Abstract
Three types of sequence variations—single-nucleotide polymorphisms (SNPs), insertions and deletions (indels), and short tandem repeats (STRs)—have been extensively reported in mammalian genomes. In this study, we discovered a novel type of sequence variation, i.e., multiple-nucleotide length polymorphisms (MNLPs) in bovine UCN3 (Urocortin 3) and its receptor CRHR2 (corticotropin-releasing hormone receptor 2) genes. Both MNLPs featured involvement of multiple-nucleotide length polymorphisms (5–18 bases), low sequence identity, and 1.7- to 11-fold changes in promoter activity between two alleles. Therefore, this novel genetic complexity would contribute significantly to the evolutionary, functional, and phenotypic complexity of genomes within or among species.
UROCORTIN 3 (UCN3) and corticotropin-releasing hormone receptor 2 (CRHR2) are members of the corticotropin-releasing hormone (CRH) family of peptides. Overall, the CRH family of peptides plays diverse roles in physiological, developmental, and behavioral events, such as activation of the hypothalamic–pituitary–adrenal axis, modulation of cardiovascular, immunological, and gastrointestinal functions, stimulation of anxiety-related behavior, control of locomotor activity, and regulation of food intake and energy balance (Richard et al. 2002; Bernier 2006). In this study, we discovered a novel type of sequence variation, multiple-nucleotide length polymorphisms (MNLPs) in the promoter regions of both bovine UCN3 and CRHR2 genes. The MNLPs involved 5–18 nucleotides in a biallelic manner, but showed length polymorphisms and low sequence identity between two alleles. The promoter activity assay detected the difference in gene expression activity between two MNLP alleles in each of these two genes. These results provide evidence that this novel type of sequence variation aids in the understanding of the evolutionary, functional, and phenotypic complexity of genomes within or among species.
MATERIALS AND METHODS
Promoter determination and MNLP recognition:
The genomic organization of bovine UCN3 and CRHR2 was determined by aligning a compiled cDNA sequence using three expressed sequence tags (BC114855, DV838893, and AB240582) with a genomic DNA contig (AAFC03043460) for the former gene and aligning the human mRNA sequence (NM_001883) with a bovine genomic DNA contig (AAFC03056271) for the latter gene. Partial promoter sequences, primer binding sites, and amplified regions of both genes are shown in Figures 1A and 2A, respectively. Initial sequencing of amplified PCR products on six Wagyu × Limousin F1 bulls showed that some animals had a lead portion of clean and readable sequences for both products, followed by a region with a series of double peaks until the sequence end. The currently known bovine sequence was used as a reference to deduce the sequence for the second peak. Interestingly, sequence deduction indicated that these animals were heterozygous with two alleles of MNLPs: a 5-bp/10-bp heterozygote for the UCN3 gene (Figure 1B) and a 12-bp/18-bp heterozygote for the CRHR2 gene (Figure 2B), respectively. We expected that each of these MNLPs should segregate in a Wagyu × Limousin F2 reference population (Jiang et al. 2005), which were genotyped by direct sequencing of the PCR products as described above. In addition to the MNLPs, a total of nine single-nucleotide polymorphisms (SNPs) were also detected in the amplified promoter regions of both UCN3 and CRHR2 genes, including five (AAFC03043460.1: g.8208C > T, g.8265C > T, g.8287T > C, g.8412A > G, and g.8426T > A) in the former gene and four (AAFC03056271.1: g.33704A > G, g.33803C > T, g.34007C > A, and g.34017G > C) in the latter gene, respectively (Jiang et al. 2006).
