Abstract
A comparative physical map of the AA genome (Oryza sativa) and the BB genome (O. punctata) was constructed by aligning a physical map of O. punctata, deduced from 63,942 BAC end sequences (BESs) and 34,224 fingerprints, onto the O. sativa genome sequence. The level of conservation of each chromosome between the two species was determined by calculating a ratio of BES alignments. The alignment result suggests more divergence of intergenic and repeat regions in comparison to gene-rich regions. Further, this characteristic enabled localization of heterochromatic and euchromatic regions for each chromosome of both species. The alignment identified 16 locations containing expansions, contractions, inversions, and transpositions. By aligning 40% of the punctata BES on the map, 87% of the punctata FPC map covered 98% of the O. sativa genome sequence. The genome size of O. punctata was estimated to be 8% larger than that of O. sativa with individual chromosome differences of 1.5–16.5%. The sum of expansions and contractions observed in regions >500 kb were similar, suggesting that most of the contractions/expansions contributing to the genome size difference between the two species are small, thus preserving the macro-collinearity between these species, which diverged ∼2 million years ago.
COMPARATIVE genome analysis is proving to be an excellent tool, not only to discover genes and understand their functions, but also to unravel the evolutionary relationships between species. Since related species are derived from recent common ancestors, it comes as no surprise that in both dicots and monocots extensive genetic collinearity was found when related species were mapped using common RFLP probe sets (Bonierbale et al. 1988; Hulbert et al. 1990; Ahn and Tanksley 1993; Jena et al. 1994). While extensive collinearity seems to be limited to the genus level among dicots (Tanksley et al.1988), sufficient collinearity exists to allow rough alignment of genetic maps across entire genomes throughout the entire cereal clade (Moore et al. 1995). The complete sequences of the model organisms Arabidopsis and rice (Arabidopsis Genome Initiative 2000; International Rice Genome Sequencing Project 2005) allow propagation of the annotation and structure of these genomes to their taxonomic relatives. The rearrangement events in the evolutionary history of the genus Arabidopsis has been outlined by examining the level of synteny between the linkage maps of the Arabidopsis species and the Arabidopsis genome sequence (Yogeeswaran et al. 2005; Hansson et al. 2006). The major chromosomal rearrangements between Arabidopsis and Asteraceae (Timms et al. 2006) and the structural divergence of the genic regions between Arabidopsis and Brassica (Suwabe et al. 2006; Town et al. 2006) also have been demonstrated by using the complete Arabidopsis reference sequence.
Although rice is considered a model plant and placed at the center of the cereal crop syntenic circle (Moore et al. 1995; Gale and Devos 1998a,b; Devos 2005), only a few genomewide comparative analyses as yet have been performed using the rice genome sequence as a reference. The sorghum genome was compared to the rice genome sequence using two sorghum physical maps integrated with genetic markers and BAC hybridization data (Bowers et al. 2005). Various local rearrangements between other cereals and rice have been reported in sequence-level comparisons using the rice genome sequence as the reference (Chen et al. 1998; Goff et al. 2002; Bennetzen and Ma 2003; Sorrells et al. 2003; Salse et al. 2004; Rice Chromosome 3 Sequencing Consortium 2005). Other studies focused on within-family divergence on the scale of 10–60 million years ago (MYA) (Gale and Devos 1998b; Keller and Feuillet 2000) and were restricted to genic regions or small homologous regions of the grass genomes. The subspecies of rice, japonica and indica, which diverged 0.44 MYA (MA and Bennetzen 2004), were compared at the DNA sequence level, illustrating the genomic microstructural polymorphisms that contribute to intraspecific phenotypic variations (Han and Xue 2003). Such comparative sequence analyses revealed the detailed composition and organizational differences between the cereal genomes, as well as the rate and modes of genomic change. To date, however, no studies in plants have focused on comparing different genome groups within a single genus at the whole-genome level.
