionary stable dispersal rate. The results suggest that the evolutionary dynamics of all these traits are affected by the variance in fecundity, although with variable intensity and under different demographic conditions.
Within-generation bet hedging:
Natural selection selects against the variance in offspring number when panmictic populations are not too large (Gillespie 1974, 1975, 1977; Demetrius and Gundlach 2000; Demetrius 2001). Our models suggest that selection against this variance in subdivided populations scales as 1/N (Equation A37) when regulation occurs before dispersal (soft selection) and scales as (1 − m)2/N (Equation 13) when regulation occurs after dispersal (hard selection). This is so because the contribution of the variance in fecundity of a focal individual to the variance in its number of offspring competing in a given deme depends on the square of the migration rate to that deme. Under soft selection, all the competition occurs in the focal deme so that migration has no effect on selection. By contrast, under hard selection, the offspring of an individual compete in different demes and, due to our infinite-island model assumptions, the contribution of the variance in fecundity to the various demes vanishes for all demes except for the focal deme (compare Equation 5 and Equation 8). These results corroborate the findings of Shpak (2005) and Shpak and Proulx (2006) and illustrate that they can be derived from the same model.
It is sometimes assumed in the literature that selection against the variance in offspring number and its impact on various life-history trait will be of limited importance in natural populations because they are of large size (e.g., Seger and Brockman 1985). Our results and those of Shpak (2005) and Shpak and Proulx (2006) demonstrate that selection against this variance can operate perfectly in large but subdivided populations in the presence of both soft and hard selection regimes. That selection can target the fecundity variance in large populations may be relevant for the evolution of bet-hedging strategies in general and for the theory of sex-ratio homeostasis (Verner 1965; Taylor and Sauer 1980) in particular. Indeed, the results suggest that the demographic parameters of the population will condition whether strategies involving probabilistic sex-ratio determination (i.e., each individual develops into a male or a female with a given probability) will resist invasion by strategies involving a rigid determination of it (i.e., a fixed proportion of offspring develop into either sex).
More generally, the fecundity variance might also play a role in the evolution of systems of phenotypic determination. Systems of adaptive polymorphism are expected in the presence of fluctuating environments or resource specialization and involve, among other mechanisms, the random determination of an individual's phenotype or its genetic determination (Grafen 1999; Leimar 2005). Since probabilistic phenotype determination can introduce additional variance in the number of offspring entering in competition, the conditions under which mixed evolutionary stable strategies are favored over pure genetic polymorphism are likely to be dependent on the variance in the number of offspring sent into competition by each strategy.
Helping behaviors:
While the preceding examples of bet-hedging strategies resulted in directional selection against the variance in fecundity, the selective pressure on other strategies can depend on the magnitude of this variance. Helping behaviors involving kin interaction fall into this category because their evolution depends on the relatedness between interacting individuals, whose dynamic is determined by the probability of common ancestry within deme, itself a function of the fecundity distribution (Ewens 2004; Rousset 2004). An increase in fecundity variance decreases the number of effective ancestors within demes and thus increases the relatedness between deme mates, possibly leading to an increase in the selective pressure on helping behaviors.
Taylor (1992a,b) has demonstrated that when demes are of constant size and when fecundity is infinite (i.e., no variance in fecundity), selection on helping is determined solely by direct fecundity benefits. Here, Taylor's assumption on fecundity was replaced by the assumption that fecundity follows a distribution with finite but large mean and with a variance exceeding the mean. Allowing for a nonvanishingly small coefficient of variation increases relatedness between deme members with the consequence that all kin selected effects, whether positive or negative, are increased. Here, this translates into helping being selected for at a direct fecundity cost to the actor with the intensity of selection on helping decreasing when the variance in fecundity decreases (Equation 18). This result suggests that selection on helping is also bound to depend on the mating system because the variance in the number of mating partners obtained by males will increase relatedness within demes.
The finding that the magnitude of the variance in fecundity qualitatively affects the outcome of selection on costly helping might help us to understand why different authors have repeatedly reached contradictory results when investigating selection on helping under apparently identical life cycles. While several authors have emphasized that unconditional costly helping is counterselected when social interactions occur between semelparous individuals living in structured populations of constant size (Wilson et al. 1992; Taylor 1992a,b; Rousset 2004; Lehmann et al. 2006), other authors have suggested that costly helping is selected for in that case (Nowak and May 1992; Killingback et al. 1999; Hauert and Doebeli 2004). The results by the former group of authors are based on analytical models assuming infinite and/or Poisson-distributed fecundities. The results by the latter group are mostly based on simulations. In this case, it remains unclear how the updating rules used by the authors to simulate the transmission of strategies from one generation to the next in the population influence relatedness and may thus result in significant departure from the Poisson-distributed fecundity assumption. For instance, copying the fittest neighbor will result in different dynamics of relatedness than copying neighbors with a probability proportional to their fitness. These specific features might help to explain why some authors observed the emergence of costly helping behaviors while others did not.
