Abstract
Vulval development in Caenorhabiditis elegans is inhibited by the redundant functions of the synthetic multivulva (synMuv) genes. At least 26 synMuv genes have been identified, many of which appear to act via transcriptional repression. Here we report the molecular identification of the class B synMuv gene lin-61, which encodes a protein composed of four malignant brain tumor (MBT) repeats. MBT repeats, domains of ∼100 amino acids, have been found in multiple copies in a number of transcriptional repressors, including Polycomb-group proteins. MBT repeats are important for the transcriptional repression mediated by these proteins and in some cases have been shown to bind modified histones. C. elegans contains one other MBT-repeat-containing protein, MBTR-1. We demonstrate that a deletion allele of mbtr-1 does not cause a synMuv phenotype nor does mbtr-1 appear to act redundantly with or in opposition to lin-61. We further show that lin-61 is phenotypically and biochemically distinct from other class B synMuv genes. Our data indicate that while the class B synMuv genes act together to regulate vulval development, lin-61 functions separately from some class B synMuv proteins in other biological processes.
AS cells divide during development, their descendants become increasingly restricted in their capacities to adopt different cell fates. These restrictions in cell fate involve the modulation of gene expression, frequently through modifications of the surrounding chromatin. Mutations in factors that control chromatin structure can lead to developmental defects in numerous organisms (reviewed by Margueron et al. 2005).
In Caenorhabiditis elegans, the regulation of vulval development involves evolutionarily conserved proteins important for signal transduction, chromatin remodeling, and transcriptional repression. The vulva of the C. elegans hermaphrodite is formed from three of six equipotent blast cells, P3.p–P8.p (Sulston and Horvitz 1977; Sulston and White 1980; Sternberg and Horvitz 1986). Although all six cells are competent in adopting a vuval cell fate, in wild-type development only P5.p, P6.p, and P7.p divide to generate the vulva. P3.p, P4.p, and P8.p normally divide once and fuse with the nonvulval syncytial hypodermis. A number of signaling pathways specify vulval development, including a receptor tyrosine kinase/Ras pathway, a Wnt pathway, and a Notch pathway (Greenwald et al. 1983; Yochem et al. 1988; Beitel et al. 1990; Han et al. 1990; Eisenmann et al. 1998; Gleason et al. 2002). Mutations affecting these pathways either can cause P3.p, P4.p, and P8.p aberrantly to adopt vulval cell fates and thereby generate a multivulva (Muv) phenotype or can cause none of the Pn.p cells to adopt a vulval cell fate, resulting in a vulvaless (Vul) phenotype (Greenwald et al. 1983; Sternberg and Horvitz 1989; Eisenmann et al. 1998).
The ectopic induction of P3.p, P4.p, and P8.p can also be caused by mutations in the synthetic multivulva (synMuv) genes, which have been placed into three classes, A, B, and C, on the basis of their genetic interactions (Ferguson and Horvitz 1989; Ceol and Horvitz 2004). Because of redundancy among the three classes, only animals with loss-of-function mutations in two synMuv classes have a highly penetrant Muv phenotype, whereas animals with a loss-of-function mutation in a single class are predominantly not Muv. Many of the synMuv genes encode proteins implicated in chromatin remodeling and transcriptional repression (Lu and Horvitz 1998; von Zelewsky et al. 2000; Ceol and Horvitz 2001, 2004; Couteau et al. 2002; Dufourcq et al. 2002; Poulin et al. 2005). The gene lin-3, which encodes an EGF-like ligand that promotes vulval induction (Hill and Sternberg 1992), appears to be transcriptionally repressed by at least some of the synMuv genes, and it has been proposed that loss of synMuv gene activity results in the ectopic expression of lin-3 and the consequent activation of the receptor tyrosine kinase/Ras pathway that induces vulval formation (Cui et al. 2006a).
The synMuv proteins likely form a number of distinct transcriptional regulatory complexes (Figure 1). EFL-1 E2F, DPL-1 DP, and LIN-54, which likely bind directly to DNA and repress transcription, are components of the evolutionarily conserved DP, Rb, and class B synMuv (DRM) complex (Ceol and Horvitz 2001; Harrison et al. 2006). The DRM complex also includes the class B synMuv proteins LIN-35 Rb, LIN-53 RbAp48, LIN-9, LIN-37, and LIN-52 (Harrison et al. 2006) and is nearly identical to two complexes shown to repress transcription in Drosophila, the Myb-MuvB and dREAM complexes (Korenjak et al. 2004; Lewis et al. 2004). The synMuv proteins LET-418 Mi2, LIN-53 RbAp48, and HDA-1 HDAC1 (histone deacetylase I) are homologous to components of the mammalian nucleosome remodeling and deacetylase (NuRD) complex (Lu and Horvitz 1998; von Zelewsky et al. 2000; Dufourcq et al. 2002). These three C. elegans proteins form a complex in vivo and associate with the zinc-finger-containing synMuv protein MEP-1 (Unhavaithaya et al. 2002). The synMuv proteins MET-2 and HPL-2 are homolgous to SETDB1 and HP1, respectively (Couteau et al. 2002; Poulin et al. 2005); SETDB1 is a methyltransferase that can methylate lysine 9 of histone H3, and HP1 binds this modified residue (Bannister et al. 2001; Schultz et al. 2002). HPL-2 associates with another class B synMuv protein, LIN-13, and the two proteins might act together in transcriptional repression (Coustham et al. 2006). The class C synMuv genes encode homologs of a Tip60/NuA4-like histone acetyltransferase complex, which might act in either transcriptional repression or activation (Ceol and Horvitz 2004). Additional synMuv proteins have been identified, including LIN-8, LIN-15A, LIN-15B, LIN-36, LIN-38, LIN-56, and TAM-1 (Clark et al. 1994; Huang et al. 1994; Hsieh et al. 1999; Thomas and Horvitz 1999; Thomas et al. 2003; Davison et al. 2005; A. Saffer, E. Davison and H. R. Horvitz, unpublished observations). While it is likely that these proteins also function in transcriptional repression, whether they interact with other identified synMuv proteins remains to be determined.
Figure 1.—
A synMuv protein interaction map. Class A synMuv proteins are shown in yellow, Class B synMuv proteins in shades of blue, and Class C synMuv proteins in green. This assignment of proteins to specific classes is based on published classifications. The synMuv protein complexes indicated have been demonstrated directly in co-immunoprecipitation experiments, have been suggested by studies of protein stability, or are based on homology to complexes identified in other organisms. Interactions that have been suggested by yeast two-hybrid and GST pull-down experiments but not demonstrated in co-immunoprecipitation experiments are shown by double-headed arrows. References for the interactions shown are as follows: LIN-15A–LIN-56 (E. Davison and H. R. Horvitz, unpublished observations); LIN-8–LIN-35 (Davison et al. 2005); DRM complex (Harrison et al. 2006); NuRD-like complex (Unhavaithaya et al. 2002; Harrison et al. 2006); HDA-1–LIN-35 (Lu and Horvitz 1998); LIN-13—HPL-2 (Coustham et al. 2006); Tip60-like complex (Ceol and Horvitz 2004).
Here we report the molecular identification and characterization of the class B synMuv gene lin-61 and the finding that lin-61 encodes a protein similar to Polycomb-group (PcG) proteins. PcG proteins were initially identified by their abilities to repress the transcription of Hox genes and have since been found to repress additional targets, including genes regulated by E2F transcription factors (Dahiya et al. 2001; Ogawa et al. 2002). PcG proteins include histone methyltransferases and proteins that bind to the histones methylated by such transferases. The Drosophila PcG proteins Sex Comb on Midleg (SCM) and Sfmbt each contain MBT repeats, which are motifs of ∼100 amino acids. MBT repeats have been found in many transcriptional repressors, including human L(3)MBT (Bornemann et al. 1996; Ogawa et al. 2002; Boccuni et al. 2003), which is in a complex with multiple other PcG-group proteins and with E2F6 (Ogawa et al. 2002).
We report that lin-61 encodes a protein that contains four MBT repeats and that localizes to chromatin. LIN-61 does not associate with either of the two known synMuv protein complexes, the pocket-protein-containing DRM complex and the NuRD-like complex, and can act separately from members of these complexes. We propose that MBT-repeat-containing proteins, such as Polycomb-group proteins, cooperate with Rb-containing complexes and histone deacetylase complexes to repress certain genes but act independently of these complexes to regulate expression of other genes.