Figure 1.—
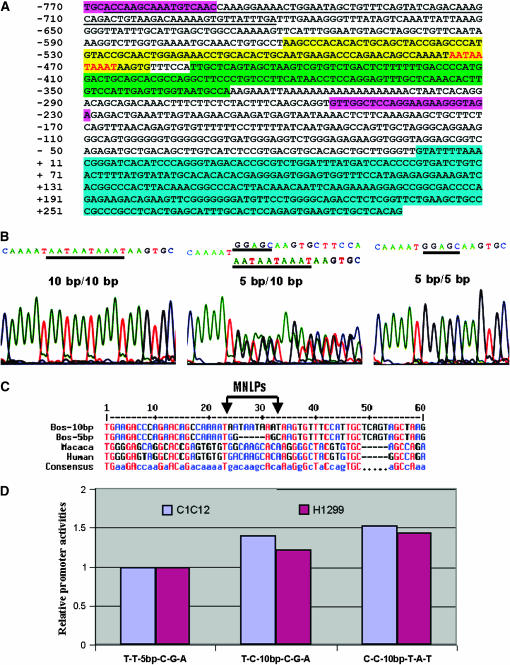
Characterization of MNLPs in the bovine UCN3 gene. (A) Nucleotide sequence of the upstream region of the bovine UCN3 gene. This sequence corresponds to a partial 5′-flanking region and exon 1. The sequence of noncoding exon 1 is highlighted in turquoise. The putative transcription site is numbered as +1. Primer binding sites are highlighted in pink. The LINE/L1 element is underlined. Two SINE elements—tRNA-Glu and BovA—are highlighted in yellow and bright green, respectively. The MNLP site is marked in red. (B) Sequence electropherograms for a 10-bp/10-bp homozygote, a 10-bp/5-bp heterozygote, and a 5-bp/5-bp homozygote. (C) Alignments of MNLPs and flanking sequences among cattle, macaca, and human. (D) MNLP effects on promoter activity of the bovine UCN3 gene.
Figure 2.—
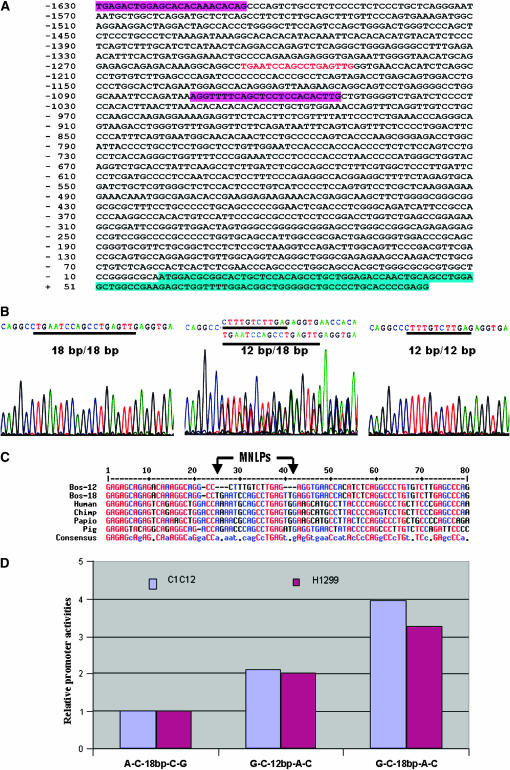
Characterization of MNLPs in the bovine CRHR2 gene. (A) Nucleotide sequence of the upstream region of the bovine CRHR2 gene. This sequence corresponds to a partial 5′-flanking region and exon 1. The sequence of coding exon 1 is highlighted in bright green. The putative transcription/translation site is numbered as +1. Primer binding sites are highlighted in pink. The MNLP site is marked in red. (B) Sequence electropherograms for an 18-bp/18-bp homozygote, an 18-bp/12-bp heterozygote, and a 12-bp/12-bp homozygote. (C) Alignments of MNLPs and flanking sequences among human, chimpanzee, olive baboon, and pig. (D) MNLP effects on promoter activity of the bovine CRHR2 gene.
Characterization of MNLPs in both genes:
The Hardy–Weinberg equilibrium was examined for both MNLPs identified in the bovine UCN3 and CRHR2 genes using Pearson's chi-square test. The RepeatMasker program was used to screen both amplified sequences for interspersed repeats (http://www.repeatmasker.org/). Each amplified sequence was initially aligned with the promoter sequence of the orthologous gene in other species at a pairwise level for detecting conserved segments using a “BLAST 2 SEQUENCES” tool developed by the National Center for Biotechnology Information. Once sequence similarity exists between cattle and other species, multiple-species sequence alignment was performed using the MultAlin program (http://prodes.toulouse.inra.fr/multalin/) for allele conservation characterization. As both MNLPs occur in the promoter region of the bovine UCN3 and CRHR2 genes, the MatInspector program (http://www.gsf.de/) was used to search for gain/loss of binding sites for transcriptional regulatory elements in both genes.