Dispersal:
This article emphasizes that the variance in the number of offspring sent into competition can be as important in determining fitness as the mean of such offspring (Equations 4 and 7). This variance will depend among other factors on the probability that an individual reproduces, its offspring disperse, migrants survive dispersal, and successful migrants survive competition. While a near endless chain of stochastic events may affect individuals as they move along their life cycle, we focused here only on the stochasticity induced by reproduction and migration. Migration has a striking and nonintuitive impact on the total variance in the number of offspring entering competition because it results in two distinct effects on this variance. First, migration redistributes the variance in fecundity of an individual over the whole population. Since the variance in the number of offspring competing in a given deme after dispersal depends on the square of the migration rate to that deme, the contribution of the variance in fecundity to the variance in competing offspring in that deme vanishes for all demes except for the focal one, whenever there is a large number of demes (compare Equation 5 and Equation 8). Second, we assumed that migration itself is a probabilistic event, which thus generates a variance in the number of competing offspring even for fixed fecundity schedules (first term in Equations 5 and 8). These two effects of migration on fitness will have two distinct consequences for the evolution of stable dispersal rates.
First, assuming that each juvenile disperses independently from each other and that the environment is constant, the evolutionary stable dispersal rate depends on a balance between two opposing forces. There is a cost of dispersal resulting from reduced survival during migration opposed by a benefit leading to local decrease in kin competition (e.g., Hamilton and May 1977; Frank 1985, 1998; Gandon and Michalakis 1999; Gandon and Rousset 1999; Perrin and Mazalov 2000). Here, this balance is displaced in favor of increased dispersal because the cost of dispersal is reduced as migration reduces the total variance in the number of offspring of an individual sent into competition. By dispersing, the offspring of the focal individual decrease the impact of the variance in fecundity on fitness. A large variance in fecundity can result in a twofold increase of the evolutionary stable dispersal rate (Figure 2). That the variance in fecundity can affect the candidate evolutionary stable strategy has already been shown for models of sex allocation (e.g., Proulx 2000, 2004). Our result that the ES dispersal rate is affected by the variance in fecundity might be of practical importance when evaluating the causes of dispersal in natural populations and it suggests that the fecundity distribution might explain a part of the observed variance in dispersal rates.
The second consequence of subdivided populations for the evolution of dispersal rates stems from the fact that if migration is a probabilistic event, it increases the variance in offspring number of an adult coming into competition. One might then wonder whether a strategy resulting in a fixed proportion of individuals dispersing would not be favored over a strategy involving probabilistic dispersal, in complete analogy with the theory of sex-ratio homeostasis (Taylor and Sauer 1980), where the production of a fixed proportion of males and females is favored over probabilistic sex-ratio determination. In the Dispersal homeostasis section in the appendix, we constructed a model precisely along these lines (see Equation A43), which demonstrates that dispersal homeostasis can indeed evolve but that the selective pressure on it is only of order 1/(fN), where f is the mean fecundity. This result is in line with work by Taylor and Sauer (1980), who showed that selection on “sex-ratio homeostasis” in panmictic populations is proportional to the inverse of the total number fN of offspring born in a group. According to our assumption that both deme size and fecundity are large, terms of such order were generally dismissed in the calculation of the various selective pressures (Equations 13, 17, and 20). However, selection on both dispersal and sex-ratio homeostasis might be effective in subdivided population when both the mean fecundity and the deme size are not too large.
Limitations and extensions of the model:
All results derived in this article are based on five main assumptions that greatly simplified the analyses and that we now discuss. First, we assumed an island model of migration. This assumption was introduced to highlight in a simple way the effects of the fecundity variance on fitness in subdivided populations, although the model derived in the appendix is more general and allows for isolation by distance (e.g., stepping-stone dispersal). In this case we expect selection on the variance in fecundity to be increased because the variance in the number of offspring competing in a given deme after dispersal depends on the square of the migration rate to that deme (Equation A16). This probability will not be vanishingly small for migration at short spatial distances so that selection against the variance will generally exceed the intensity (1 − m)2/N established here for infinite-island situations. Second, we assumed that demes are of large size (terms of order 1/N2 are neglected, see the Regulation section in the appendix). This assumption was introduced to focus primarily on selection on the variance in fecundity and to neglect selection on other measures of dispersion of the fecundity distribution that are likely to matter when demes are of small size. As can be noted by inspecting Equation A6, natural selection will not only reduce the variance in fecundity but also favor positively skewed fecundity distributions that are platykurtic: that is, distributions that are asymmetric with a fatter right tail and that are less peaked than a normally distributed variable. This is intuitively expected because it results in a shape of the fecundity distribution that reduces the likelihood of producing no offspring at all. Exactly how natural selection will shape the fecundity distribution given the constraints on reproductions remains to be investigated. Third, we assumed that the mean fecundity is finite but large. This assumption was introduced to be able to neglect demographic stochasticity and to obtain analytical expressions as simply as possible. Under ceiling regulation, demographic stochasticity is completely prevented from occurring for demes of large size as soon as individuals produce more than two offspring (Lehmann et al. 2006). However, other forms of population regulation occur in nature, such as density-dependent survival with no specific ceiling effect, which might result in wider population fluctuations. Overall, allowing for low fecundities and small deme sizes should in principle lead to situations where selection on the measures of statistical dispersions (e.g., variance, skewness) is exacerbated. Fourth, we assumed that the variance in fecundity is of larger order of magnitude than the mean so that the coefficient of variation (σv) is not vanishingly small even when the mean fecundity is large. This assumption was introduced to ensure that the variance of the fecundity distribution still influences fitness under our hypotheses of large fecundities and demes sizes. A typical example of such a fecundity distribution is given by the negative-binomial distribution, which follows from many models of reproduction (Caswell 2000, p. 455) and has been advocated to be characteristic of the fertility in humans and other species (Anscombe 1949; Cerda-Flores and Davila-Rodriguez 2000). That organisms have a variance in fecundity in excess of the mean has also been reported in studies of reproductive success (Crow and Kimura 1970, Table 7.6.4.2.; Clutton-Brock 1988). Fifth, we assumed an infinite number of demes so that the effect of drift at the level of the total population on gene frequency change is not taken into account. However, our direct fitness derivation presented in the appendix is framed within a finite population so that the inclusive fitness effect S also allows us to evaluate the first-order phenotypic effect of a mutant allele on its probability of fixation (Rousset 2004, 2006). The evaluation of the probability of fixation itself deserves further formalizations that might be achieved by following along the lines of the direct fitness approach to diffusion equations of Roze and Rousset (2003, 2004). We finally mention that we investigated here only three phenotypic traits, whose evolution depends on the magnitude of relatedness (i.e., FST) that is itself a function of the variance in fecundity. However, the evolution of many other traits including sex ratio, mate choice, harming behaviors such as spiteful acts, niche-constructing phenotypes, or resource exploitation curves depend on relatedness (e.g., Hamilton 1971; Boomsma and Grafen 1991; Taylor and Getz 1994; Gandon and Michalakis 1999; Ajar 2003; Lehmann 2006). Consequently, the expression of all these traits is likely to be indirectly influenced by the variance in fecundity.