MATERIALS AND METHODS
Strains:
Unless otherwise specified, all C. elegans strains were cultured at 20° on NGM agar seeded with Escherichia coli strain OP50 as described by Brenner (1974). The wild-type strain was N2 (Bristol). Mutant alleles used are listed below and are described by Riddle et al. (1997) unless otherwise noted: LGI—unc-14(e57), unc-15(e73), lin-61(sy223, n3442, n3446, n3447, n3624, n3687, n3736, n3807, n3809, n3922) (this study), mbtr-1(n4775) (this study), lin-65(n3441) (Ceol et al. 2006), lin-53(n3368) (Andersen et al. 2006), ccEx6188 [rol-6(su1006); myo-3∷Ngfp-lacZ] (Hsieh et al. 1999); LGII—lin-8(n2731) (Thomas et al. 2003), lin-38(n751), lin-56(n2728) (Thomas et al. 2003), trr-1(n3712) (Ceol and Horvitz 2004), dpl-1(n3316) (Ceol and Horvitz 2001), mnCI[dpy-10(e128) unc-52(e444)] (Herman 1978); LGIII—lin-13(n770) (Ferguson and Horvitz 1989), lin-37(n758), mat-3(ku233) (Garbe et al. 2004), hpl-2(n4274) (E. Andersen and H. R. Horvitz, personal communication), lin-52(n3718) (Ceol et al. 2006), qCI[dpy-19(e1259) glp-1(q339)]; LGIV—ark-1(n3701) (Ceol et al. 2006); LGV—hda-1(e1795) (Dufourcq et al. 2002), tam-1(cc567) (Hsieh et al. 1999), let-418(n3719) (Ceol et al. 2006), mep-1(n3703) (Ceol et al. 2006), lin-54(n2231, n3423) (Thomas et al. 2003; Harrison et al. 2006), qIs56 [lag-2∷gfp; unc-119(+)] (Siegfried and Kimble 2002); LGX—lin-15A(n433, n767, sy197), lin-15B(n744), mys-1(n3681, n4075) (Ceol and Horvitz 2004), sli-1(n3538) (Ceol et al. 2006), gap-1(ga133) (Hajnal et al. 1997), lin(n3542) (Ceol et al. 2006), pkIs1605 [rol-6(su1006); hsp16/2∷gfp-lacZ(out of frame)] (Pothof et al. 2003). The translocations nT1[unc(n754)] (LGIV and LGV), nT1 [qIs51] (LGIV and LGV) and hT2 [qIs48] (LGI and LGIII) and the chromosomal inversion mIn1 [dpy-10(e128) mIs14] were used as balancers; each contains an integrated gfp transgene linked to the balancer (Edgley and Riddle 2001; Mathies et al. 2003).
Isolation of the mbtr-1(n4775) deletion allele:
Genomic DNA pools from EMS-mutagenized animals were screened for a deletion using PCR, as described by Ceol and Horvitz (2001). Deletion mutant animals were isolated from a frozen stock and backcrossed to the wild type at least twice. mbtr-1(n4775) removes nucleotides 30,255–32,134 of cosmid Y48G1A. The sequence of the deletion junction is ATTTTAAAAATTGAG/AATTTTGTTGAA, with the slash indicating the deletion breakpoint.
Transgenic strains:
For rescue of the lin-61(sy223); lin-15A(n767) and lin-61(n3624); lin-15A(n767) synMuv phenotypes, cosmid or subclone DNA (5 or 10 ng/μl) was coinjected with a dominant rol-6 marker plasmid (pRF4) (80 ng/μl) as described in Mello et al. (1991). pMMH15, which was constructed by subcloning a StuI–SacII fragment of the cosmid R06C7 corresponding to bases 15,366–19,753 into pBluescript, was injected for subclone rescue. For expression of mbtr-1 and lin-61 driven by the dpy-7 promoter, constructs were injected at 25 ng/μl with sur-5∷gfp (pTG96; kindly provided by M. Han) at 20 ng/μl and a 1-kb ladder (Invitrogen, Carlsbad, CA) at 80 ng/μl.
RNA interference analysis of lin-61 and mbtr-1:
Templates for in vitro transcription reactions were made by PCR amplification of cDNAs yk732e5 or yk268b4 (kindly provided by Y. Kohara), including flanking T3 and T7 promoter regions. RNA was transcribed in vitro using T3 and T7 polymerases and was denatured for 10 min and annealed prior to injection.
Antibody preparation, immunocytochemistry, and Western blots:
Anti-LIN-61 antiserum was generated by immunizing rabbits and guinea pigs with purified GST-LIN-61 (amino acids 159–491). This region corresponds to the amino acids likely to be absent in the protein produced in lin-61(n3809) animals, allowing these animals to provide a control for antibody specificity. The antiserum was affinity purified against full-length MBP-LIN-61. The rabbits and guinea pigs were immunized and maintained by Covance (Denver, PA). Anti-LIN-61, anti-LIN-9 (Harrison et al. 2006), anti-LIN-35 (Harrison et al. 2006), anti-LIN-37 (Harrison et al. 2006), anti-LIN-52 (Harrison et al. 2006), anti-HDA-1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-HPL-2 (Coustham et al. 2006), anti-LIN-56 (E. Davison and H. R. Horvitz, personal communication), and antitubulin DM1A (Millipore, Bedford, MA) antibodies were used at a 1:1000 dilution for Western blots. Anti-LIN-8 (Davison et al. 2005), anti-LIN-53 (Harrison et al. 2006), anti-LIN-54 (Harrison et al. 2006), and anti-DPL-1 (Ceol and Horvitz 2001) were used at a 1:500 dilution for Western blots. Affinity-purified antibodies were used in all cases, except for anti-LIN-35 antibodies, for which we used unpurified serum from the third production bleed. Larvae and adults for immunostaining were fixed in 1% paraformaldehyde for 30 min, as described by Finney and Ruvkun (1990). Embryos were fixed for 20 min in 0.8% paraformaldehyde, as described by Guenther and Garriga (1996). Affinity-purified anti-LIN-61 antisera were used at a 1:100 dilution for immunocytochemistry.
Phenotypic characterization:
To score RNA interference (RNAi) hypersensitivity, we assessed the sensitivity of worms to bacteria expressing hmr-1 or cel-1 dsRNAs, which previously have been used to characterize the RNAi hypersensitivity of synMuv mutants (Wang et al. 2005). Wild-type animals are only mildly affected by the dsRNA produced by these bacterial strains, but animals that are hypersensitive to RNAi are severely affected (Wang et al. 2005). L4 larvae were grown on E. coli strain HT115 expressing either hmr-1 or cel-1 dsRNA, and 24 hr later the young adult hermaphrodites were transferred to fresh plates with E. coli expressing the same dsRNA (Kamath and Ahringer 2003). The hermaphrodites were allowed to lay eggs for 24 hr and were then removed. The progeny of hermaphrodites grown on E. coli-expressing cel-1 dsRNA were scored for developmental arrest at the L2 larval stage. The progeny of hermaphrodites grown on E. coli-expressing hmr-1 dsRNA were scored for embryonic lethality. To score ectopic PGL-1 expression, L1 larvae were permeabilized using a freeze-crack method followed by a methanol-acetone fixation, as described in Wang et al. (2005). Permeabilized larvae were incubated with OIC1D4 monoclonal anti-PGL-1 antibodies (Developmental Studies Hybridoma Bank, University of Iowa) at a dilution of 1:20 overnight followed by a 1-hr incubation with Alexa Fluor 594 goat anti-mouse IgM (Invitrogen, Carlsbad, CA) at a dilution of 1:25. To score for the Tam phenotype (Hsieh et al. 1999), hermaphrodites homozygous for the extrachromosomal array ccEx6188 [myo-3∷Ngfp-lacZ] were grown for at least two generations at 25°. Using a dissecting microscope equipped with fluorescence optics, we scored animals homozygous for the extrachromosomal array and either for lin-61(n3809), lin-61(n3992), lin-35(n745), and mbtr-1(n4775) or for both lin-61(n3809) and mbtr-1(n4775) for reduced GFP expression as compared to animals carrying only the ccEx6188 transgene. Hermaphrodites homozygous for the transgene pkIs1605 [rol-6(su1006); hsp16/2∷gfp/lacZ(out-of-frame)] were scored for expression of LacZ after being grown at 20°, heat-shocked at 31° for 2 hr, and allowed to recover at 20° for 1 hr. The hermaphrodites were then fixed and stained with X-gal for the presence of β-galactosidase. Vulval defects in a mat-3(ku233) mutant background were scored using Nomarski differential interference contrast microscopy to observe mid-L4 hermaphrodites. Vulval development was scored as abnormal if the invagination was asymmetric or if the developing vulva contained fewer than the 22 nuclei found in wild-type animals (Garbe et al. 2004).
Embyro lysates:
Embryos were harvested from liquid cultures, resuspended in 1 ml of lysis buffer (25 mm HEPES, pH 7.6, 150 mm NaCl, 1 mm DTT, 1 mm EDTA, 0.5 mm EGTA, 0.1% NP-40, 10% glycerol) with Complete EDTA-free protease inhibitors (Roche Diagnostics, Basel, Switzerland) for each gram of embryos and frozen in liquid nitrogen. The embryos were thawed at room temperature and sonicated 15 times for 10 sec using a Branson sonifier 450 at setting 5. Lysates were clarified by two 15-min 16,000 g centrifugations in a microcentrifuge at 4°. Protein concentration was determined using the Pierce Coomassie Plus Protein Assay Reagent (Pierce Biotechnology, Rockford, IL). Lysate was diluted to 5–10 mg/ml and was used immediately or stored at −80°.
Immunoprecipitation experiments:
Antibodies were crosslinked to protein A Dynabeads (Invitrogen, Carlsbad, CA) using dimethyl pimelimidate (Pierce Biotechnology) essentially as described by Harlow and Lane (1999), with the two following exceptions: reactions were stopped with 0.1 m Tris, pH 8.0, and beads were washed three times for 1 min in 100 mm glycine, pH 2.5, followed by a single wash with lysis buffer. The beads were then resuspended in PBS. A total of 500 μl of precleared lysate (2.5–5 mg of total protein) was incubated with 25 μl of affinity-purified antibody bound to 25 μl of beads at 4° for 1–2 hr for each immunoprecipitation reaction and were then washed three times for 5 min each at 4° in lysis buffer. The beads were then resuspended in 20 μl of 2× protein sample buffer, boiled for 5 min, and loaded on an SDS–polyacrylamide gel. HRP-conjugated protein A (Bio-Rad, Hercules, CA) was used for detection of antigens on Western blots following co-immunoprecipitation experiments.
