Abstract
The Ccr4-Not complex consists of nine subunits and acts as a regulator of mRNA biogenesis in Saccharomyces cerevisiae. The human ortholog of yeast NOT4, CNOT4, displays UbcH5B-dependent ubiquitin-protein ligase (E3 ligase) activity in a reconstituted in vitro system. However, an in vivo role for this enzymatic activity has not been identified. Site-directed mutagenesis of the RING finger of yeast Not4p identified residues required for interaction with Ubc4p and Ubc5p, the yeast orthologs of UbcH5B. Subsequent in vitro assays with purified Ccr4-Not complexes showed Not4p-mediated E3 ligase activity, which was dependent on the interaction with Ubc4p. To investigate the in vivo relevance of this activity, we performed synthetic genetic array (SGA) analyses using not4Δ and not4L35A alleles. This indicates involvement of the RING finger of Not4p in transcription, ubiquitylation, and DNA damage responses. In addition, we found a phenotypic overlap between deletions of UBC4 and mutants encoding single-amino-acid substitutions of the RING finger of Not4p. Together, our results show that Not4p functions as an E3 ligase by modulating Ubc4p/Ubc5p-mediated stress responses in vivo.
IN yeast, the Ccr4-Not complex is composed of nine subunits and has initially been identified as a transcriptional repressor (Collart and Struhl 1994). However, this evolutionary conserved complex is now known to regulate mRNA biogenesis at multiple levels (reviewed in Denis and Chen 2003; Collart and Timmers 2004). Several genetic and physical interactions between the Ccr4-Not complex and transcription initiation factors were described (Collart 1996; Lemaire and Collart 2000; Deluen et al. 2002). In addition, genetic interactions suggest a role for the Ccr4-Not complex in transcription elongation (Denis et al. 2001). Furthermore, the Ccr4p and Caf1p subunits of the complex are part of the major cytoplasmic mRNA deadenylase (Tucker et al. 2001; Chen et al. 2002).
Our previous work showed a requirement for the Ccr4-Not complex in transcriptional activation of genes encoding subunits of the ribonucleotide reductase (RNR) enzyme, RNR1-4. TBP, Set1p complex, and RNA polymerase II recruitment to the RNR3 promoter was shown to be dependent on NOT4 (Mulder et al. 2005). Moreover, deletion of genes encoding Ccr4-Not subunits resulted in derepression of several stress-response element (STRE)-regulated genes in an Msn2p-Msn4p-dependent manner (Lenssen et al. 2002). This function is controlled by the Glc7p-Bud14p phosphatase (Lenssen et al. 2005) and is responsible for expression of HSP genes following heat shock (Martinez-Pastor et al. 1996).
Several studies noted that NOT4 encoded a protein containing a Zn-finger motif (Cade and Errede 1994; Irie et al. 1994), which was later found to be a RING finger in its human counterpart CNOT4 (Hanzawa et al. 2001). RING-finger-containing proteins constitute a subgroup of ubiquitin protein ligases (Lorick et al. 1999). Indeed, the human ortholog of Not4p, CNOT4, displays ubiquitylation activity in vitro (Albert et al. 2002). Covalent attachment of ubiquitin to a target lysine residue of a substrate requires a three-step cascade involving an activating enzyme (E1), a conjugating enzyme (E2), and a ligase (E3) (reviewed in Glickman and Ciechanover 2002). In addition, ubiquitylation of sequence-specific activators is a critical event in transcription activation (reviewed in Muratani and Tansey 2003). The E3 ligase activity of CNOT4 was shown to be dependent on the selective and specific interaction with UbcH5B, an ubiquitin conjugating enzyme (Winkler et al. 2004). In yeast, two orthologs of UbcH5B exist, UBC4 and UBC5. These genes are redundant, but display partially distinct functions in vivo (Seufert and Jentsch 1990; Chuang and Madura 2005).
Here, we show that yeast Not4p displays E3 ligase activity in vitro and that this activity is important in vivo for Ubc4/5p-dependent stress responses. We identify residues in its RING finger that are critical for this activity by mediating interaction with Ubc4p and Ubc5p. Three independent approaches were taken to investigate the relevance of this activity in vivo. First, complementation of known phenotypes of not4Δ strains, by expressing RING-finger mutants, showed that not all processes involving Not4p require its E3 ligase activity. Second, genetic screens carried out to isolate novel synthetic lethal interactors of the not4Δ and the not4L35A allele reveal overlapping and distinct functions in transcription, ubiquitylation, and tolerance to DNA damage responses. Finally, we found that Ubc4/5p-interaction-deficient RING-finger mutants displayed sensitivity to hydroxyurea (HU), resistance to acute heat shock, and sensitivity to hygromycin B. These phenotypes are shared with deletions UBC4 and UBC5. These data suggest that the RING-finger-mediated E3 ligase activity of Not4p modulates Ubc4p/Ubc5p-dependent stress responses in vivo.
MATERIALS AND METHODS
Strains and media:
For strains, see Table 1. Cells were grown in YPD or SC lacking the appropriate amino acids and at the indicated temperatures.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| EGY48 | MATatrp1 ura3 his3 LEU2∷pLexAop1-LEU2 | Zervos et al. (1993) |
| W303-1B | MATαleu2-3,112 his3-11 trp1-1 can1-100 ade2-1 ura3-1 | Thomas and Rothstein (1989) |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| KMY58 | Isogenic to BY4741 except not4:KanMX | EUROSCARF |
| 2922 | MATαmfa1Δ∷MFA1pr-HIS3 his3Δ1 ura3Δ0 lys2Δ0 can1Δ | Gift from C. Boone (Tong et al. 2001) |
| KMY2 | Isogenic to W303-1B except not4:KanMX6 | Deluen et al. (2002) |
| KMY25 | Isogenic to W303-1B except not4:KanMX6 not5∷KanMX6 + NOT5:URA3 | This work |
| KMY40 | Isogenic to 2922 except not4:URA3 | This work |
| KMY41 | Isogenic to KMY40 except not4L35A:URA3 | This work |
| KMY48 | Isogenic to KMY40 except not4K97R:URA3 | This work |
| KMY43 | W303-1B ubc4:HIS3 | This work |
| KMY45 | W303-1B except not4:KanMX6 ubc4:HIS3 | This work |
| KMY115 | W303-1B except CAF40-TAP:TRP1 | This work |
| KMY116 | Isogenic to KMY2 except CAF40-TAP:TRP1 | This work |
Plasmids:
Plasmid pRS314, containing HA-tagged wild-type NOT4 under the control of the DED1 promoter, was used for site-directed mutagenesis to generate the RING-finger mutant alleles. pRS306-NOT4 (NOT4 nt −473 to nt +1765 relative to the ATG) was constructed using a PCR fragment obtained from genomic DNA of strain W303-1A. pRS306-not4L35A and pRS306-not4K97R were obtained by site-directed mutagenesis of pRS306-NOT4. Constructs were generated using Pfu DNA polymerase and mutations were verified by DNA sequence analysis.