On the basis of all these comments and the premise that there is as much genetic variation for the variance in fecundity as there is for the mean, we expect that the results presented in this article are of relevance for natural systems. As a final note, we can thus only reemphasize Gillespie's (1977) view that the role of the variance deserves a more prominent place in our thinking about evolutionary processes.
Acknowledgments
We thank one anonymous reviewer for very helpful comments on this manuscript. L.L. acknowledges financial support from the Swiss National Science Foundation.
APPENDIX
Fitness function:
Here we derive an expression for the direct fitness function w (Equation 2), which involves both the mean and the variance of the number of offspring produced by a focal individual and generalizes the derivation of Gillespie (1975) to subdivided populations (see also Shpak and Proulx 2006). We assume that the mean fecundity of individuals is finite but sufficiently large to prevent any demographic stochasticity. Accordingly, each deme will be treated as being of constant size N. This assumption implies that the model is not exact but provides a balance between accuracy and complexity. An exact model can be obtained by recasting the forthcoming derivation within the framework of the general metapopulation model of Rousset and Ronce (2004) by conditioning the expressions for fitness on demographic states. For completeness, we first consider that the population is constituted of a finite number nd of demes and then take the infinite island limit (nd → ∞). We now follow the events of the life cycle presented in the main text in reverse order.
Regulation:
The expectation of the random number Nijx of offspring surviving regulation in deme i and descending from individual x breeding in deme j can be written as
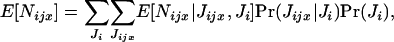 |
(A1) |
where Pr(Jijx | Ji)Pr(Ji) is the joint probability that Jijx offspring of individual x enter in competition in deme i among a total number of Ji offspring and E[Nijx | Jijx, Ji] is the conditional expectation of Nijx given Jijx and
 |
(A2) |
where each random variable Jijy is assumed to be independently distributed.
Density-dependent regulation is assumed to affect each individual independently and equally. According to this assumption, the conditional expectation of the number Nijx of recruited offspring of x given Ji and Jijx is N times the probability that a randomly sampled juvenile is an offspring of x (Gillespie 1975), and this is
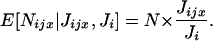 |
(A3) |
The random numbers of juveniles in this ratio can be expressed as deviations around their expectations; thereby
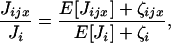 |
(A4) |
where ζijx ≡ Jijx − E[Jijx] and 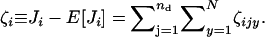 The ratio of the number of juveniles can now be expanded according to the series
The ratio of the number of juveniles can now be expanded according to the series
 |
(A5) |
which, once substituted into Equation A1, gives the fitness function
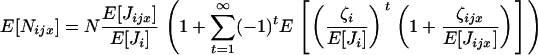 |
(A6) |
Equation A6 illustrates that the expectation of the number of recruited offspring in deme i and descending from individual x breeding in deme j is a function of all the moments 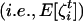 of the distribution of the number of offspring entering in competition in deme i after dispersal. It involves, among other measures of statistical dispersion, the variance, the skewness, and the kurtosis of the distribution of number of offspring entering in competition.
of the distribution of the number of offspring entering in competition in deme i after dispersal. It involves, among other measures of statistical dispersion, the variance, the skewness, and the kurtosis of the distribution of number of offspring entering in competition.
