Abstract
We determined the relative positions of the tandem-repeat molecular cytogenetic marker B77, translocation breakpoints, and telosome arms in Gossypium hirsutum cytogenetic stocks by fluorescence in situ hybridization (FISH) analysis of meiotic quadrivalents in 16 single and 2 double translocation heterozygotes and five monotelodisomics. Results delimited the B77 FISH locus to the right arm of the D-subgenome chromosome 14 (14R) and the short arm (14sh), respectively. By equating 14R with 14sh and 14L (left) with 14Lo (long), the findings established a unified nomenclature for the arms of chromosome 14. Previously reported chromosome 14 arm locations were confirmed for four of the five translocations involving chromosome 14, namely NT1L-14L (2780), NT2R-14R (2B-1), NT14L-23R (2777), and NT14R-24R (2781), whereas the location of breakpoint T6L-14L was not confirmed and was reassigned to arm 14R. When used as a probe on Southern blots, the B77 signal was associated with a terminus of the D-subgenome RFLP linkage group (LG) D04 by linkage analysis of an interspecific F2 population, now known to be chromosome 20. However, additional codominant DNA marker information in the affected region excluded the B77 polymorphism detected by Southern blot hybridization from chromosome 20 and, indeed, from the remainder of the genome.
FLUORESCENCE in situ hybridization (FISH) is a powerful technique for physical localization of DNA sequences to individual chromosomes and subchromosomal regions. Low-copy and unique DNA sequences have been successfully mapped to chromosomes in humans, animals, and plants (Ashley et al. 1994; Lemieux et al. 1994; Dong and Quick 1995). Repetitive sequences, such as ribosomal DNA sequences, have been detected and mapped in a large range of plant species, such as cotton, wheat, tomato, and others (Jiang and Gill 1994; Ji et al. 1999a, 2004). When combined with meiotic analysis of cytogenetic stocks such as translocations, in situ hybridization provides a more powerful tool for integrative mapping (Price et al. 1990; Crane et al. 1993; Stelly et al. 1996; Wang et al. 2006).
FISH to pachytene bivalents offers numerous advantages over FISH to diakinesis and metaphase chromosomes and has been utilized broadly in plants for gene localization and other studies (Zhong et al. 1999; Kim et al. 2005). Studies on cotton pachytene bivalents have been limited (Mursal and Endrizzi 1976) and, to our knowledge, there have been no reports of systematic research based on FISH to pachytene bivalents.
Cotton (Gossypium hirsutum L.) has long been regarded as an allotetraploid (2n = 4x = 52) with Ah and Dh subgenomes (Skovsted 1934; Beasley 1940, 1942). Subgenomic affiliations of the 26 chromosomes of G. hirsutum were determined by meiotic analysis of interspecific hybrids between G. hirsutum translocation homozygotes and diploid species. Individual chromosomes were identified and numbered according to meiotic analysis of crosses among translocation lines (Menzel and Brown 1978; Brown 1980; Brown et al. 1981). Chromosomes of A- and D-subgenomes were designated as chromosomes 1–13 and 14–26, respectively. On the whole, A-subgenome chromosomes are discernibly larger than D-subgenome chromosomes (Skovsted 1934), but chromosomes of the two subgenomes overlap in size, precluding reliable subgenomic assignment on the basis of size alone (Kimber 1961).
A high-resolution genetic recombination map of sequence-tagged sites for Gossypium genomes was coalesced into 26 linkage groups (LGs), 20 of which were assigned to individual chromosomes (Rong et al. 2004). The remaining six groups were eventually assigned to individual chromosomes by meiotic in situ hybridization analysis of related translocation stocks with linkage-group-specific bacterial artificial chromosome clones (Wang et al. 2006), thus leading to complete identification of the 26 cotton chromosomes. As a result, the 13 homeologous chromosome pairs have also been completely established, which were supported by numerous prior studies, including conventional cytogenetics (Endrizzi et al. 1985), molecular cytogenetics (Crane et al. 1993), and linkage mapping of various molecular markers (Reinisch et al. 1994; Lacape et al. 2003; Nguyen et al. 2004; Rong et al. 2004; Han et al. 2006).
A total of 62 translocations have been maintained in the Cotton Cytogenetics Collection (Stelly 1993). The breakpoints affect 25 of the 26 chromosomes, and most have been localized to an arm and mapped relative to each other and their respective centromeres (Menzel et al. 1985). Menzel et al. (1985) arbitrarily designated the two arms of each chromosome as “right” (R) and “left” (L). Telosomes were designated as “short” (sh) and “long” (Lo) according to their relative size (Endrizzi and Ramsay 1979, 1980; Endrizzi et al. 1985). The correspondence of “L” and “R” to “Lo” and “sh” designations was reported for a number of the chromosomes (Menzel et al. 1985), but a limited investigation subsequently revealed that some assignments were incorrect (Stelly et al. 1996). Thus, two partially independent systems of nomenclature exist for the chromosome arms of G. hirsutum, one based on translocations and the other on telosomes. Moreover, the modest map of genes governing conventional traits, the extensive molecular marker map (Reinisch et al. 1994; Rong et al. 2004), and the breakpoint map (Menzel et al. 1985) are currently independent and thus need to be integrated.
A restriction fragment length polymorphism (RFLP) detected using a tandemly repeated sequence, B77 (572 bp), as a probe, was previously loosely associated (19 cM, no flaking markers) with a single locus in D-subgenome LG D04 (Zhao et al. 1998), which was later assigned to chromosome 20 (Rong et al. 2004). Interesting features of B77 include its subgenomic specificity, tandem nature, and genetic variability (Zhao et al. 1998). The large size of the B77 locus (∼0.5 Mb) presented a facile opportunity to jointly investigate B77, LG D04, and the use of FISH for integrative mapping. At the outset of the study reported here, we endeavored (1) to identify which chromosome bears B77 and thus to identify LG D04; (2) to further localize B77 with respect to subchromosomal regions (arm and segment) defined by translocation and/or telosome breakpoints; and (3) to test previous arm assignments of cytogenetic landmarks for their respective chromosomes.