The annual type of Oryza punctata is a diploid wild rice species belonging to the BB genome group of the genus Oryza and is distributed mainly in Africa. The AA and BB genome types were distinguished on the basis of evidence derived from interspecific crossing, cytogenetics, and genomic DNA hybridization. Evolutionally, the BB genome group is the closest to the AA genome group, which contains sequenced cultivated rice (O. sativa). Ge et al. (2005) estimated that the AA and BB genome groups diverged from a common ancestor ∼2 MYA. As this species is known to possess resistance to bacterial blight and brown plant hopper, both of which are considered important traits for improving cultivated rice, information concerning the O. punctata genome is of keen interest. The close evolutionary relationship between O. sativa and O. punctata provides the potential for direct utilization of O. sativa sequence information to investigate the BB genome (or vice versa) for both crop improvement and evolutionary studies. Currently, only a limited amount of knowledge concerning the O. punctata genome, especially in terms of its structural composition, is known.
Cloned DNA “fingerprinting” is a staple of genomics (Olson et al. 1986) and has been adapted to develop whole-genome physical maps. In general, DNA from a set of clones is restriction digested and the resulting fragments sized using agarose gel or automated capillary electrophoresis. Clones containing the identical segment of a genome will generate the same set of restriction enzyme fragments. Clones that overlap will share a subset of their respective restriction fragments. To analyze large data sets of fingerprints generated from genomic libraries, a software, FPC (Soderlund et al. 2000), is deployed. Therein the concept of a shared “restriction fragment” is generalized to that of a “consensus band” (CB). FPC can assemble thousands of clone fingerprints into “contigs,” bundled together on the basis of their linear overlaps, evidenced by shared CBs. FPC maps have served crucial roles in genome research projects (Marra et al. 1997; Mozo et al. 1999; International Human Genome Mapping Consortium 2001; Chen et al. 2002) for map-based cloning, clone-by-clone sequencing, or selecting a minimal tiling path of clones (Engler et al. 2003).
Here, we report a deep coverage BAC-based, FPC physical map of O. punctata conjoined with an exhaustive set of BAC end sequences (BESs) as a resource to study the BB diploid genome of Oryza. By aligning the BES of the clones composing the FPC physical map, whole-genome-level comparisons were made between the BB diploid genome and the sequenced AA diploid genome, O. sativa (International Rice Genome Sequencing Project 2005).
MATERIALS AND METHODS
BAC plasmid DNA extraction:
A large insert BAC library of O. punctata (accession no. IRGC 105690) constructed by the Arizona Genomics Institute (Ammiraju et al. 2006) was used for generating BESs and SNaPshot fingerprints.
DNA was extracted from 36,864 BAC clones by a modified alkaline lysis method in a 96-well format. Briefly, BAC clones of O. punctata were inoculated into a deep 96-well plate (Marsh) containing 1.2 ml of 2× YT media plus 12.5 μg/ml chloramphenicol. The plates were covered with AirPore Tape Sheets (ISC BioExpress, Kaysville, UT) and shaken at 250 rpm at 37° for 18 hr. The cells were harvested by centrifugation (Jouan, Winchester, VA) at 3200 rpm (1970 × g) for 15 min, followed by decanting the media. Cell pellets were resuspended in 150 μl of solution I (50 mm Tris·Cl, 10 mm EDTA, pH 8.0, 100 μg/ml RNase A) followed by shaking using a vortex. Lysis and neutralization were achieved by adding 150 μl of solution II (0.2 m NaOH, 1% SDS) and 150 μl of solution III (3.0 m potassium acetate, pH 5.5) into the sample plates without a lysis incubation time. The lysates were then cleared and precipitated as follows. They were transferred to a stacked filter, Unifilter (Whatman, Brentford, UK), and receiver, Uniplate (Whatman), containing 270 μl of isopropanol/well. The stacked filter and receiver plate were then centrifuged at 4° for 30 min at 3200 rpm and decanted. After adding 400 μl of 70% ethanol, the plates were centrifuged at 4° for 10 min at 3200 rpm. A brief (<10 sec) upside-down spin was performed to remove the remaining liquid and the plates were air dried for 10 min or until the DNA pellets appeared transparent in the wells. Each precipitated DNA pellet was resuspended in 25 μl of 1 mm Tris (pH 8.0) and used immediately or stored at 4° for future use. All pipetting and liquid-handling steps were performed using a 96-channel liquid-handling system, Quadra 96 model 320 (Tomtec, Hamden, CT).