Promoter activity assay and statistical analysis:
The effects of MNLPs and SNPs on promoter activities were examined using a Dual-Luciferase Report Assay system (Promega, Madison, WI). The forward and reverse gene-specific primers (Figures 1A and 2A) were then engineered with a 5′ BglII and a 3′ HindIII site plus a 5′ tail of CTTC, respectively, for directional cloning into the BglII/HindIII site of pGL3-basic (Promega). Three types of haplotypes at all six polymorphic sites in the UCN3 gene, T-T-5 bp-C-G-A, T-C-10 bp-C-G-A, and C-C-10 bp-T-A-T, and three types of haplotypes at all five polymorphic sites in the CRHR2 gene, A-C-18 bp-C-G, G-C-12 bp-A-C, and G-C-18 bp-A-C, were prepared for the promoter constructs. Mouse myoblast C2C12 and human lung carcinoma H1299 cells were transfected with each of the recombinant pGL3 plasmids containing the constructs described above. pRL-CMV plasmid was also cotransfected into C2C12 and H1299 cells as a transfection control. The mouse cells were harvested after 40 hr, while the human cells were collected 28 hr post-transfection and firefly luciferase and Renilla luciferase activities were measured with the Dual Luciferase Reporter Assay system according to the manufacturer's protocol. Light emission was quantified with a Multilabel Counter (Wallace 1420 Victor 2, Turku, Finland). Triplicate data were collected from three independent experiments. The ratios of firefly luciferase activity to Renilla luciferase activity were calculated and used to compare the differences in activity among haplotypes. The data were analyzed using the SAS MIXED procedure (Version 9.1; SAS Institute, Cary, NC) on the basis of the linear model
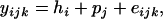 |
where 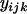 is the ratio of the ith haplotype in the kth replication of the jth experiments,
is the ratio of the ith haplotype in the kth replication of the jth experiments, 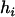 is the fixed effect of the ith haplotype,
is the fixed effect of the ith haplotype, 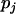 is the random effect of the jth experiment, and
is the random effect of the jth experiment, and 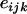 is the residual. The overall mean is not included in the model, to ensure identifiability of all unknown parameters in the model (i.e., all parameters are estimated uniquely).
is the residual. The overall mean is not included in the model, to ensure identifiability of all unknown parameters in the model (i.e., all parameters are estimated uniquely).
RESULTS AND DISCUSSION
Population genetics of MNLPs in cattle:
Sequencing of PCR products on 240 F2 progeny clearly demonstrated segregation of two MNLP alleles in each of these two genes. Pearson's chi-square test revealed that the MNLP genotype distributions in both genes fell into Hardy–Weinberg equilibrium (for the UCN3 gene, χ2 = 1.589, P > 0.05; and for the CRHR2 gene, χ2 = 1.362, P > 0.05). In addition, the segregation status of these MNLP alleles in the two genes was also examined in the Wagyu breed of cattle. Among 25 full-blood Wagyu bulls, the frequencies were 0.32 for the 5-bp allele and 0.68 for the 10-bp allele in the UCN3 gene and 0.42 for the 12-bp allele and 0.58 for the 18-bp alleles in the CRHR2 gene, respectively. In recent years, DNA sequencing has emerged as the most sensitive and automated method to identify genetic variants (Nickerson et al. 1997). When detecting SNPs by sequencing on heterozygous individuals, a double peak occurs only at the polymorphic site. However, when dealing with MNLPs, insertions and deletions (indels), and short tandem repeats (STRs) by sequencing on heterozygous individuals, a series of double peaks appear from the start of the polymorphic site to the end of the sequence. Therefore, much attention needs to be paid to recognize these three types of polymorphisms. In particular, the MNLPs could be interpreted by many investigators as “noisy” or “messy” sequences and would therefore be ignored. This could explain why no groups have previously reported such MNLPs in any known living organisms.