MATERIALS AND METHODS
Plant material:
Single and double reciprocal translocation heterozygotes used in molecular-meiotic analyses were developed from translocation lines maintained in the Cotton Cytogenetics Collection at Texas A&M University (Stelly 1993). Single translocation heterozygotes (NTs) were produced by crossing the translocation homozygotes to the genetic standard line TM-1. Double translocation heterozygotes (dNTs) were produced by intercrossing the translocation homozygotes. Monotelodisomic translocation heterozygotes (TeNTs) were produced by intermating monotelodisomics and chromosomally related translocation homozygotes, the latter serving as pollen parent. Progeny were screened phenotypically and meiotically to identify the TeNT aneuploids. A segmental duplication-deficiency (dp-df), which was produced by outcrossing NT14R-24R to TM-1 and was shown by FISH to be deficient for 14R (Ji et al. 1999b), was also used in this study. The NTs, dNTs, and TeNTs used in this study are listed in Table 1.
TABLE 1.
Single, double, and monotelodisomic translocation heterozygotes used in this study
|
NTs
|
|||
|---|---|---|---|
| Line no.a | Name | dNTs | TeNTs |
| 2780 | NT1L-14L | dNT[7L-18R, 20L-22R] | Te14LoNT1L-14L |
| 2B-1 | NT2R-14R | dNT[14L-23R, 19R-24R] | Te14LoNT2R-14R |
| AZ-7 | NT6L-14Rb | Te14LoNT6L-14L | |
| 4659 | NT7L-18R | Te14LoNT14R-24R | |
| 2767 | NT15R-16R | Te14LoNT14L-23R | |
| 6340 | NT9L-17Rb | ||
| 2772 | NT9R-20L | ||
| 2870 | NT9L-25 | ||
| 4675 | NT10L-21L | ||
| 2925 | NT13R-19R | ||
| 2777 | NT14L-23R | ||
| 2781 | NT14R-24R | ||
| SL15 | NT15R-20R | ||
| 7-3F | NT19R-21R | ||
| 2786 | NT19R-24R | ||
| DP4 | NT20L-22R | ||
From Brown et al. (1981).
Previous designation as NT6L-14L was found to be incorrect (see text).
Chromosome preparation:
Meiotic chromosome spreads were prepared according to the procedures of Crane et al. (1993) with some modifications. Briefly, upon removal of calyx and corolla, meiotic buds are fixed in two or more changes of 2:1 (v/v) acetone:acetic acid with 1% polyvinylpyrrolidone (Sigma, St. Louis; Mr 40,000) at room temperature for 24 hr, washed in distilled water, and stored in distilled water for several hours to a couple of weeks or in 70% ethanol for several months at 4°. Buds were individually macerated in 1% acetocarmine and screened for metaphase I (MI) under a microscope; selected macerates were transferred to a clean slide and squashed under a silicolized coverslip at 75–80° on a temperature-controlled hot plate. Slides were frozen in liquid nitrogen and then stored in a freezer at −135°.
Probe labeling and in situ hybridization:
A biotin-labeled probe was prepared by nick translation (BRL BioNick kit) of a plasmid containing B77 element, a 572-bp clone from a tandemly repeated (∼900 times) sequence of G. barbadense (2n = 4x = 52; Zhao et al. 1998). The probe mixture contained probe DNA (final concentration 1.2 ng/μl), Escherichia coli DNA (final concentration 240 ng/μl), 50% deionized formamide, 20% dextran sulfate, and 2× SSC. The procedures of in situ hybridization and signal detection followed Ji et al. (1997).
Fluorescence microscopy:
Slides were screened and photographed with an Olympus AX-70 microscope equipped with UV and blue and green excitation filter sets. Photographs were taken on Fujicolor 400 professional film. Prints were digitally scanned, processed, and reproduced.
Chromosomal and subchromosomal localization:
The physical association of B77 FISH signal with translocation-bearing multivalents was used to discern the chromosomal location of B77. Positions of B77 FISH signals on metaphase I multivalents were used to subchromosomally localize B77 relative to the translocation breakpoints. The numbers and positions of FISH signals on TeNT IVs were used to define relationships among B77, translocation breakpoints, and telosome-defined arms. The requisite interpretations were based on principles detailed previously (Price et al. 1990; Crane et al. 1993; Stelly et al. 1996).
Detection of major nucleolar organizing regions:
Differentially bright propidium iodide (PI; red) fluorescence of nucleolar organizing regions (NORs) in somatic and meiotic metaphase chromatin [when doubly stained with DAPI (4′, 6-diamidino-2-2-phenylindole) and PI] often allows for facile detection of major NORs (Hanson et al. 1996; Ji et al. 1997). At metaphase (when most chromosome regions are well contracted), the NORs are brighter red than other chromatin. Therefore, the three major NORs of G. hirsutum (Bergey et al. 1989) can be detected by their differential PI fluorescence. In this study, we used this technique for dual detection of the major NORs and B77 in meiocytes probed only with B77 (fluorescein isothiocyanate detection) and stained with DAPI and PI.