Yeast genetic techniques:
Yeast deletion strains (ATG to STOP) were obtained using PCR-product-mediated gene replacement and were verified by PCR analysis. Genomic not4 mutants were obtained by integrating the pRS306-not4L35A or pRS306-not4K97R plasmids into the genomic NOT4 locus in a not4Δ background using the SmaI restriction site in the NOT4 promoter region (nt −226 relative to +1 ATG).
Yeast two-hybrid analysis:
EGY48 cells were transformed with the indicated B42-NOT4 constructs (in the pJG4-5 vector) and LexA-UBC4 or LexA-UBC5 (in the pEG202 vector). Interactions were determined by spot assay on X-Gal indicator plates as described previously (Albert et al. 2000).
Protein purifications:
TAP-tag-mediated protein purifications were performed essentially as described (Logie and Peterson 1999). Briefly, 20 liters of YPD culture were grown to OD600 ∼2–3, washed, and lysed in E-buffer (20 mm HEPES-KOH pH 8, 350 mm NaCl, 10% glycerol, 0.1% Tween-20). Lysates were cleared by centrifugation in a Beckman 50.2Ti rotor (45,000 rpm, 45 min, 4°). An aliquot of lysate was used for purification over a 200-μl IgG sepharose column (IgG-sepharose fast flow; Pharmacia, Piscataway, NJ). Proteins were bound by rotating at 4° for 2 hr and subsequently washed with 35 ml E-buffer and 10 ml tobacco etch virus (TEV) protease cleavage buffer (10 mm Tris–HCl pH 8, 150 mm NaCl, 0.1% Tween-20, 0.5 mm EDTA, and 1 mm DTT). TEV protease (100 units) cleavage was performed in 1 ml at 18° for 2 hr. The TEV eluate was bound to 100 μl calmodulin affinity resin (Stratagene, La Jolla, CA) in binding buffer (10 mm Tris–HCl pH 8, 150 mm NaCl, 1 mm MgAc, 1 mm imidazole, 2 mm CaCl2, 0.1% Tween-20, 10% glycerol, and 10 mm β-mercaptoethanol) while rotating at 4° for 1 hr. The column was washed with 25 ml binding buffer and bound proteins were recovered in elution buffer (10 mm Tris–HCl pH 8, 150 mm NaCl, 1 mm MgAc, 1 mm imidazole, 2 mm EGTA, 0.1% Tween-20, 10% glycerol, and 10 mm β-mercaptoethanol). His-Ubc4p was expressed from plasmid pQE32-UBC4 (kind gift from T. K. Albert) and purified over Ni-NTA resin and eluted in lysis buffer (50 mm Tris–HCl pH 7.5, 50 mm KCl, 2.5 mm MgCl2, 0.5 mm EDTA, 0.25 mm dithiotreitol, and 10% glycerol) containing 200 mm imidazole. Peak fractions were pooled and desalted using a PD10 column (Pharmacia) to lysis buffer without imidazole.
In vitro ubiquitylation assay:
Purified Ccr4-Not complexes (∼500 ng total protein) were incubated with 250 ng his-Ubc4p, 50 ng rabbit E1 enzyme (Boston Biochem), and 500 ng ubiquitin (purified as described) (Winkler et al. 2004) in reaction buffer (50 mm Tris–HCl pH 8, 50 mm KCl, 2.5 mm MgCl2, 0.5 mm EDTA, 0.25 mm dithiotreitol, and 2 mm ATP) for 90 min at 30°. Reactions were stopped by addition of sample buffer and incubation at 95° for 5 min. Samples were separated by SDS–PAGE on a 10% gel, transferred to a nitrocellulose membrane, and analyzed using antibodies against ubiquitin (P4D1, Santa Cruz Biotechnoloy) or Not4p (rabbit polyclonal serum).
Plasmid shuffle assay:
W303not4Δnot5Δ cells expressing NOT5 from a URA3 plasmid were transformed with the indicated plasmid-based not4 alleles. Purified colonies were taken from plates, serially diluted (10-fold), and spotted onto SC-W and SC-W + 0.05% 5-fluoroorotic acid (5-FOA). Cells were grown for 3 days at 30°.
Phenotypic analysis:
W303not4Δ cells were transformed with various NOT4 constructs. Purified colonies were taken from plates, serially diluted (10-fold), and spotted onto the appropriate media. To test HU or hygromycin B sensitivity, cells were spotted on YPD plates containing the indicated concentrations of HU or hygromycin B. UV sensitivity was assessed by spotting cells on YPD plates and exposing to the indicated doses of UV light (Stratalinker, Stratagene). Cells were grown for 3–4 days at 30°. To test temperature sensitivity, cells were spotted on YPD plates and grown at 30° and 37°, respectively.
Synthetic genetic array analysis:
Synthetic genetic array (SGA) was performed essentially as described previously (Tong et al. 2001). Briefly, BY4741 knockout collection strains were crossed with 2922not4Δ or 2922not4L35A strains. Diploids were selected on media containing G418 (200 μg/ml) lacking uracil and subsequently sporulated for 8 days at 22°. Haploid MATa progeny was selected on minimal medium supplemented with uracil, lysine, and canavanine (50 μg/ml). MATa URA+ cells were subsequently selected on minimal medium supplemented with lysine and canavanine (50 μg/ml). Double deletion strains were isolated on minimal medium supplemented with lysine, canavanine (50 μg/ml), and G418 (200 μg/ml). Growth was assessed after 24, 48, and 72 hr by visual inspection.
Isolated potential genetic interactors were verified by repeating the SGA procedure as described above, except that double knockout cells were selected on SC–URA + G418 (200 μg/ml). A subset of the genetic interactions was validated using random spore analysis and/or tetrad dissection. Bar code sequencing was used to confirm the identity of the deletion strains showing synthetic growth phenotypes with the not4Δ or not4L35A alleles.
Heat-shock survival assay:
W303not4Δ cells expressing mutant not4 alleles or an empty vector (pRS314) were cultured in YPD and kept in exponential phase for at least 30 hr before heat shock at 50° for 10 min. An equivalent number of cells (∼1000) were plated on YPD medium before and after heat shock to assess the percentage of survival. Before treatment, samples were taken for Northern blot analysis of HSP gene expression. Plasmids expressing not4 alleles were not lost during growth in YPD (data not shown).