Because Equation A6 is complicated, we follow previous work (Gillespie 1975; Proulx 2000; Shpak and Proulx 2006) by establishing an expression for fitness that takes only the variance of the distributions of competing juveniles into account. To do this, we note that the sum 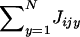 appearing in the expectation E[Ji] (see Equation A12) is about as big as NE[Jijy] when N becomes large. By contrast, the central limit theorem ascertains that the sum
appearing in the expectation E[Ji] (see Equation A12) is about as big as NE[Jijy] when N becomes large. By contrast, the central limit theorem ascertains that the sum 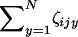 appearing in the deviation around the mean ζi is only about as big as
appearing in the deviation around the mean ζi is only about as big as 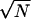 when N is large (e.g., Grimmet and Stirzaker 2001, pp. 193–194). Therefore, the ratio
when N is large (e.g., Grimmet and Stirzaker 2001, pp. 193–194). Therefore, the ratio 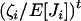 appearing in Equation A6 is approximately of order of magnitude
appearing in Equation A6 is approximately of order of magnitude 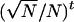 when N becomes large and rapidly shrinks with increasing t. As in Gillespie (1975, p. 114), we thus keep only leading terms of order 1/N and neglect terms of order 1/N2 and higher order in Equation A6. After rearrangement, we obtain
when N becomes large and rapidly shrinks with increasing t. As in Gillespie (1975, p. 114), we thus keep only leading terms of order 1/N and neglect terms of order 1/N2 and higher order in Equation A6. After rearrangement, we obtain
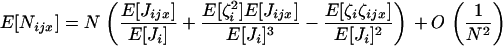 |
(A7) |
From the assumption of the independence of the distributions of juveniles we have 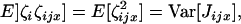 which is the variance in the number of juveniles entering in competition in deme i and descending from individual x breeding in deme j, and
which is the variance in the number of juveniles entering in competition in deme i and descending from individual x breeding in deme j, and 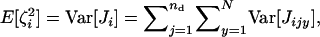 which is the total variance in the number of juveniles entering in competition in deme i. Equation A7 can now be written as
which is the total variance in the number of juveniles entering in competition in deme i. Equation A7 can now be written as
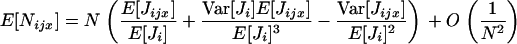 |
(A8) |
and depends only on the means and the variances of the distributions of the number of juveniles entering in competition in deme i. The means and the variances appearing in Equation A8 are for the number of juveniles entering in competition in demes after dispersal. In the next section, we express these moments in terms of means and variances of juveniles produced before dispersal.
We note that the term in parentheses in Equation A7 agrees precisely with the approximation of the expectation of a ratio of random variables as obtained by the delta method (Lynch and Walsh 1998, pp. 807–813), which for the random variables X and Y reads
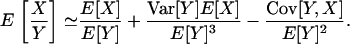 |
(A9) |
Migration:
The mean number of juveniles entering in competition in deme i and descending from individual x reproducing in deme j can be written as
 |
(A10) |
where Jjx is the number of offspring of individual x produced before the dispersal stage and Pr(Jjx) is the corresponding probability distribution. Given Jjx and the assumption that each juvenile migrates independently with probability mij from deme j to deme i, the number of dispersing juveniles of individual x to deme i follows the binomial distribution 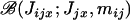 with parameters Jjx and mij. Hence, E[Jijx | Jjx] = mijJjx with the result that
with parameters Jjx and mij. Hence, E[Jijx | Jjx] = mijJjx with the result that
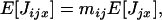 |
(A11) |
where E[Jjx] is the mean fecundity of individual x (here the mean number of offspring produced before the dispersal stage). Summing up the contribution of each individual to deme i, we obtain
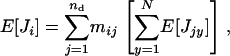 |
(A12) |
where the term in brackets is the total fecundity in deme j, which, when divided by N yields the average mean fecundity of individuals in deme j.
The variance in the number of juveniles entering in competition in deme i and descending from individual x can be written as
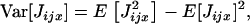 |
(A13) |
where the second moment  can be evaluated by conditioning on the number Jjx of juveniles produced before dispersal; that is,
can be evaluated by conditioning on the number Jjx of juveniles produced before dispersal; that is,
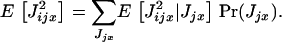 |
(A14) |
From the binomial distribution we have
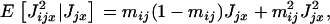 |
(A15) |
which allows us to express equation A13 as
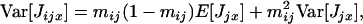 |
(A16) |
where Var[Jjx] is the variance in the number of juveniles produced before dispersal by individual x in deme j.
Remembering that the total variance in the number of juveniles entering in competition in deme i is given by 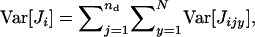 we have
we have
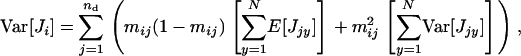 |
(A17) |
where the term in the second brackets is the total variance in the number of juveniles produced in deme j.
Selection:
We now use the foregoing results to establish the direct fitness w of a focal individual (FI) bearing a mutant allele (e.g., Taylor and Frank 1996; Frank 1998; Rousset and Billiard 2000; Rousset 2004). We consider that individual x breeding in deme j is this FI and assume that interactions are spatially homogeneous. With this assumption, the fitness of the FI can be expressed as a function of its own phenotype, of the phenotype of the individuals in its deme, of the phenotypes of individuals one step further apart, of the phenotypes of individuals two steps further apart, and so on (e.g., Rousset and Billiard 2000; Rousset 2004). For simplicity, we further consider here only the island model of dispersal where the individuals in the population can be pooled into three classes so that the fitness of the FI is expressed only as a function of its own phenotype, of the average phenotype of its deme mates, and of the average phenotype of individuals from different demes. We call m the dispersal probability of a juvenile so that its probability to disperse to a given nonnatal deme is m/(nd − 1). Designating by f• and 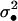 the mean and the variance of the fecundity distribution of the FI, using Equations A8 and A11–A17, we find that the mean number of offspring of the FI reaching adulthood in the focal deme can be written as
the mean and the variance of the fecundity distribution of the FI, using Equations A8 and A11–A17, we find that the mean number of offspring of the FI reaching adulthood in the focal deme can be written as
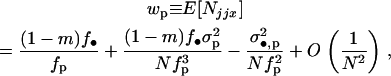 |
(A18) |
with
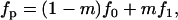 |
(A19) |
where 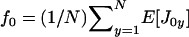 is the average mean fecundity of individuals in the focal deme (at distance 0) and
is the average mean fecundity of individuals in the focal deme (at distance 0) and 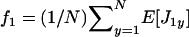 is the average mean fecundity of individuals breeding in a different deme (at distance 1). Such average fecundities can conveniently be expressed as functions of the underlying average phenotypes of individuals, which in turn are determined by their genotypes (e.g., Frank 1998; Rousset and Billiard 2000; Rousset 2003, 2004; Roze and Rousset 2003).