RESULTS
Chromosomal localization of B77:
The RFLP locus B77 was previously mapped by linkage analysis to a terminus of linkage group D04, ∼19 cM away from the nearest marker G1016 of this linkage group (Zhao et al. 1998). Although the LOD score was statistically significant, the lack of flanking markers renders the linkage of terminal markers tenuous. This uncertainty is made even greater in that the marker was “dominant”; i.e., segregation was for presence vs. absence of the B77 allele with no ability to detect heterozygotes. Association of B77 with a D-subgenome chromosome was further confirmed by FISH (Zhao et al. 1998). LG D04 was recently assigned to chromosome 20 (Lacape et al. 2003; Rong et al. 2004). Accordingly, we first subjected B77 to molecular-meiotic tests by hybridization to NTs and dNTs involving chromosome 20. However, in metaphase I spreads of NT9R-20L, NT15R-20R, NT20L-22R, and dNT[7L-18R, 20L-22R], the pair of B77 signals invariably occurred on one bivalent per cell, and the respective IV was devoid of FISH signal (data not shown), indicating that B77 is neither in chromosome 20 nor in other D-subgenome chromosomes 15, 18, and 22. To expedite subsequent identification efforts, we hybridized B77 to metaphase I spreads of dNT[14L-23R, 19R-24R], which involved four of the remaining D-subgenome chromosomes, i.e., 14, 19, 23, and 24. FISH signals were observed, both of which were associated with one IV/cell (Figure 1A), indicating that one of the four chromosomes involved in the translocations carries the B77 sequence. The presence of a NOR on chromosome 23 was then used to discriminate between the two IVs. Following PI staining, the three differentially PI-fluorescing major NORs were observed on two bivalents and one of the two IVs (Figure 1, B and C). The B77 FITC signals were invariably associated with the NOR-bearing IV, i.e., with NT14L-23R IV, suggesting that B77 lies in chromosome 14 or 23, not 19 or 24.
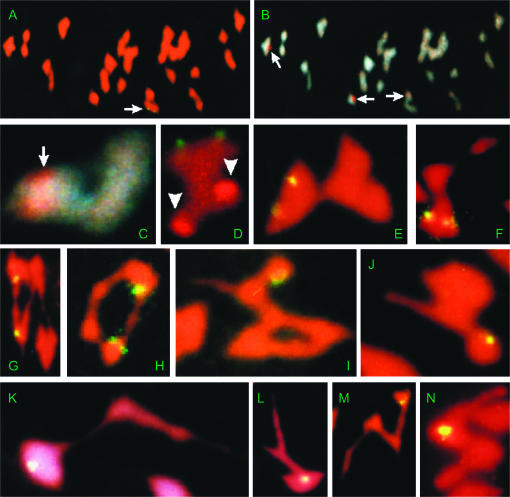
Figure 1.—
Meiotic MI chromosome spreads of single reciprocal translocation heterozygotes (NTs), double reciprocal translocation heterozygotes (dNTs), and monotelodisomic translocation heterozygotes (TeNTs) from G. hirsutum after fluorescence in situ hybridization of clone B77. (A–D) dNT[14L-23R, 19R-24R], where A–C are from the same chromosome spread. (A) A pair of hybridization signals were associated with just one (arrow) of the two IVs. (B and C) DAPI staining of the same spread at low and high magnifications, respectively. (B) The IV associated with FISH signals is shown here to be associated with one of the three major NOR sites (arrows) on chromosomes 9, 16, and 23, each demarcated by a red spot from differentially brighter PI staining (Hanson et al. 1996). (C) High magnification of IV only (rotated image). (D) NT14L-23R “barbell”-IV from a different meiotic spread, showing the pair of green B77 FISH signals to be on the “exterior” and opposite the major NOR in chromosome 23 (brighter red; arrowheads). (E) NT1L-14L “barbell”-IV, showing the pair of B77 FISH signals (yellow) on the IV “exterior.” (F) NT2R-14R “frying pan”-IV showing the pair of B77 FISH signals in the “proximal” IV region. (G) A pair of FISH signals on the same side of an NT6L-14L alternate “ring”-IV. (H) A pair of FISH signals on the same side of an NT14R-24R adjacent “ring”-IV. (I) A single unpaired FISH signal on the “knob” (unpaired arm at the nontelosomic end of the chain) of a Te14LoNT1L-14L “wheel barrow”-IV. (J) A single unpaired FISH signal on the “knob” of a Te14LoNT14L-23R “wheel barrow”-IV. (K and L) A single unpaired FISH signal was in the “exterior” of the Te14LoNT6L-14R “frying pan”-IVs. (M) A single unpaired FISH signal on the “knob” of a Te14LoNT14R-24R “N-shaped chain”-IV. (N) A single unpaired FISH signal on a Te14LoNT2R-14R “U-shaped chain”-IV.
To deduce whether B77 is located in chromosome 14 or 23, we first determined the relative physical positions of B77 and NOR sites on NT14L-23R IVs. We observed that they were located on opposite sides of NT14L-23R IVs (Figure 1D), indicating that B77 is in chromosome 14, not 23. To further test this inference, we determined whether or not B77 was associated with IVs of the other four translocations involving chromosome 14 that are maintained in the Cotton Cytogenetics Collection. Association was detected in all cases, i.e., for NT1L-14L (Figure 1E), NT2R-14R (Figure 1F), NT6L-14L (Figure 1G), and NT14R-24R (Figure 1H). Supporting evidences also came from nonassociation of B77 with the remaining D-subgenome chromosomes 16, 17, 21, and 25 (data not shown) except 26, for which no translocation stock is available in the current collection. Given that translocations constitute the basis for the existing nomenclature for chromosomes of G. hirsutum (Menzel and Brown 1978; Brown 1980; Brown et al. 1981), these data demonstrated that the B77 FISH site is in chromosome 14.