RNA extraction:
Total RNA was purified using the hot phenol extraction procedure, as described previously (Mulder et al. 2005). Briefly, 40 ml yeast cultures (OD600 = 0.5–1) in YPD were added to 40 ml YPD containing 400 mm HU or 0.02% methyl methanesulfonate (MMS) to obtain the final concentration of 200 mm HU or 0.01% MMS. Samples (7.5–10 ml) were collected by centrifugation for 2 min at 5000 rpm. Cell pellets were frozen on dry ice. Frozen cells were resuspended in 500 μl of acid hot phenol:chloroform (5:1, pH 4.7, 65°) and 500 μl of TES buffer (10 mm Tris–HCl pH 7.5, 1 mm EDTA, 0.5% SDS). Cells were incubated for 1 hr at 65° and vortexed every 10 min for 20 sec. The aqueous solution was extracted with phenol:chloroform and with chloroform:isoamyl alcohol (25:1). Finally, total RNA was collected by ethanol precipitation.
Northern blotting:
RNA (10 μg) was separated by electrophoresis on a 1% agarose gel containing 10 mm Na− phosphate pH 6.7. Subsequently, RNA was transferred to a nylon membrane and crosslinked by UV-light irradiation. PCR product probes for full-length RNR1, RNR2, RNR3, RNR4, HSP42, HSP78, HSP104, ACT1, TUB1, and 18S rRNA were radiolabeled using the RediPrime II kit (Amersham Pharmacia Biotech). After prehybridization (1–4 hr), probes were added and membranes were incubated overnight at 42°. Blots were rinsed with 2× SSC at room temperature and sequentially washed with 2× SSC, 1× SSC, 0.5× SSC, and 0.3× SSC (twice) for 15 min at 65°. Membranes were either exposed to X-ray films or subjected to quantification using a Storm 820 PhosphorImager and ImageQuant software.
RESULTS
Not4p interacts with Ubc4p/Ubc5p and exhibits ubiquitin protein ligase activity in vitro in the context of the Ccr4-Not complex:
CNOT4, the human ortholog of NOT4, displays ubiquitylation activity in vitro (Albert et al. 2002). This activity is dependent on the specific and selective interaction with UbcH5B (Winkler et al. 2004). In addition, Not4p was shown to interact with both Ubc4p and Ubc5p, the yeast orthologs of UbcH5B (Winkler et al. 2004; Krogan et al. 2006). To be able to investigate the role of this ubiquitylation activity in vivo, we took advantage of the high degree of homology between the RING-finger domains of human CNOT4 and yeast Not4p. Using NMR chemical perturbation data obtained from experiments using CNOT4 and UbcH5B (Albert et al. 2002) and the proposed structure of this complex (Dominguez et al. 2004), we designed RING-finger mutant alleles of NOT4 (Figure 1A). The encoded proteins contained single-amino-acid substitutions at the surface of Not4p and were predicted to abolish the interaction between Not4p and Ubc4p/Ubc5p. Using a yeast two-hybrid setup, we determined the interaction with Ubc4p/Ubc5p. As shown in Figure 1B, substitution of the leucine residue at position 35 with alanine (L35A) resulted in a complete loss of interaction with both Ubc4p and Ubc5p. This was similar to substitution of I37 (I37A) and I64 to alanine (I64A) or tryptophan (I64W). Interestingly, we did not find that all mutations disrupted Ubc4p/Ubc5p interaction (Y61A, N63A, R78A, and K97R) (Figure 1B). The K97R substitution was included to inactivate a potential sumoylation site (LKME) located outside of the RING-finger domain (Figure 1A). Equal expression of the proteins was verified by immunoblotting (data not shown). Together, this analysis indicated that residues L35, I37, and I64 are critical for the interaction between Not4p and Ubc4p/Ubc5p.
Figure 1.—
Isolation of NOT4 RING-finger point mutants disrupting the interaction of Not4p with Ubc4p/Ubc5p. (A) Alignment of the N-terminal region (residues 1–120) of human and yeast Not4p including the RING finger. Identical (∷) and similar (:) residues are indicated. Solid and shaded residues specify the conserved cysteines and the residues targeted for substitution, respectively. (B) Yeast two-hybrid interaction between B42-Not4p variants and LexA-Ubc4p or LexA-Ubc5p. Three independent clones were spotted on X-Gal indicator plates and grown overnight at 30°.
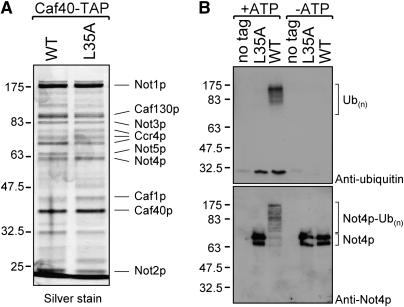
Recombinant human CNOT4 was previously shown to display ubiquitylation activity in a reconstituted in vitro system (Albert et al. 2002), but this has not been shown for yeast Not4p. Moreover, these experiments have not been performed in the context of the complete Ccr4-Not complex. To purify the Ccr4-Not complex from yeast, we fused the TAP-tag to the 3′ end of the endogenous CAF40 gene. Deletion of NOT4 from this strain enabled subsequent reintroduction of either the wild-type or a mutant (not4L35A) allele of NOT4 to its original locus (data not shown). Purification of TAP-tagged Caf40p from strains expressing either NOT4 or not4L35A yielded Ccr4-Not complexes that were identical in subunit composition, as verified by silver staining (Figure 2A) and Western blotting (data not shown). We analyzed activity of both complexes in in vitro ubiquitylation assays supplied with E1 enzyme, recombinant Ubc4p, and ubiquitin. A mock purification from a nontagged strain was included as a negative control in the experiment. We observed a robust auto-ubiquitylation activity in the lanes containing wild-type Not4p using antibodies directed against ubiquitin or Not4p (Figure 2B). In contrast, when the Ccr4-Not complexes contained the L35A form of Not4p, no auto-ubiquitylation could be observed (Figure 2B). As expected, Not4p-mediated ubiquitylation was dependent on addition of ATP and Ubc4p to the reaction (Figure 2B and data not shown). Together, these experiments show for the first time that the Ccr4-Not complex displays in vitro ubiquitylation activity. Moreover, this activity critically depends on the interaction between Ubc4p and Not4p. We note that in line with other multisubunit E3 ligase complexes Ubc4p and Ubc5p are not stable components of the Ccr4-Not complex as indicated by mass spectrometric analyses (data not shown).
Figure 2.—
The Ccr4-Not complex displays Ubc4p-dependent ubiquitylation activity in vitro, which requires an intact RING-finger domain of Not4p. (A) Tandem affinity-purified Ccr4-Not complexes containing either wild-type or L35A Not4p. Caf40-TAP strains expressing NOT4 or not4L35A were used to isolate the Ccr4-Not complex. Purified proteins were separated by 10% SDS–PAGE and visualized by silver staining. (B) Not4p-mediated ubiquitylation activity depends on the interaction with Ubc4p. Purified Ccr4-Not complexes containing WT or L35A forms of Not4p were used in in vitro ubiquitylation reactions. Reactions were analyzed by Western blotting using antibodies against ubiquitin and Not4p.