RESULTS
lin-61 is a class B synMuv gene:
Ten lin-61 alleles have been isolated in five different screens. The original lin-61 allele, sy223, was isolated in the laboratory of P. Sternberg (personal communication) on the basis of its synMuv phenotype in combination with a loss-of-function mutation in lin-15A, sy197, and sy223 and was mapped to linkage group I (LGI). We found that sy223 complemented lin-53(n833), an allele of the only identified class B synMuv gene on LGI at the time. These data suggested that sy223 defined a new class B synMuv gene. Five additional alleles, n3442, n3446, n3447, n3624, and n3736, were isolated in a screen for mutations that cause a synMuv phenotype with lin-15A(n767) (Ceol et al. 2006). Four more alleles, n3687, n3807, n3809, and n3922, were isolated in screens for mutants altered in transgene expression (H. T. Schwartz, D. M. Wendell and H. R. Horvitz, personal communication). These alleles were all mapped to LGI and shown to be allelic with sy223 in complementation tests (C. J. Ceol, H. T. Schwartz, D. M. Wendell and H. R. Horvitz, personal communication; data not shown).
None of the 10 lin-61 alleles caused a Muv phenotype in the absence of other mutations (Table 1). Each caused a synMuv phenotype in combination with a loss-of-function mutation of the class A synMuv gene lin-56 (Table 2). A putative lin-61 null allele, n3809 (see below), caused a synMuv phenotype in combination with loss of function of each of the four class A synMuv genes (Table 1) but not in combination with mutations in the class B synMuv genes lin-15B, lin-35, or lin-37 (Table 1). Loss of function of lin-61 also did not increase the Muv phenotype caused by hda-1(e1795) (Table 1). Whereas null mutations in classically defined class B synMuv genes such as lin-35 and lin-15B enhance the weak Muv phenotype of loss-of-function mutations of class C synMuv genes (Ceol and Horvitz 2004), lin-61(n3809) did not enhance the weak Muv phenotype caused by loss of function in either of the class C genes trr-1 or mys-1 (Table 1). This result might suggest that lin-61 has class C synMuv activity (Ceol and Horvitz 2004). However, whereas null mutations in class C synMuv genes cause a P8.p induction as single mutants at a penetrance of ∼15% (Table 1; Ceol and Horvitz 2004), 0/24 lin-61(n3809) animals showed P8.p induction. Furthermore, mutations in class C synMuv genes cause a Muv phenotype in combination with loss of function of class B synMuv genes, but, in combination with mutations in any of a number of class B synMuv genes, lin-61(n3809) did not cause a Muv phenotype (Table 1). We therefore suggest that lin-61 is not likely to have class C synMuv activity. The failure to enhance the Muv phenotype of animals mutant for class C synMuv genes could instead result from the fact that putative null mutations in lin-61 cause a weaker synMuv phenotype than do null mutations in other class B synMuv genes. For these reasons, we consider lin-61 to be a class B synMuv gene.
TABLE 1.
lin-61 mutations cause a class B synMuv phenotype
| Genotype | % Muv (n) |
|---|---|
| Single mutants | |
| lin-61(sy223) | 0 (260) |
| lin-61(n3442) | 0 (327) |
| lin-61(n3446) | 0 (217) |
| lin-61(n3447) | 0 (334) |
| lin-61(n3624) | 0 (242) |
| lin-61(n3687) | 0 (252) |
| lin-61(n3736) | 0 (234) |
| lin-61(n3807) | 0 (278) |
| lin-61(n3809) | 0 (269) |
| lin-61(n3922) | 0 (82) |
| lin-15B(n744) | 0 (272) |
| lin-35(n745) | 0 (104) |
| lin-37(n758) | 0 (318) |
| hda-1(e1795) | 31 (143) |
| trr-1(n3712)a | 18 (39) |
| mys-1(n3681)ab | 8 (36) |
| mys-1(n4075)ab | 15 (20) |
| lin-61 + class A synMuv double mutants | |
| lin-61(n3809); lin-8(n2731) | 72 (414) |
| lin-61(n3809); lin-38(n751) | 93 (175) |
| lin-61(n3809); lin-56(n2728) | 100 (180) |
| lin-61(n3809); lin-15A(n433) | 14 (261) |
| lin-61(n3809); lin-15A(n767) | 97 (166) |
| lin-61 + class B synMuv double mutants | |
| lin-61(n3809); lin-15B(n744) | 0 (153) |
| lin-61(n3809); lin-35(n745) | 0 (178) |
| lin-61(n3809); lin-37(n758) | 0 (165) |
| lin-61(n3809); hda-1(e1795) | 20 (96) |
| lin-61 + class C synMuv double mutants | |
| lin-61(n3809); trr-1(n3712)a | 17 (111) |
| lin-61(n3809); mys-1(n3681)a | 7 (45) |
All animals were raised at 20°. The Muv phenotype was scored using a dissecting microscope, except in the cases noted. trr-1(n3712) mutant homozygotes were recognized as the non-GFP progeny of trr-1(n3712)/mIn1[dpy-10 mIs14] heterozygous parents. hda-1(e1795) homozygotes were recognized as the non-GFP progeny of hda-1(e1795)/nT1[qIs51]; +/nT1[qIs51] heterozygous parents. lin-61(n3687) and lin-61(n3922) were also homozygous for the linked integrated transgene nIs133, which carries pkd-2:gfp and a rescuing lin-15AB construct.
Muv, more than three Pn.p cells were induced as scored using Nomarski optics.
These data are from Ceol and Horvitz (2004). mys-1(n4075) is a deletion mutation.
TABLE 2.
Sequences of lin-61 alleles and allele strengths
| Wild-type sequence | Mutant sequence | % Muv (n)
|
|||
|---|---|---|---|---|---|
| lin-61 allele | Mutation effect | With lin-56(n2728) | With lin-15A(n433) | ||
| + | — | — | — | 0 (many) | 0 (many) |
| lin-61(n3442) | agAAT | aaAAT | Exon 4 splice acceptor | 98 (154) | 14 (220) |
| lin-61(n3446) | CAA | TAA | Q412ochre | 96 (87) | 13 (241) |
| lin-61(n3809) | CAA | TAA | Q159ochre | 92 (176) | 14 (261) |
| lin-61(sy223) | agCTC | aaCTC | Exon 6 splice acceptor | 89 (255) | 11 (129) |
| lin-61(n3624) | CCG | TCG | P132S | 85 (220) | 5.6 (251) |
| lin-61(n3807) | GGA | GAA | G250E | 83 (136) | 6.9 (246) |
| lin-61(n3736) | TTT | TCT | F247S | 80 (313) | 1.3 (232) |
| lin-61(n3922) | GGA | GAA | G445R | 52 (167) | NAa |
| lin-61(n3447) | AGT | AAT | S354N | 47 (237) | 1.1 (278) |
| lin-61(n3687) | CAA | TAA | Q322ochre | 23 (305) | NAa |
Amino acid substitutions are indicated as wild-type residue, residue number, and mutant residue. Coding bases are shown as uppercase letters. Intronic bases are shown as lowercase letters. All animals were raised at 20°. The Muv phenotype was scored using a dissecting microscope.
NA, not applicable because lin-61(n3687) and lin-61(n3922) were also homozygous for the linked integrated transgene nIs133, which carries pkd-2∷gfp and a rescuing lin-15AB construct.
lin-61 encodes an MBT-repeat-containing protein:
We mapped sy223 to an interval between unc-14 and unc-15 on LGI. A pool of four cosmids (C01H6, C12E8, C12C7, and C33F11) covering the central portion of this region rescued the synMuv phenotype of lin-61(sy223); lin-15A(n767) animals, and a single cosmid from this region, R06C7, rescued the synMuv phenotype of lin-61(n3624); lin-15A(n767) animals. A subcloned StuI–SacII fragment containing R06C7.7 as the only complete predicted open reading frame was capable of rescuing the lin-61(n3624); lin-15A(n767) synMuv phenotype (Figure 2A). As reported elsewhere, RNAi directed against R06C7.7 caused a synMuv phenotype in animals mutant for the class A synMuv gene lin-15A but not in wild-type animals (Poulin et al. 2005; our unpublished data). To confirm that R06C7.7 is lin-61, we determined the sequence of R06C7.7 from lin-61(sy223) animals. sy223 encodes a G-to-A transition at the splice-acceptor site of the last predicted exon of R06C7.7 (Figure 2B and Table 2). Mutations affecting the coding region of R06C7.7 were found for all other lin-61 alleles, including three nonsense mutations, five missense mutations, and one mutation in a splice-acceptor site (Figure 2B, Table 2).
Figure 2.—
Molecular cloning of lin-61. (A) lin-61 maps between unc-14 and unc-15 on LGI. Part of cosmid R06C7 is shown below as a shaded bar. The rescuing StuI–SacII fragment of R06C7 is shown below the cosmid. Open boxes represent the exons of the predicted genes within the subclone. Arrows indicate the direction of transcription. (B) lin-61 gene structure as determined from cDNA and genomic sequences. Solid boxes indicate coding sequence. Open box indicates the 3′ untranslated region. Predicted translation initiation (ATG) and termination (TAA) codons are shown along with the site of polyadenylation [poly(A)] and of SL1 trans-splicing (SL1). The positions of the mutations found in the 10 lin-61 alleles are indicated above the gene structure. (C) Alignment of the MBT repeats from the C. elegans (Ce) proteins LIN-61 and MBTR-1 and the Homo sapiens (Hs) proteins L(3)MBT2 and L(3)MBT1, accession nos. Q969R5 and Q9Y468, respectively. Each repeat is shown separately with the repeat number indicated next to the protein name. The top portion corresponds to the N-terminal arm and the bottom portion corresponds to the contiguous β-core region of the MBT repeat, as defined by structural analysis (Sathyamurthy et al. 2003; Wang et al. 2003). Shaded residues indicate identities among >8 of the 15 MBT repeats. Circled residues indicate positions of missense mutations in LIN-61. The corresponding allele is indicated above the residue. The missense mutation n3624 is located between the first and second repeats. The boxed region indicates the 15 amino acids inserted in the second MBT repeats of LIN-61 and MBTR-1. (D) Schematic of C. elegans LIN-61, H. sapiens L(3)MBT2, and Mus musculus Mbtd1 proteins, accession nos. NP_492050, Q969R5, and AAH62907, respectively. Shaded boxes indicate the positions and relative sizes of the four MBT repeats.