Common and different features of MNLPs in both bovine UCN3 and CRHR2 genes:
Two homozygous alleles of AAFC03043460.1, g.8272-8281AATAATAAAT > GGAGC in the promoter region of the bovine UCN3 gene, and two homozygous alleles of AAFC03056271.1, g.33947-33964TGAATCCAGCCTGAGTTG > CTTTGTCTTGAG in the promoter region of the bovine CRHR2 gene, are shown in Figures 1B and 2B, respectively. Both MNLPs featured involvement of multiple nucleotides (5–18 bases) and length polymorphisms between two alleles (a 10-bp allele vs. a 5-bp allele in UCN3 and an 18-bp allele vs. a 12-bp allele in CRHR2, respectively). The two MNLP alleles in both genes are distant from each other due to low sequence identity except that both alleles in the CRHR2 gene share a core motif of TGAG (Figure 2B). The RepeatMasker program (http://www.repeatmasker.org/) revealed that the MNLP site in the UCN3 gene is located near the end of a SINE/tRNA-Glu element (Figure1A), while no repetitive sequences were detected in the promoter region of the CRHR2 gene (Figure 2A). Due to the nature of the repetitive DNA sequence, the amplified promoter sequence of bovine UCN3 had relatively low sequence similarity to the orthologous promoters in human and macaca (Figure 1C). However, the promoter region of the CRHR2 gene was relatively conserved with a high sequence similarity among cattle, humans, chimpanzees, olive baboons, and pigs (Figure 2C).
Significant impact of haplotypes on promoter activity:
The haplotypes of promoter constructs of the bovine UCN3 gene significantly affected expression activity in both C2C12 (F = 15.33, P < 0.0001) and H1299 cells (F = 10.49, P = 0.0002). Overall, the construct with T-T-5 bp-C-G-A yielded the lowest activity compared to those with T-C-10 bp-C-G-A and C-C-10 bp-T-A-T in both cell lines. When the ratio between firefly and Renilla luciferase activity for the T-T-5 bp-C-G-A haplotype was normalized to 1, promoter activities in the T-T-10 bp-C-G-A and C-C-10 bp-T-A-T haplotypes were 39 and 52% greater in the C2C12 cell line, respectively, and 21 and 43% greater in the H1299 cell line, respectively (Figure 1D). The haplotype effect on promoter activity was more pronounced among the three constructs in the bovine CRHR2 gene (in the C2C12 cells, F = 243.39, P < 0.0001; and in the H1299 cells, F = 29.26, P < 0.001). When the ratio of firefly to Renilla luciferase activity was normalized to 1 for the A-C-18 bp-C-G haplotype, promoter activities of the G-C-12 bp-A-C and G-C-18 bp-A-C haplotypes were 113 and 294% higher in C2C12 cells and 101 and 225% greater in H1299 cells, respectively (Figure 2D).
Gain/loss of potential regulatory binding sites:
Screening of nearly an entire promoter region (5148 bp for UCN3 and 3319 bp for CRHR2) using the MatInspector program (Quandt et al. 1995) revealed that MNLPs cause presence/absence of a total of 15 provisional regulatory binding sites in both genes. These regulatory binding sites are for autoimmune regulator, avian C-type LTR TATA box, Brn-3 (POU-IV protein class), glucocorticoid receptor, hepatic nuclear factor 1, homeobox transcription factor Gsh-1, LIM-homeodomain transcription factor, PBX–HOXA9 binding site, progesterone receptor binding site, prostate-specific homeodomain protein NKX3.1, and Yin and Yang 1 repressor sites in the UCN3 gene and for cAMP-response element-binding protein, HepG2-specific P450 2C factor-1, Pax-6 paired domain binding site, and ribonucleoprotein-associated zinc finger protein MOK-2 in the CRHR2 gene, respectively. In general, the short-length MNLP alleles have a higher chance to lose transcriptional regulatory binding sites than the long-length MNLP alleles so that they have potentials to reduce promoter activity more significantly.