Not4p has functions distinct from its ubiquitin protein ligase activity in vivo:
To investigate the role for the ubiquitin protein ligase (E3) activity of Not4p in vivo, we tested complementation of several known not4Δ phenotypes by our collection of RING-finger mutants. Several genetic interactions within the Ccr4-Not complex are described (Maillet et al. 2000), including synthetic lethality between deletions of NOT4 and NOT5. We hypothesized that if this function of Not4p depends on its ubiquitylation activity, the not4Δnot5Δ synthetic lethality would not be complemented by introduction of E2-interaction-deficient mutants. NOT4 RING-finger mutants were introduced into not4Δnot5Δ cells expressing NOT5 from a plasmid carrying the URA3 marker. Plasmid shuffle analysis showed that mutants disrupting the interaction between Not4p and Ubc4p/Ubc5p were capable in complementing the not4Δnot5Δ synthetic lethality, although a minor effect was observed with the R78A allele (Figure 3A). Next, complementation of the temperature sensitivity of cells lacking NOT4 was assessed. While not4Δ cells containing the control plasmid (pRS314) did not grow at the restrictive temperature, all RING-finger mutants and the wild type complemented the growth defect at 37° (Figure 3B). A third phenotype of not4Δ cells is sensitivity to UV irradiation (Westmoreland et al. 2004). We found that cells lacking NOT4 were only mildly sensitive to this stress and that all tested mutants behaved as wild type (Figure 3C). In conclusion, mutation of the RING finger did not affect the complementation potential of Not4p. This indicates that the RING-finger-mediated E3 ligase activity is not involved in the phenotypes tested, but rather involves a separate function of Not4p.
Figure 3.—
The in vivo role of Not4p is not restricted to its E3 ligase function. (A) The not4Δnot5Δ synthetic lethality is independent of the RING finger of Not4p. W303 not4Δnot5Δ containing NOT5 on a URA3 plasmid were transformed with the indicated not4 mutants on a TRP1 plasmid or the empty vector. Colonies were taken from plates, serially diluted (5-fold), and spotted on plates with or without 0.05% 5-fluoroorotic acid (5-FOA). Growth was assessed after 4 days at 30°. (B) The temperature sensitivity of not4Δ cells is complemented by RING-finger mutant alleles of NOT4. W303 not4Δ cells were transformed with plasmids expressing RING-finger mutant alleles of NOT4. Tenfold serial dilutions were spotted on YPD and grown at 30° or 37° for 4 days. (C) The not4Δ UV sensitivity is complemented by RING-finger mutant alleles of NOT4. Strains from B were serially diluted (10-fold), spotted on YPD, and exposed to the indicated doses of UV light. Growth was assessed after 3 days at 30°.
SGA analysis:
As a second approach to gain insight into the physiological role of Not4p, we initiated SGA experiments to identify novel genetic interactions with two not4 alleles. To this end, query strains were constructed that were either not4Δ or expressed the not4L35A allele integrated into the NOT4 genomic locus. These strains were subsequently used to perform SGA analysis on a collection of ∼4800 deletion mutants (Tong et al. 2001). Initially, 200 genes were identified as potential genetic interactors using the not4Δ and the not4L35A RING-finger mutant. Repetition of the screens confirmed the genetic interaction for a total of 49 genes (Tables 2 and 3) with an overlap of 48% between the not4Δ and not4L35A screens (P-value = 1.8 × 10−20; Figure 4A). Identity of the gene deletions in the library strains was determined independently by PCR analysis followed by sequencing of the “bar code.” We selected 30 of the 49 interactors for random spore analysis and/or tetrad dissection. In all cases this confirmed the identified interaction with not4Δ and not4(L35A) alleles (Tables 2 and 3). Whereas the 13 interactors exclusively found with the not4Δ allele may reflect RING-finger-independent functions of Not4p, this is less clear for the 24 interactors found exclusively with the not4L35A allele (Figure 4).
TABLE 2.
Synthetic genetic interactions with the not4L35A allele
| ORFa | Name | SL/SSb | Function/processc |
|---|---|---|---|
| YDR069Cde | DOA4 | SL | Protein deubiquitination |
| YDR313C | PIB1 | SL | Protein ubiquitination/E3 ligase activity |
| YFR010W | UBP6 | SS | Protein deubiquitination |
| YGL058We | RAD6 | SS | DNA repair/protein ubiquitination |
| YMR275Ce | BUL1 | SL | Protein ubiquitination |
| YLR285We | NNT1 | SS | Chromatin silencing |
| YNL229C | URE2 | SL | Transcription corepressor activity |
| YNL248Ce | RPA49 | SL | Transcription from Pol I promoter |
| YNL278We | CAF120 | SS | Transcription from RNA pol II promoter |
| YOL051We | MED15 | SS | Transcription from Pol II promoter |
| YOL054We | PSH1 | SL | RNA elongation from Pol II promoter |
| YOL148Ce | SPT20 | SS | Transcription/chromatin modification |
| YOR290Ce | SNF2 | SL | Chromatin remodeling |
| YJR060We | CBF1 | SS | DNA replication |
| YKL213Ce | DOA1 | SS | Double-strand break repair via NHEJ |
| YLR233C | EST1 | SL | Telomere maintenance |
| YLR320We | MMS22 | SS | DNA repair (DSBR) |
| YAL002W | VPS8 | SS | Late endosome to vacuole transport |
| YDL192We | ARF1 | SS | GTPase activity/ER to Golgi transport |
| YLR360W | VPS38 | SL | Late endosome to vacuole transport |
| YNR006We | VPS27 | SL | Protein binding |
| YOR068C | VAM10 | SS | Vacuole fusion |
| YOR332We | VMA4 | SL | Vacuolar ATPase |
| YPL234C | TFP3 | SS | Vacuolar ATPase |
| YPR173Ce | VPS4 | SL | Late endosome to vacuole transport |
| YLR200We | YKE2 | SS | Protein folding/prefoldin complex |
| YKL009Wde | MRT4 | SL | rRNA processing |
| YOR302Wde | SL | Regulation of protein biosynthesis | |
| YLR289We | GUF1 | SL | GTPase activity |
| YBR133Cde | HSL7 | SS | Protein arginine N-methyltransferase |
| YJR104Ce | SOD1 | SS | Superoxide metabolism |
| YDR452W | PHM5 | SL | Polyphosphate metabolism |
| YKL183We | LOT5 | SS | Unknown |
| YKR005C | SL | Unknown | |
| YKR035C | SS | Unknown | |
| YLR232W | SL | Unknown | |
| YPL168W | SS | Unknown |
NHEJ, nonhomologous end joining; DSBR, double-strand break repair.
ORFs identified in both not4L35A and not4Δ screens are underlined.
Synthetic lethal and synthetic sick, respectively.
Annotations derived from the Saccharomyces genome database.
These ORFs are frequently identified in SGA screens, even when performed against wild-type query strains (E. Cameroni and C. De Virgilio, unpublished results).
Interactions confirmed by random spore analysis and/or tetrad dissection.