We determined the sequence of a full-length lin-61 cDNA, yk732e5, and determined that the lin-61 transcript is SL1 spliced and comprises six exons (Figure 2B). lin-61 transcripts contain no 5′-UTR, as the SL1 leader sequence is spliced directly to the predicted ATG start codon. The next in-frame methionine codon is 373 nucleotides downstream and would produce a protein product inconsistent with the observed mobility of LIN-61 by SDS–PAGE (see below). lin-61 encodes a predicted protein of 491 amino acids composed almost exclusively of four MBT repeats (Figure 2C) as recognized by SMART and PSI-BLAST databases (Altschul et al. 1997; Schultz et al. 2000). MBT repeats were initially identified in the Drosophila protein lethal (3) malignant brain tumor [l(3)mbt] (Wismar et al. 1995) and are present in many other metazoan proteins but not in proteins from other kingdoms (Bornemann et al. 1996; Tomotsune et al. 1999; Usui et al. 2000; Boccuni et al. 2003; Markus et al. 2003; Arai and Miyazaki 2005; Klymenko et al. 2006). MBT-repeat-containing proteins include the Drosophila Polycomb-group proteins SCM and Sfmbt. In addition to their MBT repeats, l(3)mbt, SCM, and Sfmbt each contain atypical zinc fingers and a single sterile α-motif (SAM) domain (Wismar et al. 1995; Bornemann et al. 1996; Klymenko et al. 2006). The SAM domain of SCM mediates homodimerization and interaction with the Polycomb protein Polyhomeotic (Peterson et al. 1997; Kim et al. 2005). While these Drosophila proteins contain functional domains in addition to MBT repeats, proteins in other species, including human and mouse L(3)MBT2 and Mbtd1, are composed almost exclusively of four MBT repeats, similar to LIN-61 (Figure 2D). Given that LIN-61 does not contain any recognized domain apart from the MBT repeats and that it is composed almost exclusively of the four MBT repeats, the functionality of LIN-61 is likely provided by the MBT repeats.
Characterization of lin-61 alleles:
The mutation in lin-61(n3809) results in an ochre stop codon at amino acid 159, is predicted to result in a truncated LIN-61 protein, and is likely to be a null allele. lin-61(n3809); lin-56(n2728) animals have a highly penetrant synMuv phenotype (Table 2). Similarly penetrant synMuv phenotypes are caused by one other lin-61 nonsense mutation, n3446, and by either of the splice-acceptor mutations, n3442 and sy223 (Table 2). Although n3687 causes an ochre mutation at amino acid 322, animals homozygous for this lin-61 allele in combination with a loss-of-function mutation in a class A synMuv gene have a weak synMuv phenotype. The low penetrance of this synMuv phenotype suggests that a partially functional LIN-61 protein product might be made in lin-61(n3687) animals. If such a product exists, it is either reduced in abundance or not recognized by anti-LIN-61 polyclonal antibodies, as LIN-61 is not detected by immunoblotting (see below). Alternatively, the strain containing lin-61(n3687) might contain an additional mutation that partially suppresses the synMuv phenotype. As n3867 was isolated in a screen for altered transgene expression, this strain contains a closely linked integrated transgene that suppresses recombination on part of LGI, including lin-61, and drives the overexpressesion of both lin-15A and lin-15B. It is possible that this transgene suppresses the synMuv phenotype. lin-61(n3922) was isolated in the same screen and contains the same linked transgene. This transgene therefore might reduce the penetrance of the synMuv phenotype in this strain as well. As n3922 results in a missense rather than in a nonsense mutation, we cannot predict the expected penetrance of the synMuv phenotype in animals homozygous for n3922 and a mutation in a class A synMuv gene.
Four of the five missense mutations in lin-61, n3447, n3736, n3807, and n3922 alter residues in the four MBT repeats. The fifth missense mutation, n3624, causes a proline-to-serine change in a residue between the first and second MBT repeats. The five missense mutations cause weaker synMuv phenotypes than the putative null alleles based upon double mutants with the class A synMuv mutations lin-15A(n433) and lin-56(n2728). The missense mutations form an allelic series. Specifically, n3624 and n3807, the two strongest missense mutants, cause a less penetrant synMuv phenotype than the nonsense and splice-acceptor mutations, suggesting that some LIN-61 activity might remain in these mutants (Table 2). Animals homozygous for lin-61(n3736) and a class A synMuv mutation had a phenotype intermediate to the phenotypes caused by other missense mutations, as evident from the fact that lin-61(n3736) in combination with lin-15A(n433) caused a lower penetrance synMuv phenotype than did lin-61(n3807) or lin-61(n3624) at 20°. However, when these strains were raised at 23° or when the lin-61 alleles were combined with a mutation in the class A synMuv gene, lin-56, lin-61(n3676) caused a synMuv phenotype with a penetrance similar to the penetrance of the synMuv phenotypes caused by lin-61(n3624) and lin-61(n3807) (Table 2 and data not shown). lin-61(n3447) and lin-61(n3922) mutant animals had the least-penetrant synMuv phenotypes (Table 2).
The C.elegans genome encodes one additional MBT-repeat-containing protein:
Given the molecular identification of LIN-61 as a protein containing MBT repeats, we searched the C. elegans genome for additional proteins containing MBT repeats. Using BLAST (Altschul et al. 1997), Pfam (Bateman et al. 2002), and SMART (Schultz et al. 2000), we identified a single additional MBT-repeat-containing protein in the C. elegans genome, encoded by the predicted gene Y48G1A.6 (Figure 3A). We determined the sequence of a full-length cDNA for Y48G1A.6, yk268b4 (accession no. DQ904352). The GENEFINDER (WormBase at http://wormbase.org, release WS 160) prediction for the cDNA is predominantly correct, except that the predicted fourth intron is not removed in this cDNA. The incorporation of this predicted intron into the open reading frame results in a larger fourth exon than predicted but does not alter the frame of the predicted protein. Furthermore, the incorporation of this predicted intron results in a protein product that is similar to the analogous region of LIN-61 and is internal to the third MBT repeat, suggesting that this intron is likely to be an important part of the protein product. Because the protein encoded by Y48G1A.6 contains MBT repeats, we named this gene mbtr-1, for malignant brain tumor repeats.
Figure 3.—
MBTR-1 sequence and structure. (A) Alignment of MBTR-1 and LIN-61. Solid boxes indicate identities between LIN-61 and MBTR-1, and shaded boxes indicate similarities between the two proteins. Underlined regions correspond to the four MBT repeats. The solid box indicates the 15–16 amino acid insertions in the second MBT repeats of LIN-61 and MBTR-1. (B) mbtr-1 gene structure as determined from cDNA and genomic sequences. Solid boxes indicate coding sequence. Open boxes indicate 5′ and 3′ untranslated regions. Predicted translation initiation (ATG) and termination (TAA) codons are shown along with the site of polyadenylation [poly(A)]. The predicted gene within the first intron of mbtr-1, Y48G1A.2, is shown. The arrow depicts the direction of transcription. The genomic region deleted in n4775 is indicated by brackets. (C) Schematic of the MBTR-1 protein. Shaded boxes indicate the positions and relative sizes of the four MBT repeats.
The primary structure of MBTR-1 is similar to that of LIN-61 (Figure 3A). Like LIN-61, MBTR-1 is composed almost exclusively of four MBT repeats and lacks the SAM domain and zinc fingers found in many MBT-repeat-containing proteins in other organisms (Figure 3). MBTR-1 is 36% identical to LIN-61 and is more similar to LIN-61 than to proteins in any other organisms. However, according to BLAST searches (Altschul et al. 1997), LIN-61 is more similar to the five homologs in C. briggsae than it is to MBTR-1. LIN-61, MBTR-1, and the five C. briggsae homologs of these two genes share an insertion of ∼15–16 amino acids in their second MBT repeat not found in other MBT-repeat-containing proteins (Wang et al. 2003; Figure 2C and Figure 3A). This observation suggests that Caenorhabditis MBT-repeat-containing proteins might have diverged from a single ancestral protein rather than arising from multiple different MBT-repeat-containing ancestral proteins. It is unclear how these additional amino acids might alter the structure of the MBT repeat or contribute to the function of the protein.
GENEFINDER (WormBase at http://wormbase.org, release WS 160) predicts and the identification of cDNAs corresponding to Y48G1A.2 (WormBase at http://wormbase.org, release WS 160) confirm the existence of the open reading frame Y48G1A.2 within the first intron of mbtr-1 (Figure 3B). Y48G1A.2 and mbtr-1 are transcribed from different strands.
To analyze the function of mbtr-1, we identified a deletion allele, n4775, which removes exons 4 and 5 of mbtr-1 and is predicted to result in a frameshift after amino acid 165. The n4775 deletion does not remove any of the coding sequence for Y48G1A.2. It remains possible that n4775 could effect the expression of Y48G1A.2, because the deletion removes upstream sequences >1 kb from the translational start site for Y48G1A.2; these sequences might be necessary for proper expression of Y48G1A.2.