Contribution of MNLPs to genome complexity:
As indicated above, two haplotypes, T-C-10 bp-C-G-A and C-C-10 bp-T-A-T in the UCN3 gene, differed at four polymorphic sites, which resulted in 13 and 22% lower promoter activity or 3.25 and 5.50% activity per SNP for the former haplotype in the C2C12 and H1299 cells, respectively. However, the T-T-5 bp-C-G-A had 52 and 43% less activity in both cell lines compared to the C-C-10 bp-T-A-T construct. If we partition the reductions on the basis of the average effect per SNP estimated above, the short-length (5 bp) MNLP allele was estimated to cause a loss of 35.75% activity in the C2C12 cells and of 15.5% activity in the H1299 cells. In the CRHR2 gene, the construct G-C-18 bp-A-C produced the highest promoter activity among three haplotypes (Figure 2D). The short-length (12 bp) MNLP allele caused a loss of 181 and 124% activity (G-C-12 bp-A-C vs. G-C-18 bp-A-C), while each SNP was estimated to lead to an average reduction of activity by 98 and 75% (A-C-18 bp-C-G vs. G-C-18 bp-A-C) in the C2C12 and H1299 cells, respectively. These results indicated that MNLP alleles are capable of causing 1.7- to 11-fold more changes in promoter activity than SNP alleles. Therefore, MNLPs discovered in this study contribute to a major source for understanding the evolutionary, functional, and phenotypic complexity of genomes within or among species.
Potential mechanisms involved in formation of the MNLPs:
Replication slippage, recombination, and their interaction have been proposed as mutational mechanisms to explain changes in the number of short tandem repeats (Li et al. 2002), while the 5-methylcytosine deamination reactions have been widely recognized as causes of the higher level of C ↔ T (G ↔ A) SNPs, particularly at CpG dinucleotides in mammals (Holliday and Grigg 1993). However, these mutational mechanisms hardly explain the origin of the MNLPs discovered in this study. Initially, we thought that repetitive elements would cause the MNLP in the bovine UCN3 promoter region, because it is located between two SINE elements, tRNA-Glu and BovA (Figure 1A): one allele might be the ancient flanking sequence attached to the former element, while the other allele might be the flanking sequence associated with the latter element, or vice versa. However, the mechanism would never be applicable to the MNLP alleles located in the promoter region of the CRHR2 gene, because the region does not contain any kind of repetitive elements (Figure 2A). We observed that the 5-bp allele in UCN3 (Figure 1C) and the 18-bp allele in CRHR2 (Figure 2C) had a relatively higher sequence similarity to the sequences in other existing mammals. Therefore, we assume that the other two alleles, the 10-bp allele in UCN3 (Figure 1C) and the 12-bp allele in CRHR2 (Figure 2C), might be relics of common ancestral alleles or their evolutionary intermediates.
Acknowledgments
The authors appreciate the assistance of Michael MacNeil, U.S. Department of Agriculture–Agricultural Research Service, Miles City, Montana, in providing DNA for this research. This work was supported by National Institutes of Health grant RO1 CA104470 to N.S.M. and by Merial Ltd. Animal Genomics Research Fund 13Z-3031-3446 to Z.J.
References
- Bernier, N. J., 2006. The corticotropin-releasing factor system as a mediator of the appetite-suppressing effects of stress in fish. Gen. Comp. Endocrinol. 146: 45–55. [DOI] [PubMed] [Google Scholar]
- Holliday, R., and G. W. Grigg, 1993. DNA methylation and mutation. Mutat. Res. 285: 61–67. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., T. Kunej, J. J. Michal, C. T. Gaskins, J. J. Reeves et al., 2005. Significant associations of the mitochondrial transcription factor A (TFAM) promoter polymorphisms with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Biochem. Biophys. Res. Commun. 334: 516–523. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., J. J. Michal, G. A. Williams, T. F. Daniels and T. Kunej, 2006. Cross species association examination of UCN3 and CRHR2 as potential pharmacological targets for antiobesity drugs. PloS ONE 1: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., A. B. Korol, T. Fahima, A. Beiles and E. Nevo, 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11: 2453–2465. [DOI] [PubMed] [Google Scholar]
- Nickerson, D. A., V. O. Tobe and S. L. Taylor, 1997. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, K., K. Frech, H. Karas, E. Wingender and T. Werner, 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, D., Q. Lin and E. Timofeeva, 2002. The corticotropin-releasing factor family of peptides and CRF receptors: their roles in the regulation of energy balance. Eur. J. Pharmacol. 440: 189–197. [DOI] [PubMed] [Google Scholar]