TABLE 3.
Synthetic genetic interactions with the not4Δ allele
| ORFa | Name | SL/SSb | Function/Processc |
|---|---|---|---|
| YBR170Cd | NPL4 | SL | ER-associated protein catabolism |
| YDR313C | PIB1 | SL | Protein ubiquitination/E3 ligase activity |
| YFR010W | UBP6 | SS | Protein deubiquitination/26S proteasome |
| YMR275Cd | BUL1 | SL | Protein ubiquitination |
| YPL084Wd | BRO1 | SL | Ubiquitin-dependent protein catabolism |
| YGL025C | MED3 | SS | Transcription from RNA pol II promoter |
| YLR266Wd | BUR2 | SL | CDK/RNA pol II transcription |
| YOL051Wd | MED15 | SL | Transcription from RNA pol II promoter |
| YOL054Wd | PSH1 | SS | RNA elongation from Pol II promoter |
| YPR075Wd | NOT5 | SL | Transcription from RNA pol II promoter |
| YLR320Wd | MMS22 | SS | DNA repair (DSBR) |
| YAL002W | VPS8 | SS | Late endosome to vacuole transport |
| YDL192Wd | ARF1 | SL | ER to Golgi transport |
| YJL188C | BUD19 | SS | Bud site selection |
| YOR069Wd | VPS5 | SL | Retrograde transport |
| YOR332Wd | VMA4 | SL | Vacuolar ATPase |
| YPL234C | TFP3 | SL | Vacuolar ATPase |
| YPR036Wd | VMA13 | SL | Vacuolar ATPase |
| YKR076W | ECM4 | SS | Cell wall organization and biogenesis |
| YLR200Wd | YKE2 | SL | Protein folding/prefoldin complex |
| YOR302W | SS | Protein biosynthesis | |
| YDR417C | SS | Unknown | |
| YJL169W | SS | Unknown | |
| YKL183Wd | LOT5 | SS | Unknown |
| YMR085W | SS | Unknown |
DSBR, double-strand break repair.
ORFs identified in both not4L35A and not4Δ screens are underlined.
Synthetic lethal and synthetic sick, respectively.
Annotations derived from the Saccharomyces genome database.
dInteractions confirmed by random spore analysis and/or tetrad dissection.
Figure 4.—
Significant overlap between genes identified in not4L35A and not4Δ SGA screens. (A) Venn diagram displaying overlap in genes isolated in the SGA screens with not4Δ and not4L35A. (B) Genes involved in the same biological process (Gene Ontology annotations) are grouped and displayed in pie charts. Percentages of isolated genes in a functional group are given.
The genes identified as genetic interactors were arranged into various functional categories (Saccharomyces Genome Database; Figure 4B). Genes involved in transcription or the ubiquitin conjugating system constituted a large portion of the identified genes (18/49, 37%). Surprisingly, multiple genes involved in the response to DNA damage genetically interacted with a deletion of NOT4 and/or the not4L35A allele, suggesting a role for Not4p and its RING finger in this process. This is in agreement with our previous observations that the Ccr4-Not complex is involved in regulation of DNA-damage-induced gene expression (Mulder et al. 2005).
Interaction between Not4p and Ubc4p/Ubc5p is required for tolerance to hydroxyurea:
Following the observation that the not4L35A allele genetically interacts with genes involved in DNA damage responses, strains expressing the not4L35A or not4K97R allele from the original NOT4 locus were constructed and tested for HU sensitivity. The K97R mutation does not affect the RING-finger structure or the interaction with Ubc4p/Ubc5p (Figure 1B). Interestingly, cells expressing not4L35A were sensitive to HU at 200 mm, whereas the control mutation (not4K97R) was not (Figure 5A). To extend this observation, not4Δ cells were transformed with plasmids expressing other RING-finger mutants. Several mutants that could still interact with Ubc4p/Ubc5p (Y61A, N63A, and K97R) were included as controls. Cells expressing the L35A and I37A alleles, encoding proteins deficient in E2 interaction, displayed sensitivity to HU, whereas the interaction-proficient mutants behaved as wild type (Figures 5B and 1B). Interestingly, cells deleted for UBC4 showed sensitivity to high concentrations of HU comparable to the L35A and I37A mutants (compare Figure 5B and 5C). In contrast, cells lacking NOT4 are sensitive to low doses of HU. Nevertheless, a good correlation between HU sensitivity at 200 mm and interaction with Ubc4p/Ubc5p was observed, indicating that the ubiquitylation potential of Not4p is required for HU tolerance. Analysis of the not4Δubc4Δ double mutant revealed an epistatic relationship for the HU sensitivity phenotype (Figure 5D). This supports the notion that Not4p and Ubc4p constitute a functional E2–E3 pair in vivo.
Figure 5.—
The E3 ligase potential of Not4p is required for tolerance to high concentrations of HU. (A) The interaction between Not4p and Ubc4p/Ubc5p is important for tolerance to HU. W303 not4Δ cells expressing the indicated not4 alleles from its endogenous locus were serially diluted (10-fold) and spotted on YPD plates containing the indicated concentration of hydroxy urea (HU). Cells were grown for 3 days at 30°. (B) Cells expressing pRS314-based E2-interaction-deficient RING-finger mutant alleles are sensitive to high concentrations of HU. W303 not4Δ cells were transformed with the indicated not4 mutants on a CEN TRP1 plasmid or the empty vector (pRS314). Analysis was performed as in A. We note that episomal expression of the NOT4, not4L35A, and not4K97R alleles results in an identical HU sensitivity as expression from the chromosomal locus. (C) Cells lacking UBC4 are sensitive to high concentrations of HU. W303 not4Δ cells and W303 ubc4Δ cells were analyzed as in A. (D) Epistatic relationship between NOT4 and UBC4 in HU sensitivity. W303 WT, not4Δ, ubc4Δ, and not4Δubc4Δ cells were analyzed as in A. (E) The not4L35A allele-mediated HU sensitivity is independent of defects in RNR gene transcription. W303 cells expressing the indicated alleles, integrated into the NOT4 genomic locus, were grown to exponential phase and treated for 120 min with 200 mm HU or 0.01% MMS, respectively. RNA was extracted and subjected to Northern blot analysis (left section). Probes were radiolabeled PCR products. ACT1 was used as a loading control. Quantification of the Northern blots is shown in the right section.
Our previous work showed a requirement for NOT4 in efficient HU-induced transcription of RNR genes (Mulder et al. 2005). Therefore, cells lacking NOT4 or expressing WT or not4L35A from its original locus were either mock treated or incubated with HU or MMS for 2 hr. Transcript levels of RNR genes were determined by Northern blot analysis. As expected, not4Δ cells show defects in expression of RNR2, RNR3, and RNR4 mRNA. However, disruption of the interaction between Not4p and Ubc4p/Ubc5p did not significantly affect the transcriptional induction of the RNR genes (Figure 5E). Collectively, these experiments suggest that the ubiquitin protein ligase function of Not4p is involved in tolerance to high concentrations of HU in a manner distinct from transcriptional regulation of the RNR genes.