We have not identified a mutant phenotype associated with mbtr-1(n4775). mbtr-1(n4775) did not cause a synMuv phenotype in combination with either the strong class A synMuv mutant lin-15A(n767) or the strong class B synMuv mutant lin-15B(n744) (0% n > 100). Animals mutant for both mbtr-1 and lin-61 were not Muv and did not display any other obvious phenotypic defects (0% n > 100). mbtr-1 and lin-61 do not redundantly provide class A synMuv activity, as mbtr-1(n4775) lin-61(n3809); lin-15B(n744) animals were not Muv. Additionally, the deletion allele of mbtr-1 did not enhance or suppress the synMuv phenotype of lin-61(n3809); lin-15A(n767) animals: 97% of lin-61(n3809); lin-15A(n767) (n = 166) animals were Muv at 20° and 24% were Muv at 15° (n = 118), and, similarly, 94% of mbtr-1(n4775) lin-61(n3809); lin-15A(n767) (n = 157) animals were Muv at 20° and 27% were Muv at 15° (n = 145).
Expression of a lin-61 cDNA in the hypodermis under the control of the dpy-7 promoter (Myers and Greenwald 2005) rescued the synMuv phenotype of lin-61(n3809); lin-15A(n767) animals. Specifically, in seven independent lines, expression of dpy-7p∷lin-61 reduced the penetrance of the synMuv phenotype of lin-61(n3809); lin-15A(n767) animals from 97% to 27, 29, 31, 47, 48, 48, or 52% (supplemental Table 1 at http://www.genetics.org/supplemental/). However, when we used the dpy-7 promoter to drive expression of a full-length mbtr-1 cDNA in lin-61(n3809); lin-15A(n767) animals, little if any rescue of the synMuv phenotype was observed in eight independent lines. The penetrance of the synMuv phenotype for the eight lines was 78, 86, 88, 91, 93, 93, 94, and 100% (supplemental Table 1 at http://www.genetics.org/supplemental/). While we cannot be certain that in these experiments MBTR-1 was expressed at the same level and time as LIN-61was, these data suggest that, although the two proteins are closely related, MBTR-1 is unable to provide the function normally provided by LIN-61.
LIN-61 is broadly expressed in nuclei throughout development:
To determine the expression pattern and localization of LIN-61, we generated guinea pig and rabbit polyclonal antibodies against the C-terminal 332 amino acids of LIN-61. Affinity-purified antibodies recognized a band corresponding to a protein of ∼60 kDa on Western blots of protein extracts from wild type but not lin-61(n3809) animals (Figure 4A). This molecular weight is similar to the size of the predicted LIN-61 product, 57 kDa.
Figure 4.—
LIN-61 is an ubiquitously expressed nuclear protein with punctate localization. (A) Affinity-purified antibodies raised against recombinant LIN-61 were used to blot extracts from both wild-type and lin-61(n3809) mutant animals. Asterisks denote nonspecific immunoreactivity. HM4077 antibodies were raised in a guinea pig. HM4078 antibodies were raised in a rabbit. (B, D, F, H, J, and L) Whole-mount staining with anti-LIN-61 antisera. HM4077 was used for whole-mount staining of embryos, and HM4078 was used for whole-mount staining of adults. (B) LIN-61 is expressed in discrete foci in the nuclei of the developing embryo. (C) 4′,6-diamidino-2-phenylindole (DAPI) staining of the embryo shown in (B). (D) Enlargement of the boxed portion of B. (E) Enlargement of the boxed portion of C. (F) LIN-61 was absent in lin-61(n3809) embryos. (G) DAPI staining of the embryo shown in F. (H) LIN-61 was broadly expressed in the adult hermaphrodite germline and was localized to condensed chromosomes. (I) DAPI staining of the germline shown in H. (J) Enlargement of the boxed portion of H. (K) Enlargement of the boxed portion of I. (L) LIN-61 staining was absent from the germline of lin-61(n3809) adult hermaphrodites. (M) DAPI staining of the germline shown in K. WT, wild type. Bars, 10 μm.
We used both the guinea pig and rabbit polyclonal antibodies to analyze the localization of LIN-61 by immunostaining embryos, larvae, and adult hermaphrodites. Similarly to all synMuv proteins studied to date (Melendez and Greenwald 2000; Ceol and Horvitz 2001; Couteau et al. 2002; Ceol and Horvitz 2004; Davison et al. 2005; Harrison et al. 2006), LIN-61 was localized to all or almost all nuclei throughout development from the one-cell embryo to the adult (Figure 4, B and H, and data not shown). In the embryo, LIN-61 appeared to localize to discrete foci in the nucleus (Figure 4, B and D). Both HPL-2 and LIN-13 have been reported to localize to foci in the nucleus (Melendez and Greenwald 2000; Coustham et al. 2006). In addition, the human MBT-repeat-containing protein L(3)MBT and Polycomb-group proteins localize to foci in the nucleus (Buchenau et al. 1998; Saurin et al. 1998; Koga et al. 1999). In the adult hermaphrodite germline, LIN-61 was localized, at least in part, to condensed chromosomes during the diakinesis phase of meiosis, suggesting that some LIN-61 might be localized to chromatin (Figure 4, H and J). No anti-LIN-61 staining was seen in lin-61(n3809) mutant embryos, larvae, or adults (Figure 4, F and L, and data not shown).
To understand better how LIN-61 might act with other synMuv proteins to regulate vulval development, we analyzed the localization of LIN-61 in animals mutant for any of 26 genes that regulate vulval development, including four class A synMuv genes, 17 class B synMuv genes, two class C synMuv genes, and genes encoding three Ras-pathway modifiers that function to regulate vulval development (supplemental Table 2 at http://www.genetics.org/supplemental/). No change in LIN-61 localization was noted in any of these mutant backgrounds (data not shown), suggesting that these genes do not regulate vulval development by modifying LIN-61 expression or subcellular localization.
Missense mutations in LIN-61 might disrupt protein stability:
Crystal structures have been solved for both a peptide containing two MBT repeats and a peptide containing three MBT repeats (Sathyamurthy et al. 2003; Wang et al. 2003). Both structures show that individual MBT repeats consist of an N-terminal arm and a C-terminal β-barrel core region. The N-terminal arm of one repeat interacts with the β-barrel core region of the preceding repeat, resulting in a stabilized tertiary structure (Sathyamurthy et al. 2003; Wang et al. 2003). The N-terminal arm of the first repeat interacts with the core region of the last repeat, forming, in the case of three repeats, a propeller-like structure (Wang et al. 2003).
We analyzed LIN-61 protein levels in strains carrying each of the 10 mutant alleles of lin-61 to determine whether any of the mutations might result in protein misfolding and subsequent degradation. Full-length LIN-61 was absent or levels were greatly reduced in animals with any of the three nonsense mutations or two splice-acceptor mutations (Figure 5; data not shown). (It is conceivable, but unlikely, that a stable truncated protein product that does not contain the epitope recognized by either of the polyclonal antibodies was present.) lin-61(n3736), lin-61(n3807), and lin-61(n3922) animals also showed decreases in LIN-61 protein levels as compared to the wild type, while lin-61(n3447) and lin-61(n3624) animals had wild-type or nearly wild-type LIN-61 protein levels. These data were verified by analyzing LIN-61 protein levels in animals with any of the five missense mutations using both Western blots and immunocytochemistry (Figure 5 and data not shown).
Figure 5.—
Residues within the β-core region MBT repeats of LIN-61 are likely to be important for protein folding and stability. LIN-61 levels are reduced in many lin-61 mutant animals. Equivalent amounts of protein from mixed-stage cultures of each of the genotypes indicated above the lanes were loaded in each lane. Proteins were separated by SDS–PAGE and immunoblotted with the antibodies indicated at the left. Antitubulin antibodies were used to assess protein loading and transfer. The asterisk denotes nonspecific immunoreactivity.
Analysis of pleiotropies associated with loss of function of lin-61 or mbtr-1:
The class B synMuv genes have roles in many processes in addition to the regulation of vulval development, including the regulation of RNAi sensitivity, restriction of the domains of expression of PGL-1 and lag-2∷gfp, regulation of transgene expression, protection of the genome from mutations, and suppression of vulval defects caused by the mutation mat-3(ku233) (Hsieh et al. 1999; Dufourcq et al. 2002; Unhavaithaya et al. 2002; Garbe et al. 2004; Poulin et al. 2005; Wang et al. 2005; Cui et al. 2006b). While class B synMuv genes act similarly to each other in vulval development, they often do not function similarly in the aforementioned processes (Hsieh et al. 1999; Garbe et al. 2004; Poulin et al. 2005; Wang et al. 2005; Cui et al. 2006b). To understand better the biological roles of lin-61 and mbtr-1 and to compare these genes to the previously described class B synMuv genes, we investigated whether putative null mutations in either or both of these genes results in specific pleiotropies known to be affected by the synMuv genes (Table 3).
TABLE 3.
Phenotypic characterization of lin-61(n3809) and mbtr-1(n4775)
| Phenotype | lin-61(n3809) | mbtr-1(n4775) | mbtr-1(n4775) lin-61(n3809) | lin-35(n745) | lin-15B(n744) |
|---|---|---|---|---|---|
| Class A synMuv | No | No | No | No | No |
| Class B synMuv | Yes | No | Yesa | Yesb | Yesc |
| RNAi hypersensitive | No | No | No | Yesd | Yes |
| Ectopic PGL-1 staining | No | No | No | Yesd | Yes |
| Ectopic lag-2∷gfp expressione | No | No | No | Yes | Yes |
| Mutator | Yes | No | Yes | No | No |
| Tam | No | No | No | Yesf | Yesf |
| Suppressor of mat-3(ku233) | Yes | No | Yes | Yesg | Yesg |
For details concerning how each phenotype was scored, see materials and methods. Data for quantitative assays are presented in the text or in the references cited.
mbtr-1(n4775) did not enhance or suppress the synMuv phenotype of lin-61(n3809); lin-15A(n767) animals.