is the average mean fecundity of individuals breeding in a different deme (at distance 1). Such average fecundities can conveniently be expressed as functions of the underlying average phenotypes of individuals, which in turn are determined by their genotypes (e.g., Frank 1998; Rousset and Billiard 2000; Rousset 2003, 2004; Roze and Rousset 2003).
The fitness function wp also involves the variance in the number of offspring of the FI entering in competition in the focal deme that is given by
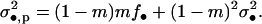 |
(A20) |
Finally, we have the average variance 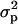 of the number of juveniles entering in competition in the focal deme, which includes juveniles from the focal deme and from the nd − 1 different demes. This variance can be written as
of the number of juveniles entering in competition in the focal deme, which includes juveniles from the focal deme and from the nd − 1 different demes. This variance can be written as
 |
(A21) |
where 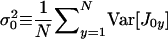 is the average variance in fecundity of individuals breeding in the focal deme and
is the average variance in fecundity of individuals breeding in the focal deme and 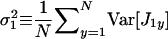 is the average variance in fecundity of individuals breeding in a different deme. These average variances can also be expressed in the direct fitness manner as a function of the phenotypes of individuals affecting them.
is the average variance in fecundity of individuals breeding in a different deme. These average variances can also be expressed in the direct fitness manner as a function of the phenotypes of individuals affecting them.
The mean number of offspring descending from the FI and reaching adulthood in a different deme can be written as
 |
(A22) |
where
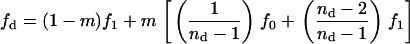 |
(A23) |
is the expected average number of offspring entering in competition in a different deme, from which a proportion m/(nd − 1) descend from the focal deme. The fitness function fd also depends on the variance in the number of offspring of the FI coming into competition in any of the nd − 1 other demes by dispersal; this is
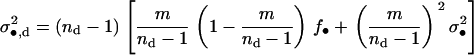 |
(A24) |
Last, to compute the fitness function we need the average variance of the number of juveniles entering in competition in a deme different than the focal deme, which is given by
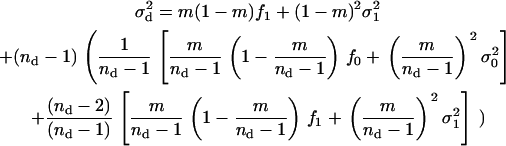 |
(A25) |
The total fitness of a focal individual is obtained by summing up Equations A18 and A22, whereby
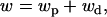 |
(A26) |
which can readily be checked to sum up to one (w = 1) when the population is monomorphic (i.e., all individuals have the same phenotype and consequently bear the same mean and variance in fecundity). By taking the infinite-island limit (nd → ∞) in the fitness functions wp and wd we obtain the equations presented in the main text.
Relatedness:
In the infinite-island model of dispersal, the probability of identity between two homologous genes sampled without replacement in the same deme after dispersal satisfies at steady state the recursion
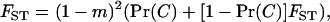 |
(A27) |
where (1 − m)2 is the probability that the two genes are of philopatric origin and Pr(C) is the probability that they descend from the same parent. This probability of coalescence can be expanded as
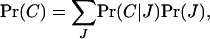 |
(A28) |
so that conditional on J juveniles being produced in a deme, the probability of coalescence is the expectation of the ratio of the total number of ways of sampling two genes from the same parent to the total number of ways of sampling two genes,
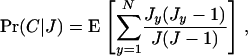 |
(A29) |
where Jy is the number of juveniles produced by individual y and where the total fecundity in a deme satisfies J = J1 + … + JN. Since relatedness is evaluated in the absence of selection, the population is assumed to be monomorphic in phenotypic effects and all individuals have the same fecundity distribution. Consequently, the probability of coalescence can be written as
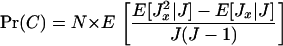 |
(A30) |
Using Equation A9 to approximate this coalescence probability, defining f ≡ E[Jx] as the mean fecundity of an individual and σ2 ≡ Var[Jx] as the variance of the fecundity of an individual, we obtain after lengthy but straightforward calculations
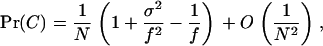 |
(A31) |
which is expressed as a function of the variance and the mean of the fecundity distribution only. When the fecundity follows the Poisson-gamma distribution with variance σ2 = f + f2/α, the coalescence probability takes the form
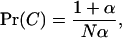 |
(A32) |
which is very close to the exact coalescence probability that can be calculated in this special case and is given by (1 + α)/(1 + Nα).