Disruption of the RING-finger-mediated interaction with Ubc4p/Ubc5p leads to resistance to acute heat shock and sensitivity to hygromycin B:
UBC4 and UBC5 are shown to be involved in stress responses, such as proteasomal degradation of short-lived and abnormal proteins (Seufert and Jentsch 1990). One of the phenotypes associated with this function is resistance to acute heat shock. Indeed, ubc4Δubc5Δ double-mutant strains display tolerance to acute heat shock, whereas single deletions of UBC4 or UBC5 do not show this phenotype (Seufert and Jentsch 1990). Interestingly, deletion of NOT4 or NOT5 results in the same phenotype (Lenssen et al. 2002). We confirmed that deletion of NOT4 induced a strong resistance to acute heat stress (Figure 6A). In addition, introduction of plasmids expressing RING-finger variants of NOT4 showed that the L35A, I37A, I64A, and I64W alleles led to an increased survival (Figure 6A). In contrast, the Y61A, N63A, and E69K alleles, encoding proteins that retain the interaction with Ubc4p/Ubc5p, resulted in survival rates comparable to wild type. This suggests that the ubiquitin protein ligase activity of Not4p, together with Ubc4p and/or Ubc5p, is involved in regulation of this stress response. Tolerance to acute heat stress in ubc4Δubc5Δ, not5Δ, and not4Δ strains has been linked with increased levels of heat-shock gene expression under normal growth conditions (Seufert and Jentsch 1990; Lenssen et al. 2002, 2005). Therefore, Northern blot analysis was performed to determine the level of heat-shock gene expression in the RING-finger mutant strains. Notably, HSP104 is the major heat-shock protein responsible for the response to acute heat stress (Sanchez and Lindquist 1990). As expected, HSP104, HSP78, and HSP42 transcript levels were increased in the absence of NOT4 (Figure 6B). Although interaction-deficient RING-finger mutants were resistant to acute heat stress, the basal levels of HSP gene expression were not affected (Figure 6B). In addition, heat-shock-induced transcription of these genes in these strains was comparable to wild type (data not shown).
Figure 6.—
Disruption of the RING-finger-mediated interaction of Not4p with Ubc4p/Ubc5p results in tolerance to acute heat shock. (A) E2 interaction-deficient RING-finger mutants are resistant to acute heat shock. W303 not4Δ cells expressing the indicated plasmid-based RING-finger mutants were grown exponentially for 2 days in YPD. Complementation of the not4Δ slow-growth phenotype by all mutant alleles prevented plasmid loss (data not shown). Identical cell numbers were plated on YPD before and after acute heat shock (10 min, 50°). Error bars indicate standard deviations of two experiments. (B) Heat-shock resistance of RING-finger mutants is independent of HSP gene expression under normal conditions. RNA was extracted before heat shock and subjected to Northern blotting. Quantification of HSP42, HSP78, and HSP104 expression (after normalization to TUB1 signals) is shown. Error bars indicate standard deviations of two experiments. (C) The E3 ligase activity of Not4p is required for tolerance to hygromycin B. Serial dilutions of W303 not4Δ strains expressing NOT4, not4L35A, or not4K97R from its original locus were spotted on YPD containing the indicated concentrations of hygromycin B.
Recently, Ubc4p was reported to associate to the 26S proteasome in the presence of hygromycin B (Chuang and Madura 2005), a compound that induces translation errors leading to misfolded proteins. Strains deleted for UBC4 and UBC5 were shown to be sensitive to hygromycin B. Interestingly, we found that the not4L35A-expressing strain was also sensitive to this drug (Figure 5D). In conclusion, strains expressing NOT4 RING-finger mutants displayed similar phenotypes to strains deleted for UBC4, supporting the model that Ubc4p and Not4p form a functional E2/E3 pair in vivo.
DISCUSSION
Previous work showed that CNOT4, the human ortholog of Not4p, displays ubiquitin protein ligase (E3) activity in vitro (Albert et al. 2002; Winkler et al. 2004). To facilitate in vivo analysis of the RING finger of Not4p we first obtained mutants preventing the interaction with Ubc4p and Ubc5p (Figure 1). Substitution of leucine 35 with alanine resulted in complete loss of in vitro E3 ligase activity of Not4p in the context of the Ccr4-Not complex (Figure 2). Second, using SGA analysis we identified various novel genetic interactions with either not4Δ or not4L35A alleles (Tables 2 and 3). Significant overlap between the screens was observed (Figure 4). Of the identified genes, several were previously suggested to play a role in DNA damage response pathways. Interestingly, we could show that disruption of the E3 ligase activity of Not4p resulted in sensitivity to HU (Figure 5), a phenotype shared by many genes involved in DNA damage responses. Finally, we found a strong correlation between the ability of Not4p to interact with Ubc4p/Ubc5p and tolerance to acute heat shock and sensitivity to hygromycin B (Figure 6). Together, our results indicate that the E3 activity of Not4p is required for an adequate response to replication stress and the presence of misfolded proteins in vivo.
Isolation of synthetic genetic interactors of not4 alleles reveals links with transcription, ubiquitylation, and DNA damage responses:
Complementation analysis of several known phenotypes of not4Δ cells did not yield insight into the physiological role of the RING finger of Not4p (Figure 3 and data not shown). Therefore, we performed genomewide screens to identify novel genetic interactions with either a null or a RING-finger mutant (L35A) allele of NOT4 (Tables 2 and 3). The synthetic interactors isolated in these screens were categorized in four major groups consisting of genes involved in transcription, ubiquitylation, DNA damage response processes (discussed below), and organelle function (Figure 4).