Ectopic expression refers to misexpression of GFP in the gut.
Hseih et al. (1999).
Mutations in a number of class B synMuv genes have been reported to cause hypersensitivity to RNAi (Wang et al. 2005; Cui et al. 2006b). We found in multiple experiments that lin-61(n3809), mbtr-1(n4775), and mbtr-1(n4775) lin-61(n3809) animals did not show enhanced sensitivity to either hmr-1 or cel-1 RNAi as compared to the wild type (data not shown). In the same experiments, lin-15B(n744), rrf-3(pk1426), and eri-1(mg366) were RNAi hypersensitive, as has previously been reported (Simmer et al. 2002; Kennedy et al. 2004; Wang et al. 2005).
PGL-1 is expressed specifically in the germline of wild-type animals (Kawasaki et al. 1998) and is misexpressed in the soma of animals with loss-of-function mutations in a number of class B synMuv genes, including lin-9, lin-13, lin-15B, lin-35, hpl-2, and dpl-1 (Unhavaithaya et al. 2002; Wang et al. 2005; Cui et al. 2006b). Using antibody staining, we did not observe any PGL-1 misexpression in lin-61(n3809), mbtr-1(n4775), or mbtr-1(n4775) lin-61(n3809) animals. Staining of lin-15B(n744) animals with the same antibody reliably showed misexpresion of PGL-1 in the soma, as previously reported (Wang et al. 2005).
In addition to repression of PGL-1 expression, class B synMuv genes also restrict the domain of lag-2∷gfp expression. A lag-2∷gfp reporter that is expressed in the distal tip cells and vulvas of wild-type hermaphrodites is misexpressed in the gut of hda-1 mutant animals (Dufourcq et al. 2002). RNAi against any of several other synMuv genes also causes lag-2∷gfp misexpression (Poulin et al. 2005). In a lin-61(n3809), mbtr-1(n4775), or mbtr-1(n4775) lin-61(n3809) genetic background, lag-2∷gfp did not display similar misexpression in the gut.
Most class B synMuv genes can prevent the silencing of repetitive transgene arrays in the soma, such as the myo-3∷gfp transgene (Hsieh et al. 1999), and loss of function of any of these genes results in the transgene array modifier (Tam) phenotype. Neither lin-61(n3809), lin-61(n3922), mbtr-1(n4775), nor mbtr-1(n4775) lin-61(n3809) resulted in silencing of an extrachromosomal array carrying myo-3∷gfp. Animals homozygous for lin-35(n745) and the same myo-3∷gfp extrachromosomal array had a Tam phenotype, as had previously been reported (Hsieh et al. 1999).
A role for lin-61 in protecting the genome from DNA instability was identified in a genomewide RNAi screen (Pothof et al. 2003). Using an out-of-frame LacZ reporter that is not expressed in wild-type animals, Pothof et al. (2003) demonstrated that RNAi directed against lin-61 or any of 60 other genes could cause mosaic expression of the transgene, suggesting that loss of function of these genes can result in insertion or deletion mutations that cause the LacZ open reading frame to be in frame with the translational start site. In addition to lin-61, the only other class B gene that was identified in this screen was hda-1. We used the same out-of-frame LacZ reporter to test whether a loss-of-function allele of lin-61 caused mosaic expression of LacZ similar to that caused by RNAi. Mosaic LacZ staining was evident in a significant proportion of lin-61(n3809) animals. Mosaic LacZ staining was also observed in mbtr-1(n4775) lin-61(n3809) animals but not in singly mutant mbtr-1(n4775) animals. In the same experiment, animals homozygous for the transgene alone showed no LacZ staining. In addition, animals homozygous for lin-35(n745) or lin-15B(n744) and the transgene did not show any LacZ staining. Thus, mbtr-1, lin-35, and lin-15B do not share with lin-61 a role in maintaining genome stability.
The partial loss-of-function allele ku233 of the gene mat-3, which encodes a member of the anaphase-promoting complex, causes a vulval defect that can be suppressed by loss of function of the class B synMuv genes lin-35, lin-15B, lin-53, dpl-1, and efl-1 (Garbe et al. 2004). No coding mutations have been reported in mat-3 animals homozygous for the ku233 allele. The vulval defect of mat-3(ku233) animals is likely caused by two adjacent nucleotide changes 400 bp upstream of the mat-3 translational start site, resulting in a 5- to 10-fold reduction in mat-3 RNA levels (Garbe et al. 2004). A likely null allele of lin-35 restores expression of mat-3 to wild-type levels, suggesting that LIN-35 represses transcription of mat-3 (Garbe et al. 2004). As reported for loss-of-function mutations in other class B synMuv genes, lin-61(n3809) suppressed the mat-3(ku233) vulval defect: 58% of mat-3(ku233) (n = 55) animals had abnormal vulvas, as compared to only 3.5% of lin-61(n3809); mat-3(ku233) (n = 57) animals. Thus, LIN-61 might act with LIN-35 to repress transcription of mat-3. Loss of mbtr-1 function did not suppress the mat-3(ku233) vulval defects and did not significantly modify the lin-61(n3809) suppression: 59% of mbtr-1(n4775); mat-3(ku233) (n = 51) had abnormal vulvas and 12% of mbtr-1(n4775) lin-61(n3809); mat-3(ku233) (n = 43) had abnormal vulvas. (In this case, 12% is not statistically different from 3.5% as determined by chi square test; P > 0.1.)
Double mutants between mbtr-1(n4775) and lin-61(n3809) appeared indistinguishable from the lin-61(n3809) single-mutant animals in all of these assays (Table 3). The inability to detect a role for lin-61 in a number of synMuv-regulated pleiotropies therefore is not the result of redundant function with the only other MBT-repeat-containing protein.
LIN-61 is not a core member of the DRM or NuRD-like complexes of synMuv proteins:
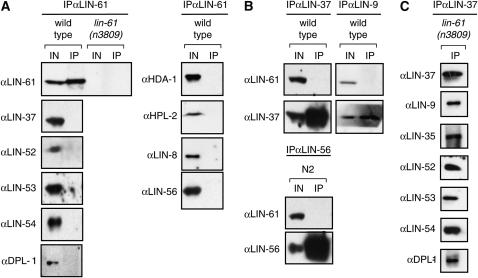
We have recently identified two complexes composed of class B synMuv proteins: the DRM complex, containing eight class B synMuv proteins including LIN-35 Rb and DPL-1 DP, and a NuRD-like complex, containing at least LET-418 Mi2, HDA-1 HDAC1, and LIN-53 RbAp48 (Unhavaithaya et al. 2002; Harrison et al. 2006). We used co-immunoprecipitation experiments to test whether LIN-61 associates with members of either of these two complexes. We demonstrated that although LIN-61 could be precipitated from wild-type but not lin-61(n3809) embryonic extracts using anti-LIN-61 antibodies, DRM complex members failed to co-immunoprecipitate with LIN-61 (Figure 6A). Reciprocally, LIN-61 failed to co-immunoprecipitate with the DRM complex members LIN-37 and LIN-9 (Figure 6B). These data indicate that LIN-61 is not a core component of the DRM complex, although LIN-61 could be weakly associated with the complex or associate at only certain stages of development or in only specific cell types. HDA-1, a component of the NuRD-like complex, similarly failed to co-immunoprecipitate with LIN-61 (Figure 6A), suggesting that LIN-61 also is not a member of the NuRD-like complex. Two class A synMuv proteins, LIN-8 and LIN-56, and the class B synMuv protein HPL-2 also did not co-immunoprecipitate with LIN-61 (Figure 6A).
Figure 6.—
LIN-61 is not a core component of the DRM or NuRD-like complexes. (A) LIN-61 does not immunoprecipitate a number of synMuv proteins, including members of the DRM and NuRD-like complexes. Extracts from either wild-type or lin-61(n3809) mutant embryos (as indicated above the lanes) were precipitated using antibodies against LIN-61 (HM4077). Immunoprecipitates were analyzed using immunoblots with antibodies specific to the antigen indicated at the left. (B) DRM complex members LIN-37 and LIN-9 co-immunoprecipitate a number of class B synMuv proteins (Harrison et al. 2006), but fail to co-immunoprecipitate LIN-61. Extracts from wild-type embryos were precipitated using antibodies indicated above the lanes and immunoblotted with antibodies specific to the antigens indicated at the left. (C) DRM complex assembly and stability is not perturbed in animals lacking lin-61 function. Extracts from lin-61(n3809) mutant embryos were precipitated using antibodies against LIN-37 and immunoblotted with antibodies specific to the antigen indicated at the left. IN, 2% of the input; IP, 100% of the immunoprecipitate.
While our data demonstrate that LIN-61 is not a core component of the DRM complex, it remained possible that LIN-61 could act to modify the formation of this complex. We therefore used co-immunoprecipitation experiments to determine whether the DRM complex is properly formed in lin-61(n3809) mutant animals. Seven members of the DRM complex that co-immunopreciptate with LIN-37 in extracts from wild-type animals also co-immunoprecipitated with LIN-37 in extracts from lin-61(n3809) mutant embryos (Figure 6C), suggesting that LIN-61 function is not required for proper DRM complex formation. However, we cannot preclude the possibility that LIN-61 might affect the activity of the DRM complex through a mechanism distinct from altering complex formation or stability.