Subchromosomal localization of B77 by analysis of NTs:
Metaphase I spreads of translocation heterozygotes were analyzed for each of the five translocations that affect chromosome 14. Two of the five NTs rarely form interstitial chiasmata, whereas the other three NTs have at least one breakpoint recombinationally distal from their respective centromere and therefore tend to form interstitial chiasmata, which lead to “barbell”-IVs and other types of IVs (Menzel et al. 1985). Interstitial chiasmata constrain the shape of IVs and provide “reference points” useful to subchromosomal localization by molecular-meiotic methods (Stelly et al. 1996).
“Barbell”-IVs were observed in NT1L-14L and NT14L-23R microsporocytes. The B77 signals on “barbell”-IVs of NT14L-23R (Figure 1D) and NT1L-14L (Figure 1E) were located on the “exterior” of the IVs, indicating that B77 is located in the arms opposite the T1L-14L and T14L-23R breakpoints in chromosome 14. According to the map of breakpoints (Menzel et al. 1985), both the T1L-14L and the T14L-23R chromosome 14 breakpoints are in the left arm (14L). Therefore, the B77 cluster must be located in the right arm of chromosome 14 (14R).
“Frying pan”-IVs were observed in NT2R-14R microsporocytes. Such IVs result when crossing over occurs in just one of the two interstitial regions. In all the observed “frying pan”-IVs, the B77 signals were always located on the inner side of the “pan” (Figure 1F), indicating that the crossing over occurs in the interstitial region in 14R and that B77 is “proximal” to the respective T2R-14R breakpoint. These results indicated that B77 is located in the interstitial region between the centromere of chromosome 14 and the T2R-14R breakpoint in chromosome 14. According to the map of breakpoints (Menzel et al. 1985), the T2R-14R breakpoint is in the right arm of chromosome 14 (14R). Therefore, B77 must be in the right arm of chromosome 14 (14R). This conclusion was consistent with the results from NT1L-14L and NT14L-23R, indicating that the relative arm assignments by Menzel et al. (1985) were internally consistent for chromosome 14 breakpoints of T1L-14L, T2R-14R, and T14L-23R.
“Ring”-IVs were observed in both NT6L-14L (alternate; Figure 1G) and NT14R-24R (adjacent; Figure 1H) metaphase I cells. In each type of NT, the B77 signals were located on just one side of the “ring”-IVs, indicating that the B77 site must be “opposite” or “distal” to the respective translocation breakpoint in chromosome 14. These data indicated that if (and only if) the T6L-14L and T14R-24R breakpoints are indeed in separate arms, as indicated by Menzel et al. (1985), then B77 must be “distal” to one breakpoint and “opposite” the other. Our findings, which indicate that B77 is proximal to neither breakpoint, are concordant with the previous report that both breakpoints are near the chromosome 14 centromere (Menzel et al. 1985). However, our NT-IV data do not indicate the arm in which B77 is located relative to these two translocation breakpoints. For this purpose, we tried a newer procedure, based on molecular-meiotic analysis of monotelodisomic translocation heterozygotes (TeNTs).
Subchromosomal localization of B77 by analysis of TeNTs:
The relationship between a breakpoint and a related telosome can be deduced from several types of “critical configurations” in which one or both interstitial regions are chiasmate (Menzel et al. 1985; Stelly et al. 1996). “Wheel barrow”-shaped TeNT IVs were observed in Te14LoNT1L-14L (Figure 1I) and Te14LoNT14L-23R metaphase I cells (Figure 1J). This type of TeNT IV configuration is critical in that it can arise only when the telosome is homologous to the breakpoint-bearing arm; it is formed if both “distal” segments, the single disomic opposite arm, and one or both of the “interstitial” regions are chiasmate (Figure 2, A and C). Therefore, the T1L-14L and T14L-23R breakpoints in chromosome 14 must be located in the arm homologous to the telosome, i.e., the long arm of chromosome 14 (14Lo). The FISH signals were observed on the highly contracted hemizygous end of the chain, which looks like a terminal “knob” from Figure 1, I and J and corresponds to region “a” in Figure 2C. The TeNT results concomitantly indicated that B77 is in arm 14sh, that 14sh = 14R, that the arm designations of the chromosome 14 breakpoints in T1L-14L and T14L-23R are internally consistent, and that B77 lies in the arm “opposite” the respective breakpoints, i.e., 14Lo.
Figure 2.—
Diagrams of monotelodisomic translocation heterozygote quadrivalents (TeNT IVs) and their corresponding MI configurations, following the nomenclature of Menzel et al. (1985) for chromosome segments. (A) A pachytene representation of a TeNT IV with the telosome and the breakpoint affecting the same arm. (B) A pachytene representation of a TeNT IV with the telosome and the breakpoint affecting different arms. (C) An MI “wheel barrow”-IV produced from A with segments b, c, d, e, and f being chiasmate. (D) An MI “frying pan”-IV produced from B with segments a, c, d, and f being chiasmate. (E) An MI “chain”-IV produced from A with segments b, c, and d being chiasmate. (F) An MI “chain”-IV produced from B with segments a, c, and d being chiasmate.