BAC end sequencing:
In 384-well plates, a 12-μl total volume reaction contained 5 μl (∼0.6–0.8 μg) BAC DNA, 0.5 μl of BigDye Terminator V.3 (ABI, Foster City, CA), 2 pmol of primer, 0.5 μl of 160 μm of each dNTP mix, and 4.3 μl of 5× sequencing buffer (ABI). The T7 primer (5′-TAA TAC GAC TCA CTA TAG GG-3′) was used as the “forward” primer and the custom BES_HR primer (5′-CAC TCA TTA GGC ACC CCA-3′) was used as the “reverse” primer. Cycle sequencing was performed using ABI thermal cyclers (ABI) in a 384-well format with the following conditions: 150 cycles of 10 sec at 95°, 5 sec at 55°, and 2.5 min at 60°. After cycle sequencing, the reaction products were purified with CleanSeq (Agencourt, Beverly, MA) magnetic beads, according to the manufacturer's instruction. Bound samples were eluted into 20 μl of water and separated on ABI 3730xl DNA capillary sequencers (50-cm capillary array) with default 2-hr run conditions. Sequences were base called using the program Phred (Ewing et al. 1998; Ewing and Green 1998), and vector and low-quality sequences were trimmed by the program LUCY (Chou and Holmes 2001). A total of 68,384 BESs were deposited in GenBank (for sequences, search using OP__Ba; for trace files, search using CENTER_PROJECT=‘OMAP_PUNCTATA’).
SNaPshot fingerprinting and FPC assembly:
The SNaPshot fingerprinting technique was used as described by Luo et al. (2003) but the restriction digest and fragment end-labeling steps were combined. Briefly, a 30-μl total volume reaction containing 10 μl BAC DNA (1.2–1.6 μg); 5 units each of BamHI, EcoRI, XbaI, XhoI, and HaeIII restriction endonucleases; 1× NEB buffer 2 (New England BioLabs, Beverly, MA); 5 μg bovine serum albumin; 1 μg DNase-free RNase A; and 1 mm MgCl2; 1 μl of SNaPshot Multiplex Ready reaction mix (ABI) was incubated in a thermocycler for 1 hr at 37° and then for 1 hr at 65°.
SNaPshot reaction products were purified and dissolved in 10 μl of Hi-Di formamide containing 0.05 μl of 500 Liz size standard (ABI), transferred into 384-well plates, and loaded into ABI 3730xl DNA sequencers. Capillary electrophoresis was performed with 36-cm capillary arrays using the ABI default GeneScan module. Peak areas, heights, colors, and fragment sizes of each peak were collected for each BAC using the ABI Data Collection program, and the data was processed by GeneMapper software (ABI) to generate files (one for each clone) containing fingerprint peaks and sizes. These files were further filtered as follows. Files containing <25 or >179 fragment peaks or missing >3 size standard peaks were discarded. Files representing cloned chloroplast DNA were also discarded. Only peaks representing fragments between 75 and 500 bases were used in subsequence FPC assembly. The initial assembly was built with a Sulston score cutoff of 1e-50 and a tolerance of 4 using FPC (Soderlund et al. 2000). The build was processed through the DQ function of FPC to break up contigs having >15% questionable clones (Q-clones). The assembly was further refined by contig end merging (with 61 CB units defining the distance from the end) with less-stringent Sulston score cutoff values (down to 1e-21) and by requiring two overlapping clone pairs for each merge. Finally, singletons were merged at a Sulston score cutoff of 1e-21. This initial FPC assembly is defined as a phase I physical map.