Poisson-distributed fecundity:
We consider here the special but relevant case when the fecundity of each individual follows a Poisson distribution and when each juvenile migrates independently from each other. With these two assumptions, the random number of dispersing and philopatric juveniles entering into competition in a given deme follows a Poisson distribution (Karlin and Taylor 1981; Grimmet and Stirzaker 2001). Substituting Equation A3 into Equation A1, we have
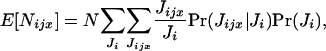 |
(A33) |
where Pr(Ji) is a Poisson distribution with mean E[Ji] and the conditional distribution Pr(Jijx | Ji) is binomial with parameters Ji and E[Jijx]/E[Ji] (Karlin and McGregor 1968; Karlin and Taylor 1981; Grimmet and Stirzaker 2001). Hence, 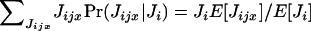 and the expected number of recruited offspring in deme i and descending from individual x breeding in deme j reduces to
and the expected number of recruited offspring in deme i and descending from individual x breeding in deme j reduces to
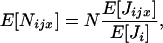 |
(A34) |
which depends only on the mean number of juveniles entering in competition and not on any other measure of statistical dispersion. Under this assumption of Poisson-distributed fecundity, the direct fitness of a focal individual in the infinite-island model of dispersal reduces to
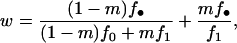 |
(A35) |
which is also equivalent to the fitness function obtained by assuming that each individual produces an infinite number of juveniles (e.g., Gandon and Michalakis 1999; Roze and Rousset 2003; Rousset 2004). Similar arguments show that the coalescence probability (equation A30) for the relatedness coefficients (equation A27) reduces to Pr(C) = 1/N under a Poisson-distributed fecundity. Note that when the fecundity has a finite and small mean, demographic stochasticity matters and one should take the demographic states of demes into account when evaluating the fitness functions (e.g., Rousset and Ronce 2004).
Within-generation bet hedging under soft selection:
Here we investigate the conditions under which an organism is prepared to decrease its mean fecundity to reduce its variance in fecundity in the presence of regulation before dispersal (soft selection). The setting is the same as the one investigated by Shpak (2005) and since competition occurs in this case only between deme mates, the direct fitness function for this model is given by
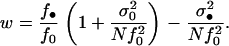 |
(A36) |
Using the means and variances of the two classes of actors given in Table 1, evaluating the selective pressure (Equation 2) at the phenotypic value of the resident allele (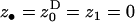 ) and taking into account only terms of leading order [i.e., neglecting terms of order 1/N2, 1/f2, 1/(Nf) and beyond], we find that the inclusive fitness effect becomes
) and taking into account only terms of leading order [i.e., neglecting terms of order 1/N2, 1/f2, 1/(Nf) and beyond], we find that the inclusive fitness effect becomes
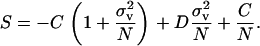 |
(A37) |
Selection on the variance in fecundity now scales as 1/N, which is consistent with the result of Shpak (2005) while the last term of the inclusive fitness effect is consistent with the soft selection models of Rousset (2004, p. 125). Note that all kin selection terms have canceled out as is usually the case under regulation before dispersal (Wade 1985; Roze and Rousset 2003).
Dispersal homeostasis:
Here we develop a model of “dispersal homeostasis.” We consider the fate of a mutant homeostatic allele, which, conditional on having produced a number J of juveniles, sends a fixed proportion of juveniles to other demes (i.e., maternal control of dispersal). By contrast, conditional on J, the number of migrant juveniles of the resident allele follows a binomial distribution as assumed in the previous sections. Accordingly, the variance in the number of juveniles entering into competition in deme i and descending from an individual x breeding in deme j and bearing a resident allele is given by Equation A16, while this variance for such an individual bearing the mutant homeostatic allele becomes
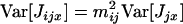 |
(A38) |
because dispersal is no longer a random variable.
It now remains to evaluate the various variance components for the fitness functions (Equations 4 and 7). Applying the same lines of reasoning as in the Selection section of this appendix, we find that the number of offspring of a FI coming into competition in the focal deme can be written as
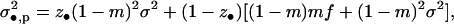 |
(A39) |
where z• is the phenotype of the focal individual, equal to one if it expresses the homeostatic allele and zero otherwise. The average variance of the number of juveniles entering into competition in the focal deme by dispersing is given by
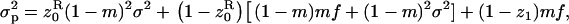 |
(A40) |
where 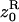 is the average phenotype of individuals in the focal deme and z1 is the average phenotype of individuals from different demes. We also need the variance in the number of offspring of the FI entering into competition in other demes; this is
is the average phenotype of individuals in the focal deme and z1 is the average phenotype of individuals from different demes. We also need the variance in the number of offspring of the FI entering into competition in other demes; this is
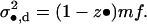 |
(A41) |
Finally, the average variance in fecundity of individuals breeding in different demes is given by
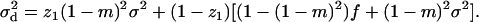 |
(A42) |
Substituting these functionals into the direct fitness function (Equation 3) and assuming that the individuals in the population are monomorphic for the mean and the variance in fecundity (given by f and σ2) allows us to calculate explicitly the selective pressure on the homeostatic allele (Equation 2). Evaluating this selective pressure at the phenotypic value of the resident allele, which does not express dispersal homeostasis (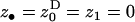 ), we find that the inclusive fitness is given by
), we find that the inclusive fitness is given by
 |
(A43) |
The selective pressure on dispersal homeostasis increases monotonically with dispersal and decreases with deme size and mean fecundity, a result that is in line with the work of Taylor and Sauer (1980), who showed that selection on sex-ratio homeostasis in panmictic populations is proportional to the inverse of the total number fN of offspring born in a group.
