Abstract
There are many types of DNA damage that are repaired by a multiplicity of different repair pathways. All damage and repair occur in the context of chromatin, and histone modifications are involved in many repair processes. We have analyzed the roles of H2A and its modifications in repair by mutagenizing modifiable residues in the N- and C-terminal tails of yeast H2A and by testing strains containing these mutations in multiple DNA repair assays. We show that residues in both tails are important for homologous recombination and nonhomologous end-joining pathways of double-strand break repair, as well as for survival of UV irradiation and oxidative damage. We show that H2A serine 122 is important for repair and/or survival in each of these assays. We also observe a complex pattern of H2A phosphorylation at residues S122, T126, and S129 in response to different damage conditions. We find that overlapping but nonidentical groups of H2A residues in both tails are involved in different pathways of repair. These data suggest the presence of a set of H2A “damage codes” in which distinct patterns of modifications on both tails of H2A may be used to identify specific types of damage or to promote specific repair pathways.
DNA is subject to a continuous assault of damaging agents. A number of factors can lead to the most serious form of DNA damage [breakage of both strands of the double helix, or double-strand breaks (DSBs)], including external agents such as ionizing radiation or chemical insult or internal mechanisms such as conversion of single-strand breaks during replication. Failure to repair DSBs can lead to gene deletion, chromosome loss or rearrangement, or cell death.
Repair of DSBs is accomplished by one of two general mechanisms. The break can be repaired via homologous recombination, usually by copying the intact information from a homologous chromosome or sister chromatid. Homologous recombination requires members of the RAD52 epistasis group; RAD52 itself is essential for all homologous recombination events in yeast (for recent reviews, see Aylon and Kupiec 2004; Dudas and Chovanec 2004; Haber et al. 2004). Alternatively, the two broken ends may be directly ligated, sometimes precisely and sometimes with loss of sequences at the breakpoint, through the process of nonhomologous end joining (NHEJ) (for reviews see Pastwa and Blasiak 2003; Aylon and Kupiec 2004; Dudasova et al. 2004). Homologous recombination is generally the preferred mechanism of DSB repair in yeast; if homologous sequences are available, the DSB will be repaired by homologous recombination 70–98% of the time, depending on mating type and other factors (Lee et al. 1999). However, in the absence of homologous sequences or in strains lacking RAD52, yeast cells can efficiently repair breaks using nonhomologous repair mechanisms. The yeast Ku proteins, yKu70p/Hdf1p and yKu80p, are DNA end-binding proteins central to NHEJ.
Both of these general mechanisms (as well as related pathways such as single-strand annealing) for repairing DSBs have been extensively studied in yeast, both genetically and biochemically (Holmes and Haber 1999; Haber 2000a,b; Haber et al. 2004; Krogh and Symington 2004; Sugawara and Haber 2006), and many additional proteins that are required for each pathway have been identified. Detection and repair of DNA damage also entails checkpoint activation; in yeast, the Mec1p and Tel1p kinases (orthologs of mammalian ATR/ATM) play critical roles in damage detection and signal transduction (Lowndes and Murguia 2000; Longhese et al. 2003; Qin and Li 2003; Harrison and Haber 2006), and these kinases are responsible for phosphorylation of histone H2A serine 129 (S129) at sites of damage (Downs et al. 2000).
In addition to DSBs, other types of DNA damage are recognized and repaired by specific factors, although different types of damage feed into common mechanisms for checkpoint activation. Damage caused by UV radiation, cyclobutane pyrimidine dimers, and (6-4) pyrimidone photoproducts is recognized and repaired by the nucleotide excision repair (NER) pathway, controlled by the RAD3 epistasis group in yeast (Prakash and Prakash 2000). Reactive oxygen species create numerous types of DNA damage, particularly DNA base modifications, apurinic/apyrimidinic sites, and single- or double-strand breaks. Most oxidative damage is repaired by the base excision repair (BER) system, although other pathways can come into play depending on the spectrum of damage (Doetsch et al. 2001; Salmon et al. 2004; Ikner and Shiozaki 2005).
DNA damage and repair occurs in the context of chromatin. Chromatin is made up of a basic repeating unit, the nucleosome, in which ∼147 bp of DNA is wrapped around a histone octamer (two copies each of histones H2A, H2B, H3, and H4). Chromatin structure plays a dynamic and central role in regulating transcription, DNA replication, repair, and recombination. A major means of regulating the structure of chromatin is through the covalent modification of histone proteins. Histone modifications can increase or decrease the higher-order folding of chromatin, and specific histone modifications present different docking surfaces for proteins interacting with the nucleosome. Many different histone modifications occur, including acetylation, phosphorylation, methylation, ubiquitylation, sumoylation, and ADP ribosylation.
Considerable recent efforts have focused on the roles of histone modification during DNA repair, particularly DSB repair. The importance of acetylation of histones H3 and H4 has been investigated. Mutation of all acetylatable lysines in histone H4 results in sensitivity to DSB-inducing agents, as do mutations in components of the NuA4 histone acetyltransferase complex (Bird et al. 2002; Choy and Kron 2002; Downs et al. 2004). Both acetylation of H4K8 (Downs et al. 2004) and deacetylation of H4K16 (Jazayeri et al. 2004) have been implicated in repair. Likewise, mutation of lysines 14 and 23 of H3 confer sensitivity to methyl methanesulfonate (MMS) or breaks induced by the EcoRI endonuclease (Qin and Parthun 2002), and deletion of the H3-specific Gcn5 acetyltransferase also causes sensitivity to double-strand breaks (Choy and Kron 2002). A recent thorough analysis mapped the acetylation levels for every lysine in the H3 and H4 tails during the formation and repair of a defined DSB (Tamburini and Tyler 2005), revealing a complex and dynamic pattern of acetylation resulting from the interplay of acetyltransferases and deacetylases recruited to DSBs. Another recent study has revealed the first indication of a role for H4 phosphorylation (on serine 1) in DSB repair (Cheung et al. 2005).
The first damage-specific histone modification to be identified was phosphorylation of H2A S129 (H2AX S139 in mammalian cells), which occurs immediately after the appearance of DSBs (Rogakou et al. 1999; Downs et al. 2000; Paull et al. 2000). Subsequent studies have revealed that H2A S129 phosphorylation is involved in the recruitment of chromatin-modifying complexes to DSBs, including the INO80 and Swr1 remodeling complexes (Downs et al. 2004; Morrison et al. 2004; van Attikum et al. 2004) and the NuA4 complex (Downs et al. 2004). One role of the INO80 complex appears to be the actual eviction of histones from the DSB to provide access for repair factors (Tsukuda et al. 2005).