Synteny Mapping and Analysis Program alignment:
Synteny Mapping and Analysis Program (SyMAP) (Soderlund et al. 2006) was used to display alignments between O. punctata BESs embedded within the FPC maps and the O. sativa reference sequence (International Rice Genome Sequencing Project 2005; O. sativa pseudomolecule version 4). To create a comparison file for SyMAP to display, a sequence-similarity search between O. punctata BES and the O. sativa reference sequence was performed using BLAT (Kent 2002) after soft repeat masking using Repeatmasker (with the parameters −xsmall −e crossmatch −nolow -no_is -norna −q) against the Arizona Genomics Institute Oryza repeat database composed of The Institute for Genomic Research Oryza repeats V3.1 (ftp://ftp.tigr.org/pub/data/TIGR_Plant_Repeats/) and Oryza LTR retroelements isolated from previous analysis (Chaparro et al. 2006). BLAT was performed with the following parameters: ‘-minIdentity=70 -tileSize=10 -minScore=30 -qMask=lower -maxIntron=10000’. The first three parameters set a low stringency: “-qMask” specifies lowercase masking on the O. punctata BES, and “-maxIntron” allows for insertions on the O. sativa reference sequence of up to 10 kb. Alignments for BES from the O. punctata FPC map onto the O. sativa reference sequence were then computed and displayed by SyMAP software (Soderlund et al. 2006).
FPC manual editing:
The phase I FPC map was manually edited to improve contiguity and to correct any discrepancies between the FPC assembly and the SyMAP alignments. Contigs that showed conflicting alignments were broken at high stringency (Sulston score cutoff: 1e-60). Contigs were merged by searching the entire FPC fingerprint database for matches to fingerprints from clones representing contig termini above the Sulston score cutoff of 1e-15. Contigs were merged if (1) more than two clones from one contig termini matched fingerprints of at least two clones from the other contig termini or (2) the fingerprint of one clone of the contig termini matched only one clone of the other termini and included overlapping evidence of those two contigs detected by SyMAP alignment. Those contigs showing inverted alignment without evidence of inversion were flipped.
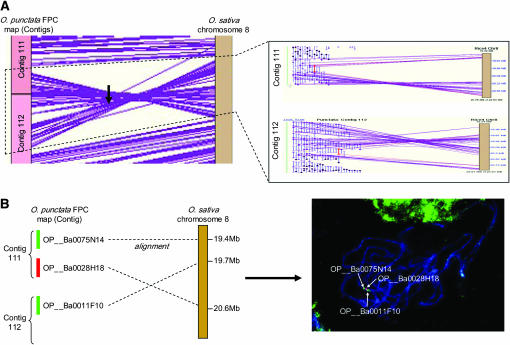
A large inversion on chromosome 8 was detected by alignment and was confirmed by fluorescence in situ hybridization (FISH) using pachytene chromosomes of O. punctata (accession no.105690) and O. punctata BAC clones (OP__Ba0075N14, OP__Ba0028H18, and OP__Ba0011F10) from each end of the breakpoints. The FISH procedure and image capture were conducted according to Walling et al. (2005).
RESULTS
The FPC-based physical map of O.punctata:
Successful fingerprints were generated from 93% (34,224 clones) of the 12-fold redundant O. punctata BAC library (Ammiraju et al. 2006) using the SNaPshot fingerprinting method (Luo et al. 2003). The remaining 7% failure represented an amalgam of library clone growth failures, SNaPshot reaction failures, and chloroplast DNA contaminants. The O. punctata BAC fingerprints were assembled using FPC (Soderlund et al. 2000) to generate a phase I FPC assembly consisting of 490 contigs and 1488 singleton clones (4.3% of total fingerprints). The average number of consensus band units (CB units) per clone was 117.4, where one CB unit was estimated to be 1.27 ± 0.2 kb on the basis of an analysis of a random sampling of 539 BAC clones of known sizes divided by the number of CB units that these clones spanned. The total number of contigs was reduced to 208 by manual editing, which represented a total of 329,435 CB units. Using 1.27 kb/CB unit, the physical map spanned ∼418 Mb and thus represented ∼98.4% of the O. punctata genome on the basis of a size of 425 Mb (Ammiraju et al. 2006).