“Frying pan”-shaped TeNT IVs were observed in Te14LoNT6L-14L (previously designated) metaphase I cells (Figure 1, K and L). This type of TeNT IV configuration is critical in that it can arise only when the telosome is opposite the breakpoint-bearing arm; it is formed if both opposite arms, the single “distal” segment, and the single “interstitial” region are chiasmate (Figure 2, B and D). Therefore, the T6L-14L breakpoint in chromosome 14 must be located in the arm opposite the telosome, i.e., the short arm of chromosome 14 (14sh). A single FISH signal was asymmetrically associated with the “pan” of each “frying pan” IV, indicating that the B77 cluster is in the hemizygous distal segment corresponding to segment “B” in Figure 2D, i.e., 14L, according to the previous arm assignments by Menzel et al. (1985). According to assignments by Menzel et al. (1985), the FISH result would indicate that B77 was in 14L and that 14sh = 14L, which is contrary to our above inference based on NT1L-14L and NT14L-23R. As shown below, arm assignments of NT2R-14R and NT14R-24R are correct for the respective chromosome 14 breakpoints. Therefore, the result indicated that the previous assignment of the T6L-14L breakpoint to arm 14L by Menzel et al. (1985) is internally inconsistent with the other four chromosome 14 translocations. On the basis of these observations, we correct the previous assignment of T6L-14L (Menzel et al. 1985) by redesignating it as T6L-14R.
“Chain”-IVs were observed in Te14LoNT14R-24R, and the FISH signal occurred at a single site on the “knob” (Figure 1M). “Chain” configurations lack interstitial chiasmata and, in themselves, do not define whether a telosome is homologous to a breakpoint-bearing arm or not (Figure 2, E and F). “Chain” configurations thus constitute “noncritical configurations.” The hemizygosity and position of the FISH signal concordantly indicated, however, that B77 lies in the arm opposite the telosome (14Lo), i.e., short arm of chromosome 14 (14sh), a finding consistent with the results from the previously mentioned TeNT critical configurations. The TeNT-based analyses indicated that B77 lies distal to or opposite the chromosome 14 breakpoint, but could not distinguish between these possibilities without additional information, e.g., interstitial chiasmata.
“Chain” IVs of Te14LoNT2R-14R were also observed to bear the B77 signal at just one location per IV, indicating hemizygosity (Figure 1N). The position of the signal, which corresponds to region “E” of Figure 2, B and F, indicated that the B77 locus is proximal to the breakpoint and that the breakpoint is in the hemizygous arm (14sh) and, thus, that 14R = 14sh. The findings for Te14LoNT2R-14R confirm the previous assignment by Menzel et al. (1985) of the T2R-14R breakpoint to arm 14R.
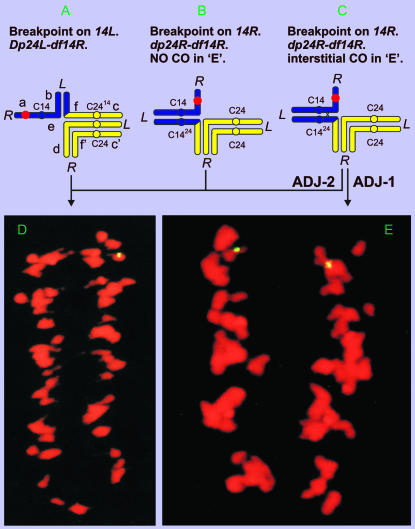
Confirmation of T14R-24R chromosome 14 breakpoint on arm 14R:
In lieu of critical configurations in Te14LoNT14R-24R meiocytes, we FISHed B77 to segmental dp-df stocks to test the arm assignment of the chromosome 14 breakpoint in T14R-24R. A dp-df plant from NT14-24 was shown by molecular cytogenetics to be hemizygous for B77 (Ji et al. 1999b). Such a plant would be adjacent-2 dp24L-df14R, if the T14-24 chromosome 14 breakpoint were in 14L (Figure 3A), or adjacent-1 dp24R-df14R, if the T14-24 chromosome 14 breakpoint were in 14R (Figure 3, B and C). The expected distribution of B77 signals at anaphase I (AI) is quite different for the two types of dp-df's. For the former, only one of the two meiosis I products would bear the B77 FISH signal, unless sister centromeres were to separate precociously (Figure 3, A and D). For the latter, one or both meiotic I products would bear the B77 signal, depending on recombination in the interstitial segment (Figure 3, B and E). The equal distribution at meiosis I of a hemizygous FISH locus (Figure 3E) of the involved dp-df can occur only under the following situations: (i) the T14R-24R breakpoint in chromosome 14 must be located in the arm carrying the FISH site, i.e., 14R; (ii) crossing over must occur in the interstitial region proximal to this breakpoint; and (iii) homologous centromeres must undergo adjacent-1 disjunction. The equal distribution of the FISH signal was observed in ∼5% of anaphase I–metaphase II spreads of the dp-df stock that was deficient for 14R, strongly indicating that the T14-24 breakpoint in chromosome 14 is located to arm 14R and thus confirming the arm assignment by Menzel et al. (1985). If the breakpoint were in the opposite arm, precocious sister-centromere separation would be required at anaphase I to produce signal in both meiotic I products. Moreover, nondisjunction would have been expected to arise occasionally from such precociously separated sister centromeres, whereas none was observed on the basis of the B77 signal.
Figure 3.—
Relationship between A–C pachytene representations of the FISH signal-adorned dp-df's (hemizygous for FISH site) from NT14-24 under two scenarios: the chromosome 14 breakpoint is located in 14R or 14L, respectively, and D and E photomicrographs of AI produced from the corresponding dp-df's. (A) NT14-24 breakpoint in chromosome 14 located on 14L [adjacent (ADJ)-2 dp24L-df14R]. (B) NT14-24 breakpoint in chromosome 14 located on 14R (ADJ-1 dp24R-df14R) with no crossing over (CO) in the interstitial region “e.” (C) NT14-24 breakpoint in chromosome 14 located on 14R (ADJ-1 dp24R-df14R) with an interstitial CO in region “e.” (D) AI spread of the dp-df's showing that the FISH signal goes to one pole, which is not diagnostic of the breakpoint's arm location. (E) AI spread of the dp-df showing the segregation of FISH signal to two poles, diagnostically indicating the T14-24 breakpoint in chromosome 14 is on 14R.