Even though modification of multiple residues within a given histone is a common feature of modification, the importance of other residues of H2A in the repair process has only begun to be elucidated. Work in the Lustig laboratory has shown that a T126A mutation alone, or complete deletion of either the N- or the C-terminal tail of H2A, results in sensitivity to bleomycin and that T126A, K21E, and the N-terminal tail deletion each result in impaired NHEJ (Wyatt et al. 2003). Work by Harvey et al. (2005) showed that H2A S122 is important for survival in the presence of a number of DNA-damage-inducing agents, and these investigators also showed that the role of H2A S122 in DSB repair is distinct from the role of S129. These results are consistent with the data shown in this study, which suggest that H2A S122 may provide a general signal for multiple kinds of DNA damage.
To understand the contributions from both the N-terminal and the C-terminal tails of H2A in multiple DNA repair pathways, we systematically mutagenized both H2A tails. We show that individual residues in both the N- and the C-terminal tails of H2A play key roles in DNA repair, including DSB repair, UV repair, and survival of oxidative damage. Furthermore, the residues in the H2A tails required for these different functions fall into overlapping but nonidentical patterns, implicating certain H2A modifications as general signals of damage and/or stress, while others play roles in distinct repair pathways. Specifically, S122 of H2A is required for repair of DSBs by either the homologous recombination (HR) or NHEJ pathways, as well as for survival of UV irradiation and oxidative damage. In contrast, S129 is important for both pathways of DSB repair, but is not essential for survival of UV or oxidative stress, although it is phosphorylated in response to all of these stresses. Other residues have even more specific roles: T126 is important for HR but dispensable for NHEJ, while S2 and K127 are critical for NHEJ but have no role in HR. Finally, we show that S129, S122, and T126 exhibit complex patterns of phosphorylation and dephosphorylation in response to different DNA-damaging agents. These data indicate that not only the type of damage but also the selected repair pathway is marked by specific H2A modifications, creating a unique histone code for each type of damage and repair.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains:
All yeast strains used in this study are shown in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype—Plasmid |
|---|---|
| FY406a | MATa(hta1-htb1)Δ∷LEU2 (hta2-htb2)Δ∷TRP1 leu2Δ1 ura3-52 lys2Δ1 lys2-128δ his3Δ200 trp1Δ63—pSAB6 (HTA1-HTB1, URA3) |
| JKY38 | Same as FY406—pJKP18 (HTA1-HTB1, HIS3) |
| JKY41 | Same as FY406—pJKP33 (hta1-S2A-HTB1, HIS3) |
| JKY42 | Same as FY406—pJKP25 (hta1-K5A-HTB1, HIS3) |
| JKY43 | Same as FY406—pJKP26 (hta1-K8A-HTB1, HIS3) |
| JKY44 | Same as FY406—pJKP27 (hta1-S11A-HTB1, HIS3) |
| JKY45 | Same as FY406—pJKP28 (hta1-K14A-HTB1, HIS3) |
| JKY46 | Same as FY406—pJKP29 (hta1-S16A-HTB1, HIS3) |
| JKY47 | Same as FY406—pJKP30 hta1-S18A-HTB1, HIS3) |
| JKY71 | Same as FY406—pJKP34 (hta1-K22Q-HTB1, HIS3) |
| JKY78 | Same as FY406—pJKP67 (hta1-K22R-HTB1, HIS3) |
| FY986a | Same as FY406—pJH161 (hta1Δ4-20-HTB1, HIS3) |
| JKY31 | Same as FY406—pJKP20 (hta1-K120A-HTB1, HIS3) |
| JKY32 | Same as FY406—pJKP21 (hta1-K121A-HTB1, HIS3) |
| JKY33 | Same as FY406—pJKP22 (hta1-S122A-HTB1, HIS3) |
| JKY72 | Same as FY406—pJKP35 (hta1-S122D-HTB1, HIS3) |
| JKY34 | Same as FY406—pJKP23 (hta1-K124A-HTB1, HIS3) |
| JKY35 | Same as FY406—pJKP24 (hta1-T126A-HTB1, HIS3) |
| JKY30 | Same as FY406—pJKP19 (hta1-K127A-HTB1, HIS3) |
| JKY73 | Same as FY406—pJKP36 (hta1-K127Q-HTB1, HIS3) |
| FHY3b | Same as FY406—pJD151 (hta1-S129A-HTB1, HIS3) |
| JKY74 | Same as FY406—pJKP37 (hta1-S122A,K127A-HTB1, HIS3) |
| JKY75 | Same as FY406—pJKP38 (hta1-S122A,S129A-HTB1, HIS3) |
| JKY112c | Same as FY406—p(hta1-T126A,S129A-HTB1, HIS3) |
| JKY79 | Same as FY406—pJKP68 (hta1-K127A,S129A-HTB1, HIS3) |
| JKY80 | Same as FY406—pJKP70 (hta1-S122A,K127A,S129A-HTB1, HIS3) |
| JKY81 | Same as FY406—pJKP69 (hta1-S122A,T126A,S129A-HTB1, HIS3) |
| T69c | Same as FY406—pJH69 (hta1-K120*-HTB1, HIS3) |
| JKY3d | Δho Δhml:ADE1 Δhmr:ADE1 lys5 leu2,3-112 trp1∷hisG ura3-52 ade3∷GAL10:HO Δrad51∷LEU2 |
| JKY5d | Δho Δhml:ADE1 Δhmr:ADE1 lys5 leu2,3-112 trp1∷hisG ura3-52 ade3∷GAL10:HO Δrad52∷TRP1 |
| JKY36 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δku70∷kanMX |
| JKY121 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δsod1∷kanMX |
| JKY130 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δrad4∷kanMX |
Asterisk indicates stop codon.
Moore and Haber (1996).