A Q-clone in FPC is defined as a clone that does not align to the CB map of its FPC contig. This can be caused by extra or missing bands of the clone's total band count against the CB map (Soderlund et al. 2000; Nelson et al. 2005). Therefore, Q-clones can be used to detect possible chimeric or low-quality contigs. The Q-clones in the O. punctata FPC map were removed by disassembling low-quality contigs containing >15% Q-clones. The physical map is available at http://www.genome.arizona.edu/symap_punctata.
Anchoring BAC contigs to chromosomes:
We generated O. punctata BESs from both ends of the fingerprinted clones (36,864 BAC clones) to use as sequence tags. A total of 68,384 BAC end sequences from 36,100 O. punctata clones were produced with an average read length of 710 bp and these were deposited in GenBank. These sequences represented 48.6 Mb of genomic sequence for O. punctata, which, on the basis of a genome size of 425 Mb (Ammiraju et al. 2006), is 11.4% of the whole genome. This is equivalent to one sequence tag/6.2 kb. Of the total BES, 94% (63,942 BES) had fingerprints in the FPC map. A high proportion of clones (89.4%/32,284 clones) had sequences from both ends, and 93.6% (30,228 clones) of those had successful fingerprints in the O. punctata FPC map.
The 63,942 BES from the successfully fingerprinted O. punctata clones were incorporated into the FPC physical map. Subsequently, BESs on the FPC physical map were aligned onto the O. sativa reference sequence (International Rice Genome Sequencing Project 2005) by sequence similarity and 60,163 BESs were assigned chromosome positions. The alignments of the O. punctata physical map to the O. sativa reference sequence using the shared sequences were generated by SyMAP (Soderlund et al. 2006) and are graphically represented in Figure 1. In total, 43% (25,572 BES) of the O. punctata BESs that were assigned genome positions on the physical map were aligned onto the O. sativa reference sequence (International Rice Genome Sequencing Project 2005) with an average e-value of 1e-132 and an identity of 93%. This alignment anchored 176 contigs onto the O. sativa chromosomes containing 94% of the total fingerprints and representing 321,533 CB units (Table 1). The chromosome-anchored contigs cover an estimated size of 408 Mb, 96% of the O. punctata genome, whereas the remaining 32 nonanchored contigs correspond to 10 Mb, 2.4% of the genome (on the basis of 1.27 kb/CB and 425 Mb of punctata genome size; Ammiraju et al. 2006) (Table 1). The number of anchored contigs for each chromosome ranges from 10 (chromosome 4) to 18 (chromosome 7) and the coverage of each chromosome ranges from 92% (chromosome 9) to 116% (chromosome 6) on the basis of the O. sativa chromosome size (International Rice Genome Sequencing Project 2005). The sizes of O. punctata chromosomes were calculated on the basis of 1.27 kb/CB and compared to that of O. sativa. Chromosomes 4 and 9 of O. punctata were slightly smaller than that of O. sativa by 1.0 and 2.5 Mb, respectively. The remaining 10 chromosomes were larger in O. punctata than in O. sativa. The increases in size range from 0.4 Mb on chromosome 8 to 5.2 Mb on chromosome 6 (Table 1).