References
- Ajar, E., 2003. Analysis of disruptive selection in subdivided populations. BMC Evol. Biol. 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anscombe, F. A., 1949. The statistical analysis of insect counts based on the negative binomial. Biometrics 5: 165–173. [PubMed] [Google Scholar]
- Bartlett, 1955. An Introduction to Stochastic Processes. Cambridge University Press, Cambridge, UK.
- Boomsma, J. J., and A. Grafen, 1991. Colony-level sex-ratio selection in the eusocial hymenoptera. J. Evol. Biol. 4: 383–407. [Google Scholar]
- Caswell, H., 2000. Matrix Population Models. Sinauer Associates, Sunderland, MA.
- Cerda-Flores, R. M., and M. I. Davila-Rodriguez, 2000. Natural fertility in northeastern Mexico. Arch. Med. Res. 31: 599–604. [DOI] [PubMed] [Google Scholar]
- Cherry, J. L., and J. Wakeley, 2003. A diffusion approximation for selection and drift in a subdivided population. Genetics 163: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesson, P. L., 1981. Models for spatially distributed populations: the effect of within-patch variability. Theor. Popul. Biol. 19: 288–325. [Google Scholar]
- Clutton-Brock, T. H. (Editor), 1988. Reproductive Success. University of Chicago Press, Chicago.
- Comins, H. N., W. D. Hamilton and R. M. May, 1980. Evolutionarily stable dispersal strategies. J. Theor. Biol. 82: 205–230. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- Demetrius, L., 2001. Mortality plateaus and directionality theory. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268: 2029–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetrius, L., and V. M. Gundlach, 2000. Game theory and evolution: finite size and absolute fitness measures. Math. Biosci. 168: 9–38. [DOI] [PubMed] [Google Scholar]
- Ewens, W. J., 1969. Population Genetics. Methuen, London.
- Ewens, W. J., 2004. Mathematical Population Genetics. Springer-Verlag, New York.
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Frank, S. A., 1985. Dispersal polymorphism in subdivided populations. J. Theor. Biol. 122: 303–309. [DOI] [PubMed] [Google Scholar]
- Frank, S. A., 1998. Foundations of Social Evolution. Princeton University Press, Princeton, NJ.
- Gandon, S., and Y. Michalakis, 1999. Evolutionary stable dispersal rate in a metapopulation with extinction and kin competition. J. Theor. Biol. 199: 275–290. [DOI] [PubMed] [Google Scholar]
- Gandon, S., and F. Rousset, 1999. Evolution of stepping-stone dispersal rates. Proc. R. Soc. Lond. Ser. B Biol. Sci. 221: 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, A., and S. A. West, 2006. Demography, altruism, and the benefits of budding. J. Evol. Biol. 19: 1707–1716. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1974. Natural selection for within-generation variance in offspring number. Genetics 76: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. H., 1975. Natural selection for within-generation variance in offspring number 2. Discrete haploid models. Genetics 81: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. H., 1977. Natural selection for variances in offspring numbers—new evolutionary principle. Am. Nat. 111: 1010–1014. [Google Scholar]
- Grafen, A., 1999. Formal Darwinism, the individual-as-maximizing-agent analogy and bet-hedging. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266: 799–803. [Google Scholar]
- Grimmet, G., and D. Stirzaker, 2001. Probability and Random Processes. Oxford University Press, Oxford.
- Haldane, J. B. S., 1927. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Hamilton, W. D., 1964. The genetical evolution of social behaviour, I. J. Theor. Biol. 7: 1–16. [DOI] [PubMed] [Google Scholar]
- Hamilton, W. D., 1971. Selection of selfish and altruistic behaviour in some extreme models, pp. 59–91 in Man and Beast: Comparative Social Behavior, edited by J. Eisenberg and W. Dillon. Smithsonian Institution Press, Washington, DC.
- Hamilton, W. D., and R. M. May, 1977. Dispersal in stable habitats. Nature 269: 578–581. [Google Scholar]
- Hauert, C., and M. Doebeli, 2004. Spatial structure often inhibits the evolution of cooperation in the snowdrift game. Nature 428: 643–646. [DOI] [PubMed] [Google Scholar]
- Irwin, A. J., and P. D. Taylor, 2001. Evolution of altruism in stepping-stone populations with overlapping generations. Theor. Popul. Biol. 60: 315–325. [DOI] [PubMed] [Google Scholar]
- Karlin, S., and J. McGregor, 1968. The role of the Poisson progeny distribution in population genetic models. Math. Biosci. 2: 11–17. [Google Scholar]
- Karlin, S., and H. M. Taylor, 1981. A Second Course in Stochastic Processes. Academic Press, New York.