DISCUSSION
At least 26 synMuv genes have been identified, 19 of which have been categorized as class B synMuv genes (Ferguson and Horvitz 1985; Hsieh et al. 1999; Thomas et al. 2003; Ceol and Horvitz 2004; Davison et al. 2005; Poulin et al. 2005; Ceol et al. 2006). Genetic and biochemical studies suggest that these class B synMuv genes are not all likely to regulate vulval cell-fate specification together through a single mechanism. For example, the genes hda-1, let-418, and lin-13 have been categorized as class B synMuv genes because loss-of-function mutations in these genes cause a strong synMuv phenotype in combination with loss of function in a class A synMuv gene. However, these loss-of-function mutations cause a weak Muv phenotype as single mutants (Ferguson and Horvitz 1985; von Zelewsky et al. 2000; Dufourcq et al. 2002). By contrast, loss of function of other class B synMuv genes does not cause a Muv phenotype in the absence of a second mutation in a class A or C synMuv gene. We have shown that some synMuv proteins likely function together in a NuRD-like complex that is biochemically distinct from the DRM complex, which contains at least 8 other class B synMuv proteins (Harrison et al. 2006).
In this article, we demonstrate that one of the two C. elegans MBT-repeat-containing proteins, LIN-61, acts with other class B synMuv proteins to regulate vulval development. We further show that the LIN-61 does not share with other class B synMuv proteins a role in RNAi hypersensitivity, pgl-1 and lag-2∷gfp repression, or modification of transgene silencing. Furthermore, LIN-61 has a role in maintaining genome stability not evident for LIN-35 or LIN-15B. These data in combination with the observation that LIN-61 does not co-immunoprecipitate with a large number of synMuv proteins suggest that LIN-61 can function independently of other class B synMuv proteins.
MBT-repeat-containing proteins are not required for C.elegans viability:
C. elegans contains only two predicted proteins containing MBT repeats, LIN-61 and MBTR-1. Both are composed almost exclusively of MBT repeats and lack both the atypical zinc fingers and the SAM domain found in many MBT-repeat-containing proteins in other organisms, including Sex Comb on Midleg, Sfmbt, and lethal (3) malignant brain tumor, proteins required for the viability of Drosophila (Wismar et al. 1995; Bornemann et al. 1996; Klymenko et al. 2006).
By contrast, the C. elegans MBT-repeat-containing proteins are not required for viability. Mutant animals that lack both LIN-61 and MBTR-1 appear superficially wild type. We showed that while LIN-61 is important for the proper regulation of vulval development in sensitized genetic backgrounds, MBTR-1 does not have a similar function. We further demonstrated that in a number of other processes there is no detectable redundancy between MBTR-1 and LIN-61.
LIN-61 is likely involved in transcriptional repression:
Studies of MBT-repeat-containing proteins in other organisms suggest that MBT repeats function in transcriptional repression. Notably, MBT repeats are found in the Drosophila Polycomb-group proteins Sfmbt and SCM (Bornemann et al. 1996; Klymenko et al. 2006), that latter of which is a substoichiometric component of the Polycomb Repression Complex 1 (PRC1) (Shao et al. 1999). PRC1 maintains transcriptional repression of genes by binding to methylated histones (Cao et al. 2002; Czermin et al. 2002; Muller et al. 2002). Genetic analysis of SCM suggests that the MBT domains are likely to be important for protein function (Bornemann et al. 1996, 1998). Human L(3)MBT1 can repress transcription when artificially recruited to promoters, and this transcriptional activity requires the MBT repeats, but not the zinc fingers or the SAM domain (Boccuni et al. 2003). In addition, Drosophila l(3)mbt is required for transcriptional repression of a number of endogenous genes (Lewis et al. 2004). Sequence and structural analyses demonstrate that MBT repeats are similar to Tudor, PWWP, and chromo domains, suggesting that, like these domains, MBT repeats might also bind to modified histones (Maurer-Stroh et al. 2003; Sathyamurthy et al. 2003; Wang et al. 2003). More recently, MBT domains from the human proteins L(3)MBT1 and CGI-72 and the Drosophila protein Sfmbt have been shown to bind histones methylated on specific residues (Kim et al. 2006; Klymenko et al. 2006). Together, these findings suggest that MBT repeats might bind to modified histones and repress transcription. Our observations—that LIN-61 is composed almost exclusively of MBT repeats, that LIN-61 functions with chromatin modifiers in vulval development, and that LIN-61 localizes to condensed chromosomes—suggest that LIN-61 functions in transcriptional repression possibly via direct interaction with modified histones.
lin-61 missense mutations identify residues of MBT repeats likely important for structure and function:
We showed that LIN-61 levels are severely reduced in n3736, n3807, and n3922 mutant animals, suggesting that the residues mutated in these animals might be necessary for proper protein folding and/or stability. Consistent with this hypothesis, we examined the crystal structures of peptides containing MBT repeats (Sathyamurthy et al. 2003; Wang et al. 2003) and observed that the residues mutated by both n3736 and n3807 are likely to be in the β-barrel core region and to be important for interaction with the N-terminal arm of the preceding repeat.
We determined a possible structure for the fourth MBT repeat of LIN-61 using homology modeling based on the crystal structure for the MBT repeats from human L(3)MBT1 (Wang et al. 2003). This structure revealed that n3922 is located in the turn between β-sheets 2 and 3 of the β-barrel core region of MBT repeat 4. In other MBT repeats, this residue is most often a neutrally charged glycine or a positively charged aspartate. The mutation in n3922 changes a glycine to a negatively charged arginine. Perhaps the protein structure can accommodate a neutral or positive charge at this turn, but is disrupted by the incorporation of a negative charge.
The missense mutations n3447 and n3624 interrupt the ability of LIN-61 to properly regulate vulval development despite having wild-type or nearly wild-type protein levels. These residues are not required for protein stability but rather are important for LIN-61 function. The missense mutation in n3447 is in the third MBT repeat and changes a serine to an asparigine. Thus, it is likely that at least the third MBT repeat is important for the function of LIN-61 in regulating vulval development and that, more specifically, the serine at residue 354 is important for function. Because the mutation in n3624 is in the region between the first and second MBT repeats, this portion of LIN-61 is also likely to be important for function.
lin-61 functions separately from the DRM and NuRD-like complexes:
In Drosophila, the MBT-repeat-containing protein l(3)mbt was identified as a substoichiometric component of the Myb–MuvB complex, named to reflect the fact that it contains both Myb and a number of proteins homologous to the C. elegans class B synMuv proteins (Lewis et al. 2004). The Myb–MuvB complex includes a fly Rb protein as well as dE2F2 and dDP and represses transcription of many E2F-responsive genes. l(3)mbt is required to mediate transcriptional repression of only a subset of these targets (Lewis et al. 2004). Thus, l(3)mbt might function with the Myb–MuvB complex only at specific promoters.
The Myb–MuvB complex and another Drosophila complex, the dREAM complex (Korenjak et al. 2004; Lewis et al. 2004), are very similar to the DRM complex that we identified in C. elegans (Harrison et al. 2006). Our co-immunoprecipitation data demonstrate that LIN-61 is not a core component of either the DRM or the NuRD-like complexes. However, since the immunoprecipitates were from embryonic protein extracts, it is possible that LIN-61 associates with these complexes at different stages in development or in specific cell types. It remains possible that, like l(3)mbt, LIN-61 functions with the DRM complex to control specific processes, such as vulval development. Nonetheless, while both l(3)mbt and LIN-61 contain MBT repeats, LIN-61 does not contain the atypical zinc fingers or the SAM domain found in l(3)mbt. The SAM domain is important for protein–protein interaction and might help to mediate the interaction of l(3)mbt with components of the Myb–MuvB complex.
hda-1, which encodes a histone deacetylase component of the NuRD-like complex, was the only synMuv gene that was identified along with lin-61 in the RNAi screen for genes with a role in protecting the genome from instability (Pothof et al. 2003). However, we detected no association between HDA-1 and LIN-61 in co-immunoprecipitation experiments. It is possible, as discussed above, that these proteins interact transiently or interact only in a subset of cells. Alternatively, HDA-1 and LIN-61 might have distinct roles in maintaining the stability of the genome. An additional 59 genes were identified in the RNAi screen (Pothof et al. 2003) and might function with HDA-1, LIN-61, or both in this capacity .
Our analysis of the pleiotropies associated with loss-of-function mutations in lin-61 further suggests that in some biological processes lin-61 functions separately from other class B synMuv genes, including components of the DRM complex. A loss-of-function mutation in lin-61 causes a class B synMuv phenotype and genomic instability and suppresses the vulval defect of mat-3(ku233) mutant animals. Putative null alleles of lin-35 and lin-15B share with lin-61 mutants only the ability to cause a class B synMuv phenotype and to suppress mat-3(ku233). However, both lin-35(n745) and lin-15B(n744) share a number of other pleiotropies that do not appear to involve the function of either MBT-repeat-containing protein, including RNAi hypersensitivity, PGL-1 somatic misexpression, the Tam phenotype, and ectopic lag-2∷gfp expression in the gut (Table 3). These data suggest that in many biological functions lin-61 does not act with lin-35 or lin-15B, although lin-35 and lin-15B might act together in these functions.
We have shown that while lin-61 shares with other class B genes the ability to regulate vulval development redundantly with the class A synMuv genes, it does not share roles in many other biological processes with other class B synMuv genes. LIN-61 is also distinguished from other class B synMuv proteins by co-immunoprecipitation experiments, which show that LIN-61 is not a core component of the DRM or NuRD-like complexes. On the basis of these observations, we suggest that other MBT-repeat-containing proteins, such as Polycomb-group proteins, act with Rb-containing and HDAC-containing complexes to control certain biological processes but act independently of these complexes to regulate other biological processes.