DISCUSSION
In this study, we localized the B77 FISH locus to the right arm of chromosome 14 (14R), demonstrated that 14R is the short arm of chromosome 14 (14sh), and confirmed the previous arm designations (R vs. L) for all the chromosome 14 translocation breakpoints, except T6L-14L, for which the breakpoint was redesignated to 14R.
Relationship between RFLP and physical maps:
In constructing a detailed RFLP map of cotton, Reinisch et al. (1994) associated a linkage group (∼149 cM) with chromosome 14 by deficiency analysis using an interspecific F1 monotelodisomic lacking the G. hirsutum chromosome arm 14sh. A recent study concatenated LG U09 to chromosome 14, bringing its length up to ∼165 cM (Rong et al. 2004). Chromosome 14 was initially hypothesized to be homeologous to the A-subgenome chromosome 2 on the basis of monosomic plant description (Endrizzi et al. 1985). Their homeology was further confirmed by duplicated DNA markers on both chromosomes (Reinisch et al. 1994; Lacape et al. 2003; Rong et al. 2004). The tandem repeat family (B77), isolated from G. barbadense, was mapped to the terminal of LG D04, now assigned to chromosome 20 (Zhao et al. 1998; Rong et al. 2004). However, our findings cytologically associate B77 FISH signals with chromosome 14. Given the strong evidence supporting the identity of chromosome 14, we proposed the following hypothesis for the conflicting data.
We hypothesized that B77 was incorrectly assigned to LG chromosome 20, previously known as LG D04. As noted above, B77 was mapped to a terminus of the linkage group, ∼19 cM away from the nearest marker (Zhao et al. 1998). Although the linkage was statistically significant, the lack of flanking markers renders the linkage of terminal markers speculative. This uncertainty is made even greater in that the marker was “dominant”; i.e., segregation was for presence vs. absence of the B77 allele with no ability to detect heterozygotes. A few additional markers were recently mapped to the terminus of the same linkage group; B77 was slightly closer (∼16 cM) to G1016, but did not fit with the other nearby markers or anywhere else in the genome (A. H. Paterson, unpublished data). The best fit for B77 is still chromosome 20, but it is no longer statistically significant. One possible explanation may be that there are small groups of B77 repeats at multiple locations in the genome, in addition to the primary array on chromosome 14. The polymorphism tenuously associated with chromosome 20 may be confounded with B77 alleles resulting from loss of restriction sites at some other locus as well. In any case, we must conclude that if there is a B77 locus on chromosome 20, it is composed of a relatively small number of elements and that the primary locus appears to be on chromosome 14.
Position of B77 relative to translocation breakpoints in chromosome 14:
The association of B77 with chromosome 14 was revealed by association of B77 FISH signal with the IVs of five different euploid chromosome 14 translocation heterozygotes and the five respective Te14Lo-bearing monotelodisomic translocation heterozygotes. B77 was subchromosomally localized by more detailed analysis of the position of B77 signal(s) on the respective multivalents and allowed placement of B77 relative to the centromere and respective breakpoints. For the three translocations with high frequencies of interstitial chiasmata (T1L-14L, T2R-14R, and T14L-23R), the heterozygotes were sufficient for mapping, whereas, for the other two (T6L-14L, T14R-24R), the monotelodisomic translocation heterozygotes and chromosomal segmental dp-df's were used for mapping.
The NT and TeNT data for these five translocations indicated that the relative arm assignments by Menzel et al. (1985) were correct for at least three of the five chromosome 14 breakpoints (T1L-14L, T2R-14R, and T14L-23R), but incorrect for T6L-14L, which was redesignated as T6L-14R. The incorrect assignment of the T6L-14L breakpoint to 14L was also indicated as a footnote in a previous report, but data were not shown (Menzel and Dougherty 1987). The analysis of the segregation of B77 on the hemizygous segment of a dp-df stock (dp24R-df14R) supported the original assignment of the T14R-24R chromosome 14 breakpoint to arm 14R (Menzel et al. 1985), but did not support its later reassignment to 14L, which was noted as a footnote in a previous report (Menzel and Dougherty 1987). Our data and those of Menzel et al. (1985) concordantly indicate that the T6L-14R and T14R-24R breakpoints are recombinationally very close to the centromeres. Two of the five translocation breakpoints in chromosome 14, i.e., the T1L-14L and T14L-23R breakpoints, affect arm 14L, whereas the other three, i.e., the T2R-14R, T6L-14R, and T14R-24R breakpoints, affect the opposing arm, 14R. In addition, our data show that 14sh = 14R and 14Lo = 14L.