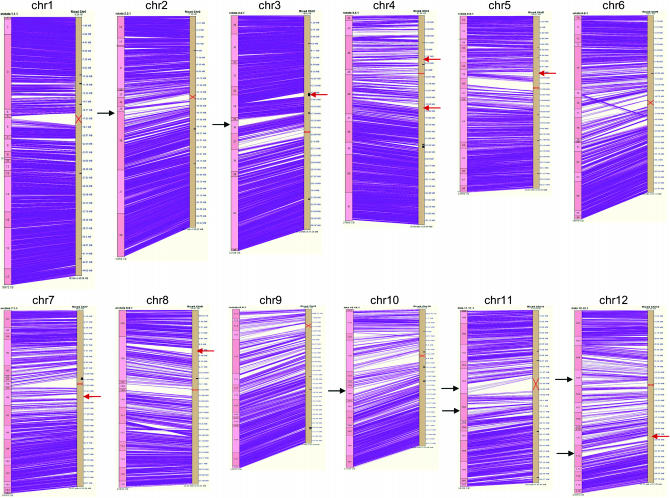
Figure 1.—
Detailed alignment view of the comparative physical map of O. punctata (BB genome type) and O. sativa (AA genome type) using SyMap (Soderlund et al. 2006). Alternating light and dark pink boxes on the left indicate O. punctata contigs. Beige bars on the right indicate O. sativa genome sequence (International Rice Genome Sequencing Project 2005). A red “X” on beige bars indicates a CentO position of O. sativa (International Rice Genome Sequencing Project 2005). Each purple line denotes the best alignment between an O. punctata BES onto the O. sativa genome sequence. The small black boxes on the beige bars indicate a physical gap in the O. sativa genome sequence (International Rice Genome Sequencing Project 2005). Black arrows indicate a position of a punctata expansion (details in Figure 2); red arrows indicate a position of a punctata contraction (details in Figure 2).
TABLE 1.
Summary of FPC map, BAC end sequences, and alignments of O. punctata
| Chromosome | Total no. contigs | Total no. of BACs | Total CB length (CB) | Estimated size (kb) (1.27 kb/CB)a | No. of OP BES | BES succ ratio | Genome representationb | % of OP BES (normalized) | No. of aligned OP BES (%) | O. sativa chromosome sizec (kb) | O. sativa pseudomolecules sizec (kb) | Covered length by alignment (kb) | % coverage of OP by the alignment | % coverage of O. sativa pseudomolecule by the aligment | % coverage of O. sativa chromosome by the alignment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 17 | 3,698 | 36,689 | 46,595 | 6,922 | 94% | 10.5% | 11.5 (8.4) | 3,594 (52) | 45,050 | 43,261 | 42,211 | 91 | 98 | 94 |
| 2 | 11 | 3,121 | 32,965 | 41,866 | 5,872 | 94% | 10.0% | 9.8 (7.9) | 2,880 (49) | 36,780 | 35,954 | 36,392 | 87 | 101 | 99 |
| 3 | 13 | 3,043 | 32,197 | 40,890 | 5,716 | 94% | 9.9% | 9.5 (7.9) | 3,058 (53) | 37,370 | 36,190 | 35,424 | 87 | 98 | 95 |
| 4 | 10 | 2,823 | 27,678 | 35,151 | 5,317 | 94% | 10.7% | 8.8 (8.5) | 2,235 (42) | 36,150 | 35,489 | 35,185 | 100 | 99 | 97 |
| 5 | 18 | 2,456 | 24,014 | 30,498 | 4,606 | 94% | 10.7% | 7.7 (8.5) | 2,039 (44) | 30,000 | 29,733 | 29,723 | 97 | 100 | 99 |
| 6 | 17 | 2,813 | 28,987 | 36,813 | 5,308 | 94% | 10.2% | 8.8 (8.1) | 2,056 (39) | 31,600 | 30,731 | 28,960 | 79 | 94 | 92 |
| 7 | 18 | 2,338 | 24,781 | 31,472 | 4,377 | 94% | 9.9% | 7.3 (7.8) | 1,927 (44) | 30,280 | 29,644 | 29,536 | 94 | 100 | 98 |
| 8 | 12 | 2,324 | 22,838 | 29,004 | 4,360 | 94% | 10.7% | 7.3 (8.5) | 1,768 (41) | 28,570 | 28,435 | 29,205 | 101 | 103 | 102 |
| 9 | 16 | 2,271 | 22,088 | 28,052 | 4,281 | 94% | 10.8% | 7.1 (8.6) | 1,520 (36) | 30,530 | 22,693 | 22,442 | 80 | 99 | 74 |
| 10 | 15 | 2,240 | 21,413 | 27,195 | 4,206 | 94% | 11.0% | 7.0 (8.7) | 1,411 (34) | 23,960 | 22,684 | 20,366 | 75 | 90 | 85 |
| 11 | 14 | 2,582 | 24,798 | 31,493 | 4,814 | 93% | 10.9% | 8.0 (8.6) | 1,726 (36) | 30,760 | 28,358 | 27,401 | 87 | 97 | 89 |
| 12 | 15 | 2,326 | 23,085 | 29,318 | 4,384 | 94% | 10.6% | 7.3 (8.4) | 1,358 (31) | 27,770 | 27,562 | 25,800 | 88 | 94 | 93 |
| No chromosome | 32 | 701 | 7,902 | 10,000 | 1,293 | 92% | — | — | — | — | — | — | — | — | — |
| Singleton | — | 1,488 | — | — | 2,486 | 84% | — | — | — | — | — | — | — | — | — |
| FP failure | — | 3,640 | — | — | 4,442 | 54% | — | — | — | — | — | — | — | — | — |
| Total/average | 208 | 37,864 | 329,435 | 418,347 | 68,384 | 93% | 11.4% | 88.0 | 25,572 (43) | 388,820 | 370,733 | 362,645 | 87 | 98 | 93 |
FP, fingerprinting.