Plasmids and site-directed mutagenesis:
Plasmid JKP18 was created by inserting a BamHI/SacII fragment from pAB6 (Hirschhorn et al. 1995), containing the HTA1-HTB1 locus, into the plasmid pRS413 (HIS3) cut with the same restriction enzymes. Mutagenic oligonucleotides were designed to contain the altered nucleotides necessary for a single amino acid substitution flanked by sequence complementary to the plasmid template. Mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Mutagenized plasmids were confirmed by sequencing and transferred into JKY29 (which contains the wild-type HTA1-HTB1 locus on a URA3 plasmid) using a plasmid shuffle. Mutation-containing plasmids are transformed into JKY29 and plated on His− media. His+ colonies are then plated on media containing 5-FOA to select for cells that have lost the URA3-containing wild-type plasmid. Numbering of amino acids in H2A follows the convention of counting the initial methionine as position 1, even though this methionine is cleaved post-translationally.
MMS and bleomycin sensitivity:
The double-strand break-inducing agents MMS and bleomycin were added to YPD agar as it cooled to final concentrations of 0.02% and 15 units/liter, respectively. Logarithmically growing cells were diluted to an OD600 of 0.2 and 5- or 10-fold serial dilutions were plated on YPD and either MMS- or bleomycin-containing media and then incubated for 2–3 days at 30°.
Plasmid end-joining assay:
The plasmid pRS413 (URA3) was linearized with SmaI, which generates a blunt end. Equal numbers of competent cells are transformed with equal concentrations of either linear or circular plasmid. Following transformation with linear plasmid, the cell must repair the URA3-containing plasmid to survive subsequent plating on Ura− media. After plating onto Ura− media, each strain's repair efficiency is assayed quantitatively, relative to the isogenic wild type, by counting colonies that survive on Ura− media. Circular plasmid is also transformed and assayed to assess the overall transformation efficiency. Each mutant is assayed a minimum of three times in triplicate and the averaged results and standard errors are reported.
UV sensitivity:
Midlog cultures were diluted to an OD600 of 0.2 and 10-fold serial dilutions were spotted on YPD plates. Five duplicate sets of plated cells were exposed to doses of 0, 100, 150, 200, or 250 J/m2 UV irradiation in a Stratalinker (Stratagene) and incubated at 30° for 3–4 days.
Oxidative damage sensitivity:
Cultures were grown to an OD600 of 0.5 and menadione was added to a final concentration of 2 mm. Cell growth with and without menadione was monitored by OD600 from 0 to 210 min after menadione addition. The effect of menadione on cell growth is expressed as the ratio of the difference in OD600 in the presence vs. absence of menadione at each time point. Each strain was assayed a minimum of three times, and the average results and standard errors are presented.
Antibodies and Western blot analysis:
Early log-phase cultures (OD600 ∼ 0.2) were treated with 0.1% MMS, 5 μg/ml phleomycin, or 10 mm menadione for 6 hr. For UV treatment, cells were spread onto YPD plates, exposed to UV (200 J/m2) in a Stratalinker, incubated for 2 hr at 30°, and collected by rinsing the surface of the plates with water. Yeast extracts were prepared by boiling cells in SDS sample buffer for 8 min and were separated on 15% polyacrylamide–SDS gels (Nextgel, Amresco M258). Proteins were transferred onto nitrocellulose membrane and blocked with Licor blocking buffer (Licor Biosciences). Primary antibodies were diluted in Licor blocking buffer as indicated: anti-H2AphosphoS122 (Abcam ab3599) 1:250; rabbit monoclonal anti-unmodified H4 (Upstate 05-858) 1:5,000; anti-H2A phosphoT126 (Abcam ab3598) 1:500; or anti-H2AphosphoS129 (Aves Labs) 1:30,000. Infrared dye-conjugated secondary antibodies (IR-680 anti-rabbit from Molecular Probes, and IR-800 anti-chicken from Licor Biosciences) were used to detect the signal using an Odyssey Infrared Imaging System.
RESULTS
Both H2A N- and C-terminal tails are required for efficient nonhomologous end joining:
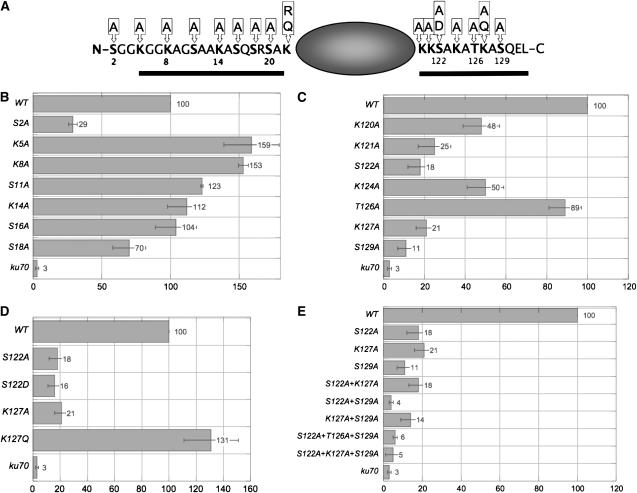
To test whether H2A residues other than the previously characterized S129 (Downs et al. 2000) are involved in DSB repair, we systematically mutagenized all the potentially modifiable residues in both the N-terminal and the C-terminal tails of yeast H2A. We initially created a series of single alanine substitutions of all lysine, serine, and threonine residues in both tails. Subsequently, we made double- and triple-mutant combinations of residues of particular interest and also created mutations intended to mimic modified or unmodified states; specifically, aspartate was used to mimic phosphorylated serine, and arginine and glutamine were used to mimic unacetylated and acetylated lysine, respectively. Figure 1A shows a schematic of H2A in which all the mutagenized positions are indicated, as well as the mutations made for each position. We also obtained N- or C-terminal tail deletions of H2A (Hirschhorn et al. 1995); the deleted regions are indicated by solid bars in Figure 1A.
Figure 1.—
Both the N- and C-terminal tails of H2A are involved in nonhomologous end joining. (A) Schematic summarizing the H2A mutations used in this study. Bars indicate the deleted regions in the ΔN and ΔC mutants. (B–E) Indicated strains were transformed with circular or linear URA3-containing plasmids. Repair efficiency of linear plasmids is indicated by the number of successful transformants on Ura− media. Transformation efficiency relative to the isogenic wild type is graphed; all values represent averages of a minimum of three independent assays. Standard errors are shown.
We were interested in whether distinct modifications of H2A might distinguish the two major pathways of DSB repair, NHEJ and HR. We therefore tested our H2A mutant strains in assays specific for either pathway, first using a plasmid end-joining assay that measures NHEJ. For this assay, URA3-containing plasmids were linearized with SmaI, which produces blunt ends, and linear plasmids were transformed into cells, which were then plated onto Ura− media. Cells must successfully repair the linear plasmids to grow; repair efficiency is read out by the number of colonies formed after transformation, relative to wild type.