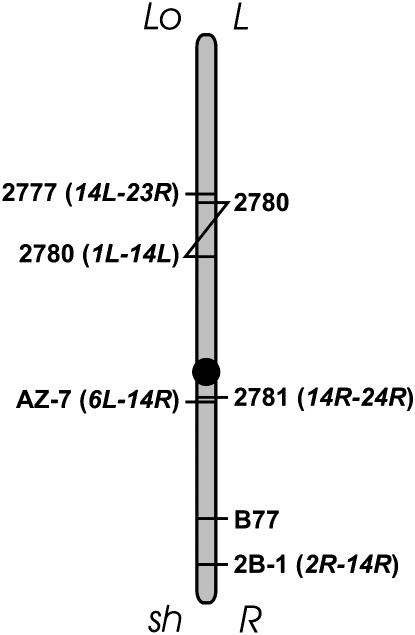
Our analysis allows placement of B77 relative to the translocation breakpoints, providing a seminal integrated map. B77 was mapped to 14R, which was opposite the two translocation breakpoints in 14L. Analysis of NT2R-14R and Te14LoNT2R-14R placed B77 in the interstitial region of NT2R-14R in chromosome 14, i.e., between the chromosome 14 centromere and the T2R-14R breakpoint. Analyses of NT6L-14R, NT14R-24R, Te14LoNT6L-14R, and Te14LoNT14R-24R placed B77 distal to T6L-14R and T14R-24R breakpoints. According to Menzel et al. (1985), T6L-14R and T14R-24R breakpoints in chromosome 14 were ∼4.5 and 2.7 cM from the centromere, respectively. High-resolution mapping with a dp-df stock indicated that the latter was 2.57 cM (our unpublished data). Our analyses place the breakpoints in the same arm; thus the data from Menzel et al. (1985) and the map from the dp-df suggest that the T6L-14R breakpoint in chromosome 14 may be farther from the centromere relative to the T14R-24R breakpoint in chromosome 14. The combined data suggest that B77 is between the NT2R-14R breakpoint (2B-1) and the NT6L-14R breakpoint in 14R or 14sh. A revised map for chromosome 14 breakpoints and B77 is shown in Figure 4.
Figure 4.—
Chromosome map of chromosome 14 breakpoints and B77. L, left arm; R, right arm; sh, short arm; Lo, long arm.
Perspectives:
Our findings have several ramifications. By anchoring B77 to the chromosome 14 map, we have rendered it a useful molecular genetic/cytogenetic marker for that specific chromosome and segment. Most significantly, the results demonstrate the feasibility of integrative mapping, where one or more unknown(s) can be mapped relative to other types of known loci, centromeres, translocation breakpoints, and telomeres (Reyes-Valdés and Stelly 1995; Reyes-Valdés et al. 1996). A skeletal map of molecular cytogenetic loci will facilitate subsequent mapping of repetitive sequences, and mature integrated maps will improve genome comparisons, interspecific introgression, analysis of transformant gene activity (position effects), and rapid assessment of karyotypic variation in wild germplasm.
Acknowledgments
The authors gratefully acknowledge the support of this work in part by the U. S. Department of Agriculture (USDA) grant 91-37300-8819 and Texas Advanced Technology Research Program (TATRP) grant 999902148 for cloning, recombination mapping, and characterization of B77; USDA grant NR1 CGP 92-37300-7655 (classical cytogenetics) for development of cytogenetic stocks; TATRP grant 999902090 (molecular cytogenetics) for physical mapping by FISH; and a Tom Slick Senior Graduate Fellowship at Texas A&M University awarded to the senior author.
The authors acknowledge and express their appreciation of the late H. James Price for his unending enthusiasm for science, collaboration, and the positive things in life.
References
- Ashley, T., T. Ried and D. C. Ward, 1994. Detection of nondisjunction and recombination in meiotic and post meiotic cells from XY-Sxr (XY, Tp(Y)1Ct) mice using multicolor fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 91: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley, J. O., 1940. The origin of American tetraploid Gossypium species. Am. Nat. 74: 285–286. [Google Scholar]
- Beasley, J. O., 1942. Meiotic chromosome behavior in species, species hybrids, haploids and induced polyploids of Gossypium. Genetics 27: 25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey, D. R., D. M. Stelly, H. J. Price and T. D. McKnight, 1989. In situ hybridization of biotinylated DNA probes to cotton meiotic chromosomes. Stain. Technol. 64: 25–37. [DOI] [PubMed] [Google Scholar]
- Brown, M. S., 1980. The identification of chromosomes of Gossypium hirsutum L. by means of translocations. J. Hered. 71: 266–274. [Google Scholar]
- Brown, M. S., M. Y. Menzel, C. A. Hasenkampf and S. Naqi, 1981. Chromosome configurations and orientations in 58 heterozygous translocations in Gossypium hirsutum. J. Hered. 72: 161–168. [Google Scholar]
- Crane, C. F., H. J. Price, D. M. Stelly and D. G. Czeschin, Jr., 1993. Identification of a homeologous chromosome pair by in situ DNA hybridization to ribosomal RNA loci in meiotic chromosomes of cotton (Gossypium hirsutum). Genome 36: 1015–1022. [DOI] [PubMed] [Google Scholar]
- Dong, H., and J. S. Quick, 1995. Detection of a 2.6 kb single/low copy DNA sequence on chromosomes of wheat (Triticum aestivum) and rye (Secale cereale) by fluorescence in situ hybridization. Genome 38: 246–249. [DOI] [PubMed] [Google Scholar]
- Endrizzi, J. E., and G. Ramsay, 1979. Monosomes and telosomes for 18 of the 26 chromosomes of Gossypium hirsutum. Can. J. Genet. Cytol. 21: 531–536. [Google Scholar]
- Endrizzi, J. E., and G. Ramsay, 1980. Identification of ten chromosome deficiencies of cotton. J. Hered. 71: 45–48. [Google Scholar]
- Endrizzi, J. E., E. L. Turcotte and R. J. Kohel, 1985. Genetics, cytology, and evolution of Gossypium. Adv. Genet. 23: 271–375. [Google Scholar]
- Han, Z. G., C. B. Wang, X. L. Song, W. Z. Guo, J. Y. Gou et al., 2006. Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor. Appl. Genet. 112: 430–439. [DOI] [PubMed] [Google Scholar]