A transcription function for NOT4:
Mutant alleles of NOT1, NOT3, NOT5, and CAF1 were previously found as suppressors of srb4-138, a temperature-sensitive mutant of SRB4 (Lee et al. 1998), which suggested opposing roles for the Ccr4-Not and Srb/Mediator complexes. However, like the Ccr4-Not complex, the Srb/Mediator complex regulates transcription both positively and negatively (van de Peppel et al. 2005). Strikingly, disruption of the tail module of the Srb/Mediator complex (by deletion of MED15 or MED3), involved in interaction with transcription activators, results in synthetic lethality with a deletion of NOT4 or the not4L35A allele (Tables 2 and 3). This suggests a functional overlap between the E3 ligase activity of Not4p and activation of transcription by the Srb/Mediator complex. Interestingly, the Med8p subunit, which resides in the tail module, has been shown to be part of an E3 ligase complex in mammals (Brower et al. 2002). A recent report showed that cells lacking MED3 or the N terminus of Srb7 are defective in transcriptional induction of RNR3 after MMS treatment (Zhang and Reese 2004). In agreement with this, we found that deletion of MED15 or MED3 resulted in sensitivity to HU (data not shown), a phenotype shared by cells lacking NOT4 or expressing E2-interaction-deficient RING-finger mutants (Figure 4). We have tested other nonessential genes of the Srb/Mediator (MED9, MED2, PGD1, ROX3, SIN4, SRB2, SRB5, NUT8, SRB10, and SRB11), SAGA (ADA1, ADA2, ADA3, GCN5, SPT3, SPT7, SGF73, and SGF29), or SWI/SNF (SNF5, SNF6, SNF11, SWI3, and TAF14) complexes for genetic interaction with the not4Δ and not4(L35A) alleles, but we failed to observe any synthetic growth phenotypes (data not shown). In addition, several genes involved in transcription elongation showed a synthetic growth phenotype when deleted in combination with NOT4. Among these were BUR2, PSH1, and YJL169W (a dubious ORF partially overlapping with SET2). Psh1p (Pob3p-Spt16p-binding protein) was found to copurify with the FACT complex, which facilitates transcription elongation through chromatin (Ho et al. 2002; Krogan et al. 2002). It is noteworthy that our SGA analysis could not uncover interactions with POB3, SPT16, or NHP6A/B since the former two genes are essential (and are not represented in the collection of single-gene knockouts) and the latter two (NHP6A and NHP6B) are functionally redundant. This suggests that isolation of PSH1/YOL054W could reflect further functional interactions between the Ccr4-Not and FACT complexes in transcription elongation. In agreement with this, it was recently found that the genes encoding the FACT complex subunit Nhp6p (Formosa et al. 2001) genetically interact with components of the Ccr4-Not complex (Biswas et al. 2006). Finally, NOT4 and BUR2 display synthetic lethality, as does a deletion of NOT4 combined with a temperature-sensitive allele of BUR1 (K. W. Mulder, A. Inagaki and H. Th. M. Timmers, unpublished results).
NOT4 is linked to the ubiquitin-26S proteasome pathway:
As expected, a second group of genes displaying synthetic growth defects or lethality with not4 alleles is linked to the ubiquitin proteasome pathway, confirming a role for the RING finger of Not4p in this process in vivo. For instance, Pib1p has been described to contain RING-finger-dependent E3 ligase activity in vitro (Shin et al. 2001). Possibly, Pib1p constitutes a redundant E3 ligase for Not4p under certain conditions. However, this redundancy is not likely to be relevant for the HU sensitivity of not4Δ and not4L35A, since pib1Δ cells are not sensitive to HU (data not shown). DOA4 is a member of the family of deubiquitinating enzymes and interacts with the 26S proteasome (Papa and Hochstrasser 1993; Papa et al. 1999). Interestingly, deletion of DOA4 results in sensitivity to HU in a manner epistatic to rad9Δ (Fiorani et al. 2004). Intriguingly, recent work shows that the HU sensitivity of cells lacking CCR4 or CAF1 is also epistatic to the pathway containing Rad9p (Traven et al. 2005), suggesting that the Ccr4-Not complex and Doa4p might be involved in this pathway in parallel to each other. In addition, Ubc4p has recently been shown to interact with the 26S proteasome in the presence of hygromycin B, inducing translational misreading (Chuang and Madura 2005). This is in concert with the observation that UBC4 and UBC5 are required for degradation of abnormal and short-lived proteins (Seufert and Jentsch 1990).
NOT4 and DNA damage responses:
Interestingly, 11% (not4L35A) and 4% (not4Δ) of the genetic interactors are involved in DNA damage responses (Tables 2 and 3 and Figure 4). Our screens revealed interactions with mms22Δ and rad6Δ, suggesting a role for the E3 ligase activity of Not4p in the cellular response to DNA damage. Notably, expression of RING-finger mutant alleles complemented the mild UV sensitivity phenotype of not4Δ cells (Figure 3). Subsequent phenotypic analysis showed HU sensitivity of strains expressing RING-finger variants of Not4p that were disrupted in their interaction with Ubc4p and Ubc5p (Figures 1 and 4). Moreover, it has been shown that components of the Ccr4-Not complex are required for cell cycle progression after ionizing radiation in a manner epistatic to the RAD9-dependent DNA-damage checkpoint (Westmoreland et al. 2004). Recent data suggest that MMS22 forms a genetic module (MMS22m) together with MMS21, RTT101, and RTT107 (Pan et al. 2006). In addition, RAD6 is part of both the BRE1m and the postreplication repair (PRR) pathway. Both these modules are genetically distinct from the RAD9m, supporting the hypothesis that NOT4 functions in the RAD9 pathway (Westmoreland et al. 2004; Traven et al. 2005).
The E3 ligase activity of Not4p is involved in regulation of Ubc4p/Ubc5p-mediated stress tolerance:
Previous work showed that integrity of the Ccr4-Not complex is required for survival in the presence of HU (Mulder et al. 2005). This drug is known to directly target the RNR complex. RNR is responsible for conversion of NDPs to dNDPs, which is the rate-limiting step in dNTP production (Jordan and Reichard 1998). HU treatment results in a decreased replication rate by limiting the dNTP pools. Deletion of genes involved in DNA-damage repair pathways also leads to sensitivity to HU. The observation that disruption of the interaction between Ubc4p/Ubc5p and Not4p confers HU sensitivity indicates a role for its E3 ligase activity in tolerance to replication stress. Interestingly, deletion of UBC4 resulted in a similar HU sensitivity (Figure 5). In addition, deletion of both UBC4 and UBC5 leads to a severe slow-growth phenotype (Seufert and Jentsch 1990), indicating functional redundancy between these genes. However, the regulation of expression of these genes differs significantly (Seufert and Jentsch 1990). For instance, UBC4 transcript levels drop significantly during stationary phase, whereas UBC5 levels are increased. Differential regulation of transcription might provide an explanation for the observation that some phenotypes that are associated with deletion of UBC4 are not evident in ubc5Δ cells, although these genes are functionally redundant during normal growth. Furthermore, UBC4 and UBC5 expression is increased by heat shock. Interestingly, strains deleted for both UBC4 and UBC5 display tolerance to acute heat shock (Seufert and Jentsch 1990). This treatment is thought to produce aberrantly folded proteins in vivo. Interestingly, deletion of NOT4 also gives rise to resistance to heat shock, which is most likely to result from derepression of HSP gene expression under noninducing conditions (Lenssen et al. 2005). We found that the role of the RING finger of Not4p in tolerance to acute heat shock is independent of HSP gene expression (Figure 6). The functional redundancy between UBC4 and UBC5 in mediating tolerance to acute heat shock and the observation that RING-finger mutants of Not4p display an identical phenotype indicate that Not4p is the critical E3 ligase in this process. Strikingly, not4Δ as well as ubc4Δubc5Δ cells are sensitive to hygromycin B (Chuang and Madura 2005). This drug induces misreading of the mRNA template in the ribosome, resulting in misfolding of the newly synthesized proteins. This sensitivity is, at least in part, dependent on the E3 ligase activity of Not4p, since the not4L35A allele could not fully complement this phenotype of not4Δ cells (Figure 6D). This suggests that the Ubc4p/Ubc5p-mediated response to abnormal and translationally damaged proteins is dependent on the interaction of Not4p with Ubc4p/Ubc5p. In agreement with this suggestion, we found a genetic interaction between not4Δ, not4L35A, and a deletion of YKE2 (Tables 2 and 3). This gene encodes a subunit of the prefolding/GimC cytoplasmic chaperone complex, involved in cellular protection against protein aggregation.