Specific mutations in both the N- and the C-terminal tails of H2A resulted in significant reduction in repair efficiency by NHEJ. While most mutations in the N terminus resulted in either wild type (or better) levels of repair, mutation of the first serine of H2A, S2, reduced end joining to <30% of wild-type levels (Figure 1B). Even more severe phenotypes were observed in the C-terminal tail, in which alanine substitutions at K121A, S122, K127, and S129 resulted in repair efficiencies of only 25, 18, 21, and 11%, respectively, of wild-type levels (Figure 1C). Other mutations in the C terminus had mild (K120A, K124A) or no (T126A) effect on end joining. While hta1-S129A mutants have been previously shown to have an end-joining defect (Downs et al. 2000), another study failed to observe this defect and instead saw an end-joining defect for hta1-T126A mutants (Wyatt et al. 2003). The Wyatt et al. study used plasmids with 4-base 3′ overhangs rather than blunt ends. However, when we repeated our study using plasmids digested with BamHI, we still did not observe an end-joining defect for the hta1-T126A mutant (D. Robinson and J. E. Krebs, unpublished results).
We next wanted to address whether the requirement for specific H2A residues in end joining reflects a role for modification of these residues. To test this, we selected the two residues with the severest phenotypes when mutated, S122 and K127 (S129 was not chosen, as it was already known to be phosphorylated) and created mutations intended to mimic potential modified states of these residues. The resulting mutant strains, hta1-S122D (mimicking phospho-serine) and hta1-K127Q (mimicking acetyl-lysine) were tested in the end-joining assay. As shown in Figure 1D, mutation of K127 to a neutral glutamine results in complete rescue of the NHEJ phenotype of the K127A mutation. This supports the idea that acetylation of this lysine is necessary for efficient NHEJ. In contrast, the S122D mutation has a phenotype indistinguishable from S122A, indicating either that this serine is not phosphorylated or that aspartate is not an adequate mimic of phospho-serine in this context. We believe that the latter is the more likely explanation, as S122 was previously shown to be phosphorylated in vivo (Wyatt et al. 2003), and we are able to detect S122 phosphorylation by Western blotting (see Figure 5 and discussion below).
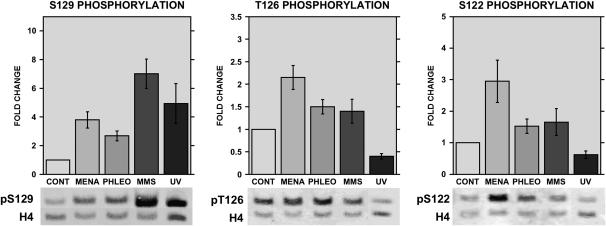
Figure 5.—
Complex patterns of H2A phosphorylation result from different DNA-damaging treatments. Western blot analysis of wild-type cells either untreated (CONT) or treated with 2 mm menadione (MENA), 5 μg/ml phleomycin (PHLEO), 0.1% MMS (MMS), or 200 J/m2 UV irradiation (UV). Blots were probed using antisera recognizing histone H2A phosphorylated at positions 129 (left), 126 (middle), or 122 (right). The levels of unmodified histone H4 were also detected as a loading control. Bar graphs represent average values and standard deviations for quantitation of four to five blots; representative blots are shown below.
We also tested the effects of combining specific H2A mutations in NHEJ (Figure 1E). We constructed double mutants between the three most impaired single mutations in this assay: S122A+K127A, S122A+S129A, and K127A+S129A. Combining S122A+K127A did not create a phenotype any more severe than either single mutation, suggesting that these residues may serve similar functions in NHEJ. The NHEJ phenotype of the hta1-K127A,S129A strain (14% of wild type) is not significantly different from the values for the single mutants (21% for K127A and 11% for S129A). However, the hta1-S122A,S129A strain was more severely impaired in end joining than either single mutant alone; in fact, this strain was as deficient as ku70Δ or ku80Δ strains in this assay. This indicates that S122 and S129 play nonredundant roles in NHEJ, as in other repair pathways (Harvey et al. 2005).
Unsurprisingly, addition of the K127A mutation to the S122A+S129A double mutant did not further enhance the end-joining phenotype, as end joining is virtually eliminated in the hta1-S122A,S129A strain already. Addition of the T126A mutation also fails to either enhance or rescue the effect of S122A+S129A.
Multiple residues in the H2A C-terminal tail are involved in survival of chemically induced double-strand breaks:
We next tested the ability of strains containing H2A mutations to survive in the presence of the DSB-inducing agents MMS and bleomycin. Both of these drugs create predominantly double-strand breaks at the concentrations used (0.02% MMS and 15 units/liter bleomycin), and these breaks are repaired primarily via homologous recombination. We confirmed this by also assaying strains deficient in homologous recombination (rad51Δ, rad52Δ, rad54Δ) and strains deficient in nonhomologous end joining (ku70Δ, ku80Δ). As expected, the rad mutant strains are hypersensitive to MMS and bleomycin, while ku deletion strains are only modestly affected (Figure 2 and data not shown).
