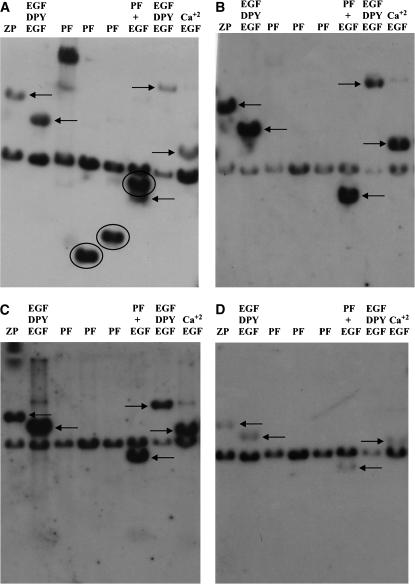
Figure 2.—
Reverse Southern analysis of dumpy gene evolution. Restriction enzyme fragments from the dumpy chromosomal walk (Wilkin et al. 2000) encoding different domains in the protein and subcloned in the plasmid vector, pBluescript (Stratagene), were subjected to electrophoresis and blotted to nylon filters as described in materials and methods. The subclones and their encoded domains are as follows: lane 1, subclone 13F encoding the zona pellucida domain; lane 2, subclone 5D encoding EGF–DPY–EGF modules; lanes 3–5, subclones 9A, 9B, and 9C encoding PF repeats; lane 6, subclone 26A encoding PF repeats and several adjacent EGF modules; lane 7, subclone 14A encoding EGF and DPY repeats; and lane 8, subclone 56D encoding predominately calcium-binding EGF domains. A filter was probed with radiolabeled genomic DNA from D. simulans (A), D. pseudoobscura (B), D. mettleri (C), and the Colorado potato beetle L. decemlineata (D). In each case, the 2.9-kb band containing the plasmid DNA hybridizes to contaminating bacterial DNA in the insect genomic DNA used as the probe. PF-containing sequences are detected only by D. simulans genomic DNA. Circled hybridizing bands contain PIGSFEAST repeats. Arrows point to hybridizing bands containing other parts of the dumpy gene indicated above each lane. Hybridizing bands, although very faint, are indicated in D by arrows. These specify, from left to right, the ZP domain (13F), EGF–DPY–EGF triplet motifs (5D and 26A), and calcium-binding EGF modules (56A).