1 .27 kb/CB was obtained using the formula [real size of the clone/CB unit of the clone] by sizing 539 O. punctata BAC clones selected randomly.
Based on estimated O. punctata chromosome size.
Comparative physical mapping between O. punctata and O.sativa:
By aligning the O. punctata FPC physical map onto the O. sativa genome sequence, a comparative physical map between the two species was constructed (Figure 1). The distribution of BES on each O. punctata chromosome was fairly even, ranging from 7.8 to 8.7% (Table 1) after normalization based on individual chromosome sizes in O. punctata. In contrast, the ratio of aligned BES of O. punctata to the O. sativa genome varied by chromosome. Overall, the 25,572 O. punctata BES that mapped onto the O. sativa reference sequence represented 87% of the O. punctata FPC physical map and 98% of the O. sativa reference sequence (Table 1), which suggests that the O. punctata genome is larger than the O. sativa genome. The O. sativa chromosome least represented in the alignment was chromosome 12 and the most represented was chromosome 3, aligning 31 and 53% of each chromosome BES, respectively (Table 1). However, the portion of the chromosome covered by the alignment was highest in chromosome 8 and lowest in chromosome 10.
Cheng et al. (2001) characterized the 12 chromosomes of cultivated rice cytogenetically by DAPI staining and was able to determine the positions of major euchromatic and heterochromatic blocks as well as the centromeres. Chromosomes 4 and 10, in particular, were noted for their distinct patterns, in which one-third of each chromosome, including both entire short arms and parts of the long arms, was highly heterochromatic (Cheng et al. 2001). The density of BES alignments shown in Figure 1 reflect the level of structural conservation between the two species. In general, dense alignments mirror the placement of euchromatic regions, and areas of few alignments represent heterochromatic regions, thus depicting the distribution patterns of euchromatic and heterochromatic regions on the chromosomes. The FPC contigs corresponding to highly heterochromatic regions on chromosomes 4 and 10 in the comparative physical map appear to encompass approximately one-half of each chromosome with less density of the alignments (Figure 1). Thus, the cytologically compressed DNA in the heterochromatin regions was less conserved between the species. In this study, no alignments were detected between the centromeric regions of these two species.
The comparative map not only was able to detect structural conservation between O. punctata and O. sativa, but also was able to find gross size differences (>500 kb) and rearrangements between the species. We identified 16 locations on the O. punctata chromosomes containing expansions, contractions, inversions, or transpositions relative to the O. sativa genome. Five chromosomes displayed an increase in size at seven regions, and six chromosomes displayed a decrease in size at seven other regions, accounting for 4.4 and 4.9 Mb of O. sativa sequence, respectively (Table 2, Figure 2). All putative expansions and contractions in the contigs were examined to determine whether the regions were artificially created by improper manual merging of contigs during FPC editing. Aside from the contraction on contig 34, all expanded or contracted regions were determined to be not involved in manual contig merges and were assembled at the more stringent (>1e-25) cutoff than that of other general contigs (>1e-15).