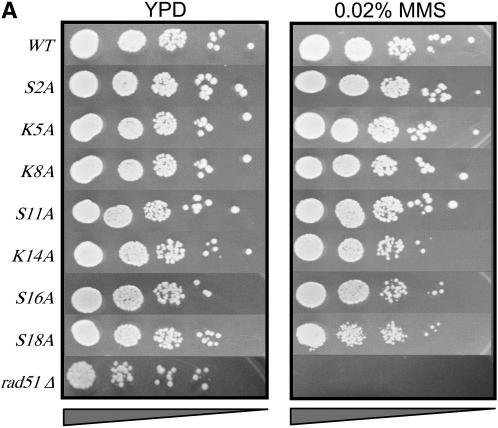
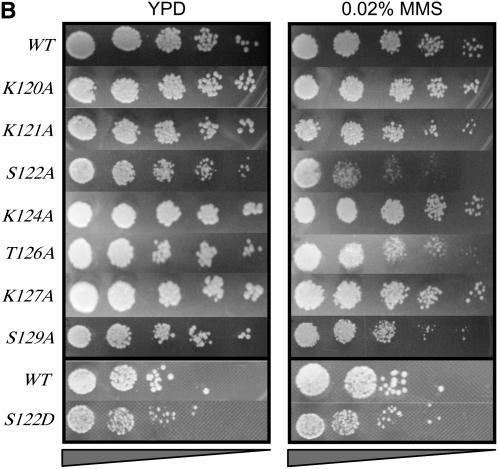
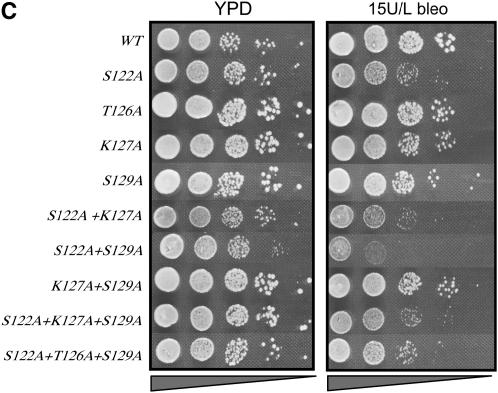
Figure 2.—
Multiple phosphorylatable residues in H2A are required for survival on MMS or bleomycin. (A) Survival in the presence of MMS of strains containing single alanine substitutions in the H2A N-terminal tail. (B) Survival in the presence of MMS of strains containing various substitutions in the H2A C-terminal tail. (C) Survival in the presence of bleomycin for selected strains containing H2A C-terminal mutations. Serial dilutions of the indicated strains were plated on YPD (left in A–C), YPD + 0.02% MMS (right in B and C), or YPD + 15 units/liter bleomycin (right, C). All survival assays were repeated a minimum of three times; representative plates are shown.
When we assayed the survival of single alanine substitutions in the H2A N terminus, we observed little to no effect of MMS (Figure 2A). The strains hta1-K14A, hta1-S16A, and hta1-S18A showed very mild reductions in growth. Strains containing a deletion of H2A from K5 to A21 (“ΔN”) are sensitive to bleomycin (Wyatt et al. 2003); this suggests that this region may indeed be important for DSB repair, but no single residue (or its modification) is key.
In contrast, several residues in the C terminus, when changed to alanine, exhibit a growth phenotype on MMS (Figure 2B). The strongest phenotype is observed for hta1-S122A strains, consistent with a recent report (Harvey et al. 2005). We also observe a mild MMS phenotype for hta1-T126A strains and the expected hta1-S129A phenotype (Downs et al. 2000). Intriguingly, while both T126 and S129 appear to be important for survival on MMS, neither appears to be required for growth on bleomycin (Figure 2C). S122, however, is needed for both. The reasons for these differences are not clear, but may reflect the different spectrum of DNA damage caused by MMS vs. bleomycin. MMS, a methylating agent, can cause abasic sites and single-strand breaks and may generate mostly double-strand breaks by conversion of single-strand breaks during replication (which are preferentially repaired by HR using the sister chromatid). On the other hand, the radiomimetic drug bleomycin causes free radical attacks on the deoxyribose sugar on both strands cooperatively, resulting in double-strand breaks with either blunt ends or 1-base overhangs (as well as some abasic sites).
We also tested combinations of mutations on both MMS and bleomycin. As was the case for NHEJ, the hta1-S122A,S129A strain showed a more severe phenotype on both MMS and bleomycin than either single mutant alone (Figure 2C and data not shown; also see Harvey et al. 2005), consistent with these residues performing different functions in repair of this DNA damage.
Both N- and C-terminal H2A tails contribute to survival of UV irradiation:
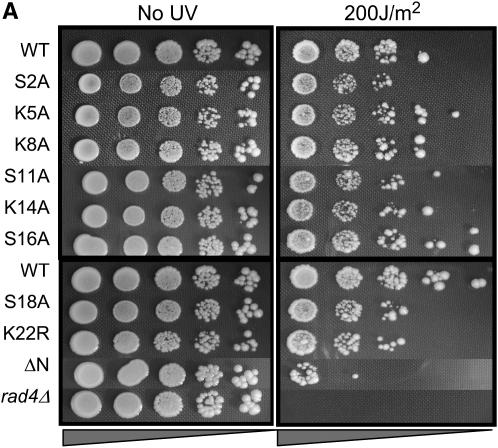
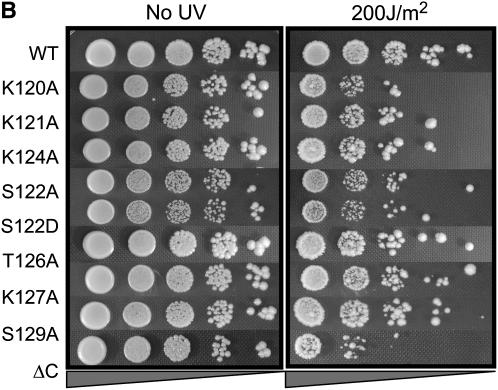
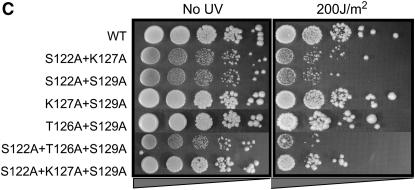
The results presented in the previous sections indicate that different pathways of DSB repair require different patterns of modification/binding sites on histone H2A. We wanted to ask whether the requirements for different specific residues of H2A extended to other types of DNA damage. Accordingly, we subjected the different hta1 mutant strains to varying doses of UV irradiation. In these experiments, midlog cultures of each strain were diluted to the same concentration and plated in 10-fold serial dilutions on YPD. Open plates were then subjected to doses of 0, 100, 150, 200, or 250 J/m2 UV and growth was monitored at 30°. The results for only the 200 J/m2 dose are shown for simplicity (Figure 3; results for exposures at 150 and 250 J/m2, bracketing the dose shown in Figure 3, can be found in supplemental Figure S1 at http://www.genetics.org/supplemental/). As a control for UV doses, we included a strain deleted for RAD4 (the homolog of human XPC); Rad4p is involved in recognition of UV-induced lesions and is required for NER in yeast.
Figure 3.—
Both N- and C-terminal mutations in H2A render cells sensitive to UV radiation. (A–C) Serial dilutions of the indicated strains were plated on YPD and either left untreated (left) or subjected to varying doses of UV radiation (right). The 200 J/m2 dose is shown. All survival assays were repeated a minimum of three times; representative plates are shown.