Acknowledgments
We thank Paul Sternberg for generously providing us with the original lin-61 allele, sy223; Daniel Denning, Niels Ringstad, Adam Saffer, and Hillel Schwartz for comments about the manuscript; Beth Castor, Na An, and Andrew Hellman for technical assistance; members of the Horvitz laboratory for construction of the deletion library; John Doll for the initial observation that LIN-61 shares sequence similarity with a Polycomb-group protein; Dawn Wendell and Hillel Schwartz for isolation of n3687, n3807, n3809, and n3922; Alan Coulson for cosmid clones; Yuji Kohara for cDNA clones; and the C. elegans Genome Sequencing Consortium for genomic sequence. M.M.H. was a Howard Hughes Predoctoral Fellow. H.R.H. is the David H. Koch Professor of Biology at the Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health grant GM24663.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. DQ904352.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, E. C., X. Lu and H. R. Horvitz, 2006. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133: 2695–2704. [DOI] [PubMed] [Google Scholar]
- Arai, S., and T. Miyazaki, 2005. Impaired maturation of myeloid progenitors in mice lacking novel Polycomb group protein MBT-1. EMBO J. 24: 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel, G. J., S. G. Clark and H. R. Horvitz, 1990. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509. [DOI] [PubMed] [Google Scholar]
- Boccuni, P., D. MacGrogan, J. M. Scandura and S. D. Nimer, 2003. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J. Biol. Chem. 278: 15412–15420. [DOI] [PubMed] [Google Scholar]
- Bornemann, D., E. Miller and J. Simon, 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122: 1621–1630. [DOI] [PubMed] [Google Scholar]
- Bornemann, D., E. Miller and J. Simon, 1998. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics 150: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchenau, P., J. Hodgson, H. Strutt and D. J. Arndt-Jovin, 1998. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J. Cell Biol. 141: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage et al., 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7: 461–473. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6: 563–576. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., F. Stegmeier, M. M. Harrison and H. R. Horvitz, 2006. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics 173: 709–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. G., X. Lu and H. R. Horvitz, 1994. The Caenorhabditis elegans locus lin–15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham, V., C. Bedet, K. Monier, S. Schott, M. Karali et al., 2006. The C. elegans HP1 homologue HPL-2 and the LIN-13 zinc finger protein form a complex implicated in vulval development. Dev. Biol. 297: 308–322. [DOI] [PubMed] [Google Scholar]
- Couteau, F., F. Guerry, F. Muller and F. Palladino, 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., J. Chen, T. R. Myers, B. J. Hwang, P. W. Sternberg et al., 2006. a SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10: 667–672. [DOI] [PubMed] [Google Scholar]
- Cui, M., E. B. Kim and M. Han, 2006. b Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof et al., 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Dahiya, A., S. Wong, S. Gonzalo, M. Gavin and D. C. Dean, 2001. Linking the Rb and polycomb pathways. Mol. Cell 8: 557–569. [DOI] [PubMed] [Google Scholar]
- Davison, E. M., M. M. Harrison, A. J. Walhout, M. Vidal and H. R. Horvitz, 2005. lin-8, which antagonizes Caenorhabditis elegans Ras-mediated vulval induction, encodes a novel nuclear protein that interacts with the LIN-35 Rb protein. Genetics 171: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq, P., M. Victor, F. Gay, D. Calvo, J. Hodgkin et al., 2002. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol. 22: 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M. L., and D. L. Riddle, 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266: 385–395. [DOI] [PubMed] [Google Scholar]
- Eisenmann, D. M., J. N. Maloof, J. S. Simske, C. Kenyon and S. K. Kim, 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin–39 during Caenorhabditis elegans vulval development. Development 125: 3667–3680. [DOI] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney, M., and G. Ruvkun, 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63: 895–905. [DOI] [PubMed] [Google Scholar]
- Garbe, D., J. B. Doto and M. V. Sundaram, 2004. Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics 167: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, J. E., H. C. Korswagen and D. M. Eisenmann, 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I. S., P. W. Sternberg and H. R. Horvitz, 1983. The lin–12 locus specifies cell fate in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Guenther, C., and G. Garriga, 1996. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development 122: 3509–3518. [DOI] [PubMed] [Google Scholar]
- Hajnal, A., C. W. Whitfield and S. K. Kim, 1997. Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev. 11: 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., R. V. Aroian and P. W. Sternberg, 1990. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126: 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane, 1999. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Harrison, M. M., C. J Ceol, X. Lu and H. R. Horvitz, 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA 103: 16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, R. K., 1978. Crossover suppressors and balanced recessive lethals in Caenorhabditis elegans. Genetics 88: 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R. J., and P. W. Sternberg, 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358: 470–476. [DOI] [PubMed] [Google Scholar]
- Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al., 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., and J. Ahringer, 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kawasaki, I., Y. H. Shim, J. Kirchner, J. Kaminker, W. B. Wood et al., 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94: 635–645. [DOI] [PubMed] [Google Scholar]
- Kennedy, S., D. Wang and G. Ruvkun, 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649. [DOI] [PubMed] [Google Scholar]
- Kim, C. A., M. R. Sawaya, D. Cascio, W. Kim and J. U. Bowie, 2005. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J. Biol. Chem. 280: 27769–27775. [DOI] [PubMed] [Google Scholar]
- Kim, J., J. Daniel, A. Espejo, A. Lake, M. Krishna et al., 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7: 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko, T., B. Papp, W. Fischle, T. Köcher, M. Schelder et al., 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine binding activities. Genes Dev. 20: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, H., S. Matsui, T. Hirota, S. Takebayashi, K. Okumura et al., 1999. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene 18: 3799–3809. [DOI] [PubMed] [Google Scholar]
- Korenjak, M., B. Taylor-Harding, U. K. Binne, J. S. Satterlee, O. Stevaux et al., 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193. [DOI] [PubMed] [Google Scholar]
- Lewis, P. W., E. L. Beall, T. C. Fleischer, D. Georlette, A. J. Link et al., 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18: 2929–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., and H. R. Horvitz, 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991. [DOI] [PubMed] [Google Scholar]
- Margueron, R., P. Trojer and D. Reinberg, 2005. The key to development: Interpreting the histone code? Curr. Opin. Genet. Dev. 15: 163–176. [DOI] [PubMed] [Google Scholar]
- Markus, J., S. Feikova, M. Sramko, L. Wolff and J. Bies, 2003. Proliferation-linked expression of the novel murine gene m4mbt encoding a nuclear zinc finger protein with four mbt domains. Gene 319: 117–126. [DOI] [PubMed] [Google Scholar]
- Mathies, L. D., S. T. Henderson and J. Kimble, 2003. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development 130: 2881–2892. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber et al., 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28: 69–74. [DOI] [PubMed] [Google Scholar]
- Melendez, A., and I. Greenwald, 2000. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics 155: 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta et al., 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Myers, T. R., and I. Greenwald, 2005. lin-35 Rb acts in the major hypodermis to oppose ras-mediated vulval induction in C. elegans. Dev. Cell 8: 117–123. [DOI] [PubMed] [Google Scholar]
- Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston and Y. Nakatani, 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136. [DOI] [PubMed] [Google Scholar]
- Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock et al., 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17: 6683–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof, J., G. van Haaften, K. Thijssen, R. S. Kamath, A. G. Fraser et al., 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 17: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin, G., Y. Dong, A. G. Fraser, N. A. Hopper and J. Ahringer, 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24: 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Sathyamurthy, A., M. D. Allen, A. G. Murzin and M. Bycroft, 2003. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2). J. Biol. Chem. 278: 46968–46973. [DOI] [PubMed] [Google Scholar]
- Saurin, A. J., C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte et al., 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D. C., K. Ayyanathan, D. Negorev, G. G. Maul and F. J. Rauscher, III, 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting and P. Bork, 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu et al., 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46. [DOI] [PubMed] [Google Scholar]
- Siegfried, K. R., and J. Kimble, 2002. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development 129: 443–453. [DOI] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet et al., 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., and H. R. Horvitz, 1986. Pattern formation during vulval development in C. elegans. Cell 44: 761–772. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., and H. R. Horvitz, 1989. The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell 58: 679–693. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and J. G. White, 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78: 577–597. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., and H. R. Horvitz, 1999. The gene lin-36 acts cell autonomously in the lin–35 Rb pathway. Development 126: 3449–3459. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., C. J. Ceol, H. T. Schwartz and H. R. Horvitz, 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164: 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomotsune, D., Y. Takihara, J. Berger, D. Duhl, S. Joo et al., 1999. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation 65: 229–239. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama et al., 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002. [DOI] [PubMed] [Google Scholar]
- Usui, H., T. Ichikawa, K. Kobayashi and T. Kumanishi, 2000. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene 248: 127–135. [DOI] [PubMed] [Google Scholar]
- von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal et al., 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127: 5277–5284. [DOI] [PubMed] [Google Scholar]
- Wang, D., S. Kennedy, D. Conte, Jr., J. K. Kim, H. W. Gabel et al., 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597. [DOI] [PubMed] [Google Scholar]
- Wang, W. K., V. Tereshko, P. Boccuni, D. MacGrogan, S. D. Nimer et al., 2003. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure 11: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wismar, J., T. Loffler, N. Habtemichael, O. Vef, M. Geissen et al., 1995. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech. Dev. 53: 141–154. [DOI] [PubMed] [Google Scholar]
- Yochem, J., K. Weston and I. Greenwald, 1988. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 335: 547–550. [DOI] [PubMed] [Google Scholar]