- Hanson, R. E, M. N. Islam-Faridi, E. A. Percival, C. F. Crane, Y. Ji et al., 1996. Distribution of 5S and 18S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105: 55–61. [DOI] [PubMed] [Google Scholar]
- Ji, Y., W. A. Raska, T. D. McKnight, M. N. Islam-Faridi, C. F. Crane et al., 1997. Use of meiotic FISH for identification of a new monosome in Gossypium hirsutum L. Genome 40: 34–40. [DOI] [PubMed] [Google Scholar]
- Ji, Y., M. De Donato, C. F. Crane, W. A. Raska, M. N. Islam-Faridi et al., 1999. a New ribosomal RNA gene locations in Gossypium hirsutum mapped by meiotic FISH. Chromosoma 108: 200–207. [DOI] [PubMed] [Google Scholar]
- Ji, Y., W. A. Raska, M. De Donato, M. N. Islam-Faridi, J. H. Price et al., 1999. b Identification and distinction among segmental duplication-deficiencies by fluorescence in situ hybridization (FISH)-adorned multivalent analysis. Genome 42: 763–771. [Google Scholar]
- Ji, Y., R. Pertuze and R. T. Chetelat, 2004. Genome differentiation by GISH in interspecific and intergeneric hybrids of tomato and related nightshades. Chromosome Res. 12: 107–116. [DOI] [PubMed] [Google Scholar]
- Jiang, J., and B. S. Gill, 1994. New 18S.26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 103: 179–185. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., M. N. Islam-Faridi, P. E. Klein, D. M. Stelly, H. J. Price et al., 2005. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimber, G., 1961. Basis of the diploid-like meiotic behavior of polyploid cotton. Nature 191: 98–100. [Google Scholar]
- Lacape, J. M., T. B. Nguyen, S. Thibivilliers, B. Bojinov, B. Courtois et al., 2003. A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46: 612–626. [DOI] [PubMed] [Google Scholar]
- Lemieux, N., B. Malfoy, R. Fetni, M. Muleris, N. Vogt et al., 1994. In situ hybridization approach at infragenic level on metaphase chromosomes. Cytogenet. Cell Genet. 66: 107–112. [DOI] [PubMed] [Google Scholar]
- Menzel, M. Y., and M. S. Brown, 1978. Reciprocal chromosome translocations in Gossypium hirsutum. J. Hered. 69: 383–390. [Google Scholar]
- Menzel, M. Y., and B. J. Dougherty, 1987. Transmission of duplication-deficiencies from cotton translocations is unrelated to map lengths of the unbalanced segments. Genetics 116: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, M. Y., K. L. Richmond and B. J. Dougherty, 1985. A chromosome translocation breakpoint map of the Gossypium hirsutum genome. J. Hered. 76: 406–414. [Google Scholar]
- Mursal, I. E. J., and J. E. Endrizzi, 1976. Re-examination of diploid-like meiotic behavior of polyploid cotton. Theor. Appl. Genet. 47: 171–178. [DOI] [PubMed] [Google Scholar]
- Nguyen, T. B., M. Giband, P. Brottier, A. M. Risterucci and J. M. Lacape, 2004. Wide coverage of the tetraploid cotton genome using newly developed microsatellite markers. Theor. Appl. Genet. 109: 167–175. [DOI] [PubMed] [Google Scholar]
- Price, H. J., D. M. Stelly, T. D. McKnight, C. F. Scheuring, D. Raska et al., 1990. Molecular cytogenetic mapping of a nucleolar organizer region in cotton. J. Hered. 81: 365–370. [Google Scholar]
- Reinisch, A. J., J. Dong, C. L. Brubaker, D. M. Stelly, J. F. Wendel et al., 1994. A detailed RFLP map of cotton, Gossypium hirsutum × Gossypium barbadense: chromosome organization and evolution in a disomic polyploid genome. Genetics 138: 829–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Valdés, M. H., and D. M. Stelly, 1995. A maximum likelihood algorithm for genome mapping of cytogenetic loci from meiotic configuration data. Proc. Natl. Acad. Sci. USA 92: 9824–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Valdés, M. H., Y. Ji, C. F. Crane, J. F. Taylor, M. N. Islam-Faridi et al., 1996. ISH-facilitated analysis of meiotic bivalent pairing. Genome 39: 784–792. [DOI] [PubMed] [Google Scholar]
- Rong, J. K., C. Abbey, J. E. Bowers, C. L. Brubaker, C. Chang et al., 2004. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166: 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovsted, A., 1934. Cytological studies of cotton. II. Two interspecific hybrids between Asiatic and New World cotton. J. Genet. 28: 407–424. [Google Scholar]
- Stelly, D. M., 1993. Interfacing cytogenetics with the cotton genome effort, pp. 1545–1550 in Proceedings of the Beltwide Cotton Conferences, edited by D. J. Herber and D. A. Richter. National Cotton Council of America, Memphis.
- Stelly, D. M., C. F. Crane, R. E. Hanson, T. D. McKnight, and H. J. Price, 1996. Molecular-meiotic analysis of cotton, pp. 99–111 in Methods of Genome Analysis in Plants: Their Merits and Pitfalls, edited by P. P. Jauhar. CRC Press, Boca Raton, FL.
- Wang, K., X. L. Song, Z. G. Han, W. Z. Guo, J. Z. Yu et al., 2006. Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor. Appl. Genet. 113: 73–80. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Y. Ji, X. Ding, D. M. Stelly and A. H. Paterson, 1998. Macromolecular organization and genetic mapping of a rapidly-evolving chromosome-specific tandem repeat family (B77) in cotton (Gossypium). Plant Mol. Biol. 38: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Zhong, X. B., J. Bodeau, P. F. Fransz, V. M. Williamson, A. van Kammen et al., 1999. FISH to meiotic pachytene chromosomes of tomato locates the root knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor. Appl. Genet. 98: 365–370. [Google Scholar]