Figure 3 shows that residues in both tails of H2A are important for survival of UV irradiation and that these important residues overlap with, but are not identical to, those required for MMS/bleomycin survival or NHEJ. In the N terminus, survival of strains containing the S2A mutation is moderately impaired, and strains containing the S18A mutation exhibit significantly reduced survival after UV treatment (Figure 3A). The hta1ΔN strain is even more severely sensitive to UV damage.
In the C terminus, the hta1-S122A strain also exhibits reduced survival after UV treatment, consistent with a universal role for S122 in damage signaling or repair (Figure 3B). This phenotype is not rescued by an S122D mutation. We also observe impaired survival for the hta1-K120A strain. Other than S122, neither of the other phosphorylatable residues, T126 and S129, is important for survival of UV irradiation. Furthermore, addition of K127A, S129A, or both K127A+S129A mutations to the S122A mutant does not alter the phenotype of S122A alone (Figure 3C). However, the hta1-S122A,T126A,S129A triple mutant has reduced survival after UV treatment compared to hta1-S122A or hta1-S122A,S129A. One explanation for this may be that while S122 phosphorylation or dephosphorylation is normally involved in UV damage recognition or repair, modification of T126 may partially substitute for the function of S122 in the absence of a serine at this position.
H2A S122 is also required in the presence of oxidative damage:
Oxidative stress, produced by intracellular reactive oxygen species, is a ubiquitous source of cellular damage, which can cause injury to most cellular macromolecules, including an array of types of DNA damage. We used the reactive quinone menadione to generate reactive oxygen species in cells. Menadione redox cycling results in the generation of hydroxyl radicals, which create predominantly base oxidation products and single-strand breaks in DNA. The BER pathway is believed to be the major repair pathway required for survival of oxidative DNA damage (Memisoglu and Samson 2000).
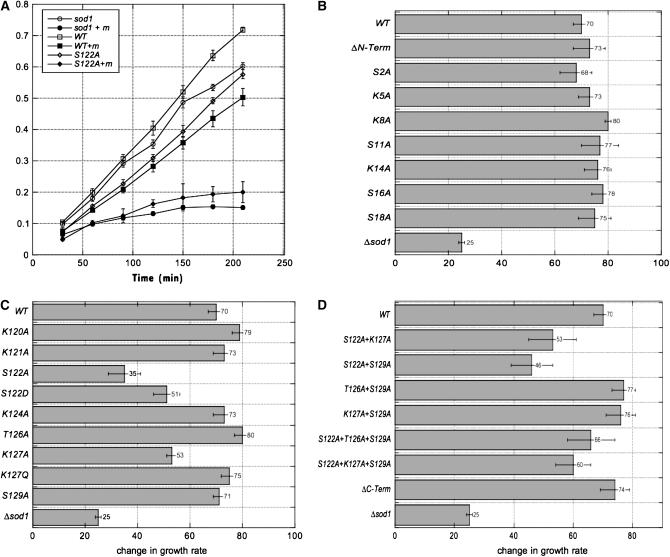
We measured the effects of menadione-induced oxidative damage on the growth of strains bearing hta1 mutations (Figure 4). We initially tested a range of menadione concentrations and chose a 2-mm final concentration of menadione, which has only moderate effects on the growth of wild-type cells, but arrests the growth of sod1Δ mutants (Figure 4A). SOD1 codes for a Cu/Zn-superoxide dismutase and is critical for protecting cells from oxidant toxicity (Jamieson 1998). We compared the growth of hta1 mutant strains over time in the presence or absence of menadione (Figure 4A and data not shown). For simplicity, we report the results for only the endpoints of these time courses for most strains.
Figure 4.—
Strains with mutations in the H2A C terminus are sensitive to menadione. (A) Growth (change in OD600) over time for selected strains in the presence (solid symbols) or absence (open symbols) of 2 mm menadione. Only wild type (squares), hta-S122A (diamonds), and sod1Δ (circles) are shown for simplicity. (B–D) Relative growth in the presence vs. absence of 2 mm menadione at 210 min after menadione addition. All values represent averages of a minimum of three independent assays and standard errors are shown.
The N terminus of H2A is entirely dispensable for survival of oxidative stress; as shown in Figure 4B, neither the hta1-ΔN mutant nor any of the N-terminal single mutants exhibit any impairment in comparison to wild type in the presence of menadione. On the other hand, both the S122A and K127A mutations in the C terminus result in sensitivity to menadione (Figure 4, A and C), with the hta1-S122A strain nearly as impaired as the sod1Δ mutant. An aspartate in position 122 partially rescues the S122A phenotype, and the K127Q mutation completely restores wild-type growth (Figure 4C), suggesting that S122 phosphorylation and K127 acetylation per se are important in the response to oxidative damage.
Surprisingly, mutation of other residues in combination with the S122A mutation, as well as complete deletion of the C terminus, partially or fully rescues the S122A defect (Figure 4D). One possible explanation for this result is that other residues in the H2A C terminus actually play opposing roles to that of S122 in the response to oxidative stress; similar complex interactions within the H2A C terminus have been observed previously for transcriptional silencing at telomeres (Wyatt et al. 2003). Alternatively, the different C-terminal residues could be important for different aspects of the oxidative stress response; for example, S122 could be involved in signaling the BER pathway, while other mutations such as hta1-ΔC could affect transcription of critical factors in the oxidative stress response, such as SOD1 itself. The hta1-S122A strain exhibits normal induction of SOD1 transcription (data not shown); possible overexpression or derepression of SOD1 in other hta1 mutants has not yet been tested. Experiments to explore genetic interactions between hta1 mutants and mutations in the BER pathway are also underway.
Complex H2A phosphorylation in response to DNA damage:
Because H2A S129 is a known target of Mec1p-dependent phosphorylation in response to DNA damage (Downs et al. 2000), and it has been shown that H2A S122 and T126 are phosphorylated in vivo in the absence of damage (Wyatt et al. 2003), we wanted to directly test whether these H2A residues are also specifically modified in response to damage. We therefore generated antibodies against modified peptides containing phosphorylated S122, T126, or S129. All of these antibodies were affinity purified using the relevant phosphopeptides and were also subtracted against the unphosphorylated peptides to remove antibodies not specific for the modified target. Finally, each antibody was tested against the peptides phosphorylated at the other residues to ensure that there was no cross-reaction between these nearby modifications. Data showing the specificity of these antibodies are shown in supplemental Figure S2 at http://www.genetics.org/supplemental/.