It is clear that the RING finger of Not4p is involved in tolerance to various stresses, including tolerance to misfolded proteins in vivo. However, its exact role in this process and identification of its substrates require further investigation. Collectively, our results provide the first evidence that Not4p functions as a RING-finger-dependent E3 ligase in vitro and identified involvement of this activity in several processes in vivo.
Acknowledgments
We thank C. Boone, T. K. Albert, F. C. P. Holstege, and D. van Leenen for sharing reagents; J. van de Peppel for sharing unpublished data; and W. W. Pijnappel for advice on the TAP purifications. We are grateful to members of the Timmers and Collart laboratories for helpful discussions. This work was supported by grants from the Netherlands Organization for Scientific Research (NWO-CW no.700-50-034) and the European Union (Improving Human Potential RTN2-2001-00026 and STREP LSHG-CT-2004-502950) to H.Th.M.T., from the European Molecular Biology Organization (ASTF 184.00-02) to K.W.M., from the Swiss National Science Foundation (SNSF) (grant no. 3100A0-100793) to M.A.C, and from the SNSF (grant no. 631-062731.00) and the Canton of Geneva to C.D.V.
References
- Albert, T. K., M. Lemaire, N. L. van Berkum, R. Gentz, M. A. Collart et al., 2000. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 28: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, T. K., H. Hanzawa, Y. I. Legtenberg, M. J. de Ruwe, F. A. van den Heuvel et al., 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, D., Y. Yu, D. Mitra and D. J. Stillman, 2006. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics 172: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower, C. S., S. Sato, C. Tomomori-Sato, T. Kamura, A. Pause et al., 2002. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc. Natl. Acad. Sci. USA 99: 10353–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade, R. M., and B. Errede, 1994. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3139–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Y. C. Chiang and C. L. Denis, 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, S. M., and K. Madura, 2005. Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics 171: 1477–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M. A., 1996. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 16: 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M. A., and K. Struhl, 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8: 525–537. [DOI] [PubMed] [Google Scholar]
- Collart, M. A., and H. T. Timmers, 2004. The eukaryotic Ccr4-not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77: 289–322. [DOI] [PubMed] [Google Scholar]
- Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler et al., 2002. The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 22: 6735–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, C. L., and J. Chen, 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73: 221–250. [DOI] [PubMed] [Google Scholar]
- Denis, C. L., Y. C. Chiang, Y. Cui and J. Chen, 2001. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, C., A. M. Bonvin, G. S. Winkler, F. M. van Schaik, H. T. Timmers et al., 2004. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12: 633–644. [DOI] [PubMed] [Google Scholar]
- Fiorani, P., R. J. Reid, A. Schepis, H. R. Jacquiau, H. Guo et al., 2004. The deubiquitinating enzyme Doa4p protects cells from DNA topoisomerase I poisons. J. Biol. Chem. 279: 21271–21281. [DOI] [PubMed] [Google Scholar]
- Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu et al., 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M. H., and A. Ciechanover, 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Hanzawa, H., M. J. de Ruwe, T. K. Albert, P. C. van Der Vliet, H. T. Timmers et al., 2001. The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem. 276: 10185–10190. [DOI] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Irie, K., K. Yamaguchi, K. Kawase and K. Matsumoto, 1994. The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-pheromone-responsive genes. Mol. Cell. Biol. 14: 3150–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, A., and P. Reichard, 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67: 71–98. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor et al., 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22: 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo et al., 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- Lee, T. I., J. J. Wyrick, S. S. Koh, E. G. Jennings, E. L. Gadbois et al., 1998. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18: 4455–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, M., and M. A. Collart, 2000. The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4-Not complex and physically interacts with the Not5 subunit. J. Biol. Chem. 275: 26925–26934. [DOI] [PubMed] [Google Scholar]
- Lenssen, E., U. Oberholzer, J. Labarre, C. De Virgilio and M. A. Collart, 2002. Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43: 1023–1037. [DOI] [PubMed] [Google Scholar]
- Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni et al., 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation–via a newly identified Glc7/Bud14 type I protein phosphatase module–and TFIID promoter distribution. Mol. Cell. Biol. 25: 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie, C., and C. L. Peterson, 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304: 726–741. [DOI] [PubMed] [Google Scholar]
- Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama et al., 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96: 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster and M. A. Collart, 2000. The essential function of Not1 lies within the Ccr4-Not complex. J. Mol. Biol. 303: 131–143. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis et al., 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15: 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Mulder, K. W., G. S. Winkler and H. T. Timmers, 2005. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 33: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani, M., and W. P. Tansey, 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4: 192–201. [DOI] [PubMed] [Google Scholar]
- Pan, X., P. Ye, D. S. Yuan, X. Wang, J. S. Bader et al., 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Papa, F. R., and M. Hochstrasser, 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366: 313–319. [DOI] [PubMed] [Google Scholar]
- Papa, F. R., A. Y. Amerik and M. Hochstrasser, 1999. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell 10: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, Y., and S. L. Lindquist, 1990. HSP104 required for induced thermotolerance. Science 248: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Seufert, W., and S. Jentsch, 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, M. E., K. D. Ogburn, O. A. Varban, P. M. Gilbert and C. G. Burd, 2001. FYVE domain targets Pib1p ubiquitin ligase to endosome and vacuolar membranes. J. Biol. Chem. 276: 41388–41393. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., and R. Rothstein, 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Traven, A., A. Hammet, N. Tenis, C. L. Denis and J. Heierhorst, 2005. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 169: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- van de Peppel, J., N. Kettelarij, H. van Bakel, T. T. Kockelkorn, D. van Leenen et al., 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522. [DOI] [PubMed] [Google Scholar]
- Westmoreland, T. J., J. R. Marks, J. A. Olson, Jr., E. M. Thompson, M. A. Resnick et al., 2004. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell 3: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G. S., T. K. Albert, C. Dominguez, Y. I. Legtenberg, R. Boelens et al., 2004. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J. Mol. Biol. 337: 157–165. [DOI] [PubMed] [Google Scholar]
- Zervos, A. S., J. Gyuris and R. Brent, 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72: 223–232. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and J. C. Reese, 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279: 39240–39250. [DOI] [PubMed] [Google Scholar]