- Killingback, T., M. Doebeli and N. Knowlton, 1999. Variable investment, the continuous prisoner's dilemma, and the origin of cooperation. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266: 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, L., 2006. The evolution of trans-generational altruism: kin selection meets niche construction. J. Evol. Biol. 20: 181–189. [DOI] [PubMed] [Google Scholar]
- Lehmann, L., N. Perrin and F. Rousset, 2006. Population demography and the evolution of helping behaviors. Evolution 60: 1137–1151. [PubMed] [Google Scholar]
- Leimar, O., 2005. The evolution of phenotypic polymorphism: randomized strategies versus evolutionary branching. Am. Nat. 165: 669–681. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Nowak, M. A., and R. M. May, 1992. Evolutionary games and spatial chaos. Nature 359: 826–829. [Google Scholar]
- Perrin, N., and L. Lehmann, 2001. Is sociality driven by the costs of dispersal or the benefits of philopatry? A role for kin discrimination mechanisms. Am. Nat. 158: 471–483. [DOI] [PubMed] [Google Scholar]
- Perrin, N., and V. Mazalov, 2000. Local competition, inbreeding and the evolution of sex-biased dispersal. Am. Nat. 155: 116–127. [DOI] [PubMed] [Google Scholar]
- Proulx, S. R., 2000. The ESS under spatial variation with application to sex allocation. Theor. Popul. Biol. 58: 33–47. [DOI] [PubMed] [Google Scholar]
- Proulx, S. R., 2004. Sources of stochasticity in models of sex allocation in spatially structured populations. J. Evol. Biol. 17: 924–930. [DOI] [PubMed] [Google Scholar]
- Rousset, F., 2003. A minimal derivation of convergence stability measures. J. Theor. Biol. 221: 665–668. [DOI] [PubMed] [Google Scholar]
- Rousset, F., 2004. Genetic Structure and Selection in Subdivided Populations. Princeton University Press, Princeton, NJ.
- Rousset, F., 2006. Separation of time scales, fixation probabilities and convergence to evolutionarily stable states under isolation by distance. Theor. Popul. Biol. 69: 165–179. [DOI] [PubMed] [Google Scholar]
- Rousset, F., and S. Billiard, 2000. A theoretical basis for measures of kin selection in subdivided populations: finite populations and localized dispersal. J. Evol. Biol. 13: 814–825. [Google Scholar]
- Rousset, F., and O. Ronce, 2004. Inclusive fitness for traits affecting metapopulation demography. Theor. Popul. Biol. 65: 127–141. [DOI] [PubMed] [Google Scholar]
- Roze, D., and F. Rousset, 2003. Selection and drift in subdivided populations: a straightforward method for deriving diffusion approximations and applications involving dominance, selfing and local extinctions. Genetics 165: 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze, D., and F. Rousset, 2004. The robustness of Hamilton's rule with inbreeding and dominance: kin selection and fixation probabilities under partial sib mating. Am. Nat. 164: 214–231. [DOI] [PubMed] [Google Scholar]
- Seger, J., and H. J. Brockman, 1985. What is bet-hedging?, pp. 182–211 in Oxford Surveys in Evolutionary Biology, edited by R. Dawkins and M. Ridley. Oxford Unversity Press, Oxford.
- Shpak, M., 2005. Evolution of variance in offspring number: the effects of population size and migration. Theory Biosci. 124: 65–85. [DOI] [PubMed] [Google Scholar]
- Shpak, M., and S. R. Proulx, 2006. The role of life cycle and migration in selection for variance in offspring number. Bull. Math. Biol. (in press). [DOI] [PubMed]
- Taylor, P., 1990. Allele-frequency change in a class-structured population. Am. Nat. 135: 95–106. [Google Scholar]
- Taylor, P., and A. Sauer, 1980. The selective advantage of sex ratio homeostasis. Am. Nat. 116: 305–310. [Google Scholar]
- Taylor, P. D., 1988. An inclusive fitness model for dispersal of offspring. J. Theor. Biol. 130: 363–378. [Google Scholar]
- Taylor, P. D., 1992. a Altruism in viscous populations—an inclusive fitness model. Evol. Ecol. 6: 352–356. [Google Scholar]
- Taylor, P. D., 1992. b Inclusive fitness in a homogeneous environment. Proc. R. Soc. Lond. Ser. B Biol. Sci. 240: 299–302. [Google Scholar]
- Taylor, P. D., and S. A. Frank, 1996. How to make a kin selection model. J. Theor. Biol. 180: 27–37. [DOI] [PubMed] [Google Scholar]
- Taylor, P. D., and W. M. Getz, 1994. An inclusive fitness model for the evolutionary advantage of sibmating. Evol. Ecol. 8: 61–69. [Google Scholar]
- Taylor, P. D., and A. J. Irwin, 2000. Overlapping generations can promote altruistic behavior. Evolution 54: 1135–1141. [DOI] [PubMed] [Google Scholar]
- Verner, J., 1965. Selection for the sex ratio. Am. Nat. 99: 419–421. [Google Scholar]
- Wade, M. J., 1985. Soft selection, hard selection, kin selection, and group selection. Am. Nat. 125: 61–73. [Google Scholar]
- Whitlock, M. C., 2003. Fixation probability and time in subdivided populations. Genetics 164: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmot, G., 1986. Mixed compound Poisson distributions. Astin Bull. 16: 59–79. [Google Scholar]
- Wilson, D., G. Pollock and L. Dugatkin, 1992. Can altruism evolve in purely viscous populations. Evol. Ecol. 6: 331–341. [Google Scholar]