Figure 5 shows Western blot analysis testing the effects of MMS, phleomycin, UV, and menadione treatment on phosphorylation of S122, T126, and S129 of H2A. S129 is phosophorylated in response to all four damaging treatments, exhibiting a threefold increase in phosphorylation levels in response to phleomycin (5 μg/ml), fourfold increases in response to UV (200 J/m2) or menadione (10 mm) treatment, and a sevenfold increase in signal after treatment with 0.1% MMS (consistent with other published results). Phosphorylation patterns for T126 and S122 are quite different, however. T126 shows a 50% increase in phosphorylation in response to phleomycin and MMS, while S122 phosphorylation increases by only a small but consistent 20% in response to phleomycin or MMS, even though the S122A mutant exhibits the strongest growth defect in the presence of these drugs (Figure 2). The highest phosphorylation levels at both T126 and S122 (twofold and threefold, respectively) are observed in the presence of 10 mm menadione. Again, these phosphorylation patterns do not necessarily correlate with the requirements of these residues for survival of oxidative damage: while the S122A mutant has a significant growth defect in menadione, T126A has no defect at this level of menadione treatment (Figure 4). This does not necessarily rule out a role for T126 at higher levels of oxidative stress, however.
Most surprisingly, both T126 and S122 phosphorylation levels decrease upon exposure to UV, in contrast to the increased phosphorylation at S129. This appears to be a true reduction in S122 and T126 phosphorylation, as overall levels of H2A do not change with these treatments (data not shown), and S129 phosphorylation clearly increases in these same samples.
These results indicate a complex pattern of phosphorylation events in the H2A C terminus in response to different classes of DNA damage: significant phosphorylation at all three residues in the presence of oxidative stress, high levels of S129 phosphorylation accompanied by moderate-to-low levels of T126 and S122 phosphorylation in the presence of MMS, and S129 phosphorylation contrasted with reduced phosphorylation levels of T126 and S122 after UV treatment. These analyses reveal individual phosphorylation events; however, they do not indicate the occurrence of specific combinations of modifications on individual histone tails. Future experiments using two-dimensional gel separation of H2A isoforms and the development of antibodies against multiply modified tails will further our understanding of the complex interplay of H2A tail modifications.
DISCUSSION
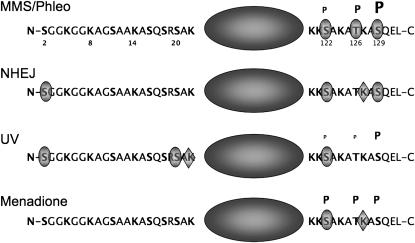
We have shown that histone H2A plays a central role in the survival of multiple forms of DNA damage. Different residues (or modification of different residues) are important for distinct repair pathways, indicating that different patterns of modification of H2A may distinguish forms of DNA damage or target different repair machineries appropriate to the specific damage. We also observe, consistent with results from the Downs laboratory (Harvey et al. 2005), that H2A S122 is required for the responses to all the diverse forms of DNA damage tested. This suggests that S122 and its dynamic phosphorylation or dephosphorylation may represent a general signal of DNA damage, while modification of other residues in both H2A tails serves to identify the specific type of damage, ensuring the recruitment of the correct repair machinery. Figure 6 summarizes the roles of different H2A residues in the response to various damage conditions.
Figure 6.—
Summary of H2A residues important for different DNA damage responses. Schematics of H2A indicating the residues shown to be important for the four general types of damage and repair discussed in this work. Shaded ovals indicate phosphorylatable residues shown to be important; shaded diamonds indicate important lysines. Residues known to be phosphorylated are indicated with a “P,” and the size of the “P” indicates the relative level of phosphorylation under the conditions shown. See text for detailed discussion. Note that the phosphorylation state of H2A has not been mapped during NHEJ, so no phosphorylation levels are indicated.
We have also demonstrated that all three phosphorylatable residues in the H2A C terminus are in fact phosphorylated and/or dephosphorylated in response to DNA damage; we also intend to determine whether important serine residues in the N terminus are similarly modified. The kinases that may target these residues are not known, although in Drosophila H2A a threonine equivalent to S122 (T119) has been shown to be phosphorylated during mitosis by nucleosomal histone kinase-1 (NHK1), and a partially purified kinase from yeast, a presumed NHK1 homolog, can also phosphorylate Drosophila H2A at this position (Aihara et al. 2004). It would be interesting to know whether DmH2A T119 plays any role in repair; the Drosophila H2A variant that contains the equivalent of S129 (DmH2Av S137) does not have a serine or threonine in a position analogous to S122 in ScH2A, but DmH2Av has been clearly implicated in DSB repair (Madigan et al. 2002; Kusch et al. 2004). Perhaps different types of damage will require functions of different H2A variants in Drosophila.
It is very likely that H2A K127 is acetylated in response to certain types of damage on the basis of the results from the K127Q mutation. Given the importance of NuA4 in repair (Bird et al. 2002; Downs et al. 2004; Utley et al. 2005), it is possible that this residue is a substrate for the Esa1p HAT. Unfortunately, we have thus far been unable to generate an antibody specific for K127 acetylation in vivo. Given the proximity of relevant residues in the H2A C terminus, it would not be surprising if modification of multiple residues in the same tail could perturb the interaction of antibodies generated against single modifications. Antibodies raised against multiply modified peptides may resolve some of these questions.
What are the actual roles of H2A modifications/specific residues in repair? H2A could facilitate repair processes at numerous levels. Specific modifications could recruit other chromatin-modifying activities, resulting in histone exchange, nucleosome remodeling, or nucleosome displacement, as has been shown to occur at double-strand breaks in response to H2A S129 phosphorylation (Downs et al. 2004; Morrison et al. 2004; van Attikum et al. 2004; Tsukuda et al. 2005). Different modifications could recruit or stabilize binding of proteins involved in repair itself or specifically exclude incorrect repair factors to facilitate repair pathway choice. Phosphorylation of S139 in human H2AX (equivalent to ScH2A S129) has been shown to increase retention of repair factors at DSBs (Paull et al. 2000; Celeste et al. 2003), although there may be only partial overlap of S129 phosphorylation and repair proteins (Rad51 and Mre11) at a DSB in yeast (Shroff et al. 2004). In fission yeast, S129 phosphorylation is required for recruitment of the checkpoint protein Crb2 to DSBs and for checkpoint maintenance (Nakamura et al. 2004), although neither S129 nor S122 appear to be required for checkpoint activation in budding yeast (Downs et al. 2000; Redon et al. 2003; Harvey et al. 2005).
In addition to potential roles in the physical repair process or in checkpoint functions, some H2A modifications instead could be involved in transcription of factors required for repair or other protection from damage. H2A and its potential modifications have been previously implicated in transcription of a number of different genes (Hirschhorn et al. 1995; Recht et al. 1996; Wyatt et al. 2003; Wang et al. 2004; Zhang et al. 2004; Kuo et al. 2005), but the effects of H2A mutations on expression of genes relevant to different repair pathways has not been explored. We have shown that H2A, particularly S122, is required for the normal transcriptional response to toxic copper levels (Kuo et al. 2005) and that specific H2A residues are also important for the normal transcriptional response to heat shock (S. Uffenbeck and J. E. Krebs, unpublished results). This suggests that S122 (or its phosphorylation levels) not only may be generally required for DNA damage responses, but also may be a universal signal for many types of cellular stress.
Acknowledgments
The authors thank Jessica Downs for yeast strains and plasmids and Art Lustig for strains and helpful discussions. This work was supported by National Science Foundation awards MCB-0315816 and EPS-0346770.
References
- Aihara, H., T. Nakagawa, K. Yasui, T. Ohta, S. Hirose et al., 2004. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 18: 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon, Y., and M. Kupiec, 2004. DSB repair: the yeast paradigm. DNA Repair 3: 797–815. [DOI] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt et al., 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5: 675–679. [DOI] [PubMed] [Google Scholar]
- Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn et al., 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15: 656–660. [DOI] [PubMed] [Google Scholar]
- Choy, J. S., and S. J. Kron, 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22: 8215–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch, P. W., N. J. Morey, R. L. Swanson and S. Jinks-Robertson, 2001. Yeast base excision repair: interconnections and networks. Prog. Nucleic Acid Res. Mol. Biol. 68: 29–39. [DOI] [PubMed] [Google Scholar]
- Downs, J. A., N. F. Lowndes and S. P. Jackson, 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger et al., 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16: 979–990. [DOI] [PubMed] [Google Scholar]
- Dudas, A., and M. Chovanec, 2004. DNA double-strand break repair by homologous recombination. Mutat. Res. 566: 131–167. [DOI] [PubMed] [Google Scholar]
- Dudasova, Z., A. Dudas and M. Chovanec, 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28: 581–601. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., 2000. a Lucky breaks: analysis of recombination in Saccharomyces. Mutat. Res. 451: 53–69. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., 2000. b Partners and pathways repairing a double-strand break. Trends Genet. 16: 259–264. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., G. Ira, A. Malkova and N. Sugawara, 2004. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. C., and J. E. Haber, 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Harvey, A. C., S. P. Jackson and J. A. Downs, 2005. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics 170: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse and F. Winston, 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15: 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, A., and J. E. Haber, 1999. Physical monitoring of HO-induced homologous recombination. Methods Mol. Biol. 113: 403–415. [DOI] [PubMed] [Google Scholar]
- Ikner, A., and K. Shiozaki, 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 569: 13–27. [DOI] [PubMed] [Google Scholar]
- Jamieson, D. J., 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14: 1511–1527. [DOI] [PubMed] [Google Scholar]
- Jazayeri, A., A. D. McAinsh and S. P. Jackson, 2004. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101: 1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, B. O., and L. S. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Kuo, H. C., J. D. Moore and J. E. Krebs, 2005. Histone H2A and Spt10 cooperate to regulate induction and autoregulation of the CUP1 metallothionein. J. Biol. Chem. 280: 104–111. [DOI] [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser et al., 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., F. Paques, J. Sylvan and J. E. Haber, 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9: 767–770. [DOI] [PubMed] [Google Scholar]
- Longhese, M. P., M. Clerici and G. Lucchini, 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532: 41–58. [DOI] [PubMed] [Google Scholar]
- Lowndes, N. F., and J. R. Murguia, 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10: 17–25. [DOI] [PubMed] [Google Scholar]
- Madigan, J. P., H. L. Chotkowski and R. L. Glaser, 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30: 3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu, A., and L. Samson, 2000. Base excision repair in yeast and mammals. Mutat. Res. 451: 39–51. [DOI] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt et al., 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- Nakamura, T. M., L. L. Du, C. Redon and P. Russell, 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24: 6215–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastwa, E., and J. Blasiak, 2003. Non-homologous DNA end joining. Acta Biochim. Pol. 50: 891–908. [PubMed] [Google Scholar]
- Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert et al., 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10: 886–895. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13–24. [DOI] [PubMed] [Google Scholar]
- Qin, J., and L. Li, 2003. Molecular anatomy of the DNA damage and replication checkpoints. Radiat. Res. 159: 139–148. [DOI] [PubMed] [Google Scholar]
- Qin, S., and M. R. Parthun, 2002. Histone h3 and the histone acetyltransferase hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22: 8353–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht, J., B. Dunn, A. Raff and M. A. Osley, 1996. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon, C., D. R. Pilch, E. P. Rogakou, A. H. Orr, N. F. Lowndes et al., 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou, E. P., C. Boon, C. Redon and W. M. Bonner, 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, T. B., B. A. Evert, B. Song and P. W. Doetsch, 2004. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 32: 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner et al., 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14: 1703–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., and J. E. Haber, 2006. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 408: 416–429. [DOI] [PubMed] [Google Scholar]
- Tamburini, B. A., and J. K. Tyler, 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25: 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda, T., A. B. Fleming, J. A. Nickoloff and M. A. Osley, 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley, R. T., N. Lacoste, O. Jobin-Robitaille, S. Allard and J. Cote, 2005. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol. Cell. Biol. 25: 8179–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum, H., O. Fritsch, B. Hohn and S. M. Gasser, 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst et al., 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. [DOI] [PubMed] [Google Scholar]
- Wyatt, H. R., H. Liaw, G. R. Green and A. J. Lustig, 2003. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., K. Griffin, N. Mondal and J. D. Parvin, 2004. Phosphorylation of histone H2A inhibits transcription on chromatin templates. J. Biol. Chem. 279: 21866–21872. [DOI] [PubMed] [Google Scholar]