TABLE 2.
Rearrangement details between O. punctata and O. sativa based on the comparative physical map
| Positions on O. punctata
|
Positions on O. sativab
|
e-Value of the alignment of both termini of the rearrangement
|
Identity of the alignment (%)
|
Size difference (kb) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Chromosome | Contig | BES (start to end) | CB position | Sizea (kb) | Start (kb) | End (kb) | Size (kb) | Start | End | Start | End | |
| Expansion | 2 | 26 | a0052L01f–a0077J24f | 200–651 | 574 | 15,483 | 15,545 | 62 | 1E-12 | 1E-1 | 89 | 95 | 512 |
| 3 | 35, 36 | a0057A01r–a0092P04f | 364 (ctg35)– 780 (ctg36) | 1,139 | 16,795 | 17,076 | 281 | 1E-63 | 1E-1 | 95 | 93 | 858 | |
| 10 | 140 | a0023E16f–a0078A12f | 161–787 | 796 | 11,189 | 11,443 | 254 | 1E-117 | 1E-1 | 95 | 94 | 542 | |
| 11 | 153 | a0011F03f–a0042D02r | 2088–3218 | 1,436 | 11,500 | 12,132 | 632 | 1E-154 | 1E-4 | 92 | 98 | 804 | |
| 11 | 154 | a0067I22r–a0026J19f | 1434–2041 | 772 | 16,410 | 16,668 | 258 | 1E-111 | 1E-1 | 92 | 97 | 514 | |
| 12 | 165 | a0024O03f–a0079M20f | 634–1316 | 867 | 10,969 | 11,196 | 226 | 1E-195 | 1E-1 | 95 | 93 | 641 | |
| 12 | 172 | a0089B01f–a0070F24r | 968–1541 | 729 | 21,182 | 21,391 | 209 | 1E-37 | 1E-8 | 91 | 95 | 520 | |
| Contraction | 3 | 34 | a0096I09r–a0079J22r | 1210–1260 | 65 | 13,378 | 14,142 | 764 | 1E-185 | 1E-1 | 95 | 97 | 699 |
| 4 | 44 | a0076E21f–a0022J10r | 3434–3550 | 149 | 7,501 | 8,342 | 840 | 1E-174 | 1E-1 | 94 | 91 | 692 | |
| 4 | 46 | a0016H16r–a0072M08r | 3241–3317 | 98 | 15,148 | 15,915 | 766 | 1E-162 | 1E-1 | 96 | 92 | 669 | |
| 5 | 59 | a0096F07r–a0080E21r | 390–525 | 173 | 9,659 | 10,557 | 899 | 1E-91 | 1E-7 | 88 | 90 | 726 | |
| 7 | 96 | a0094P20f–a0091N22f | 1178–1300 | 156 | 14,383 | 15,228 | 845 | 1E-120 | 1E-7 | 90 | 96 | 689 | |
| 8 | 106 | a0049O07f–a0037I06r | 808–1222 | 527 | 6,258 | 7,505 | 1,247 | 1E-149 | 1E-2 | 94 | 97 | 720 | |
| 12 | 172 | a0002F15r–a0067E02r | 506–666 | 204 | 20,247 | 21,108 | 861 | 1E-186 | 1E-1 | 96 | 92 | 656 | |
| Inversion | 8 | 111, 112 | a0040E21f–a0073K20r | 1820(ctg111)– 784 (ctg111) | 1,396 | 19,648 | 20,705 | 1,057 | 1E-180 | 1E-1 | 97 | 93 | — |
| Transportation | 6 | 75, 76 | a0027B15f–a0018I09r | 744(ctg75)– 41(ctg76) | 544 | 18,443 | 18,716 | 274 | 1E-126 | 1E-1 | 97 | 95 | — |
Based on 1.27 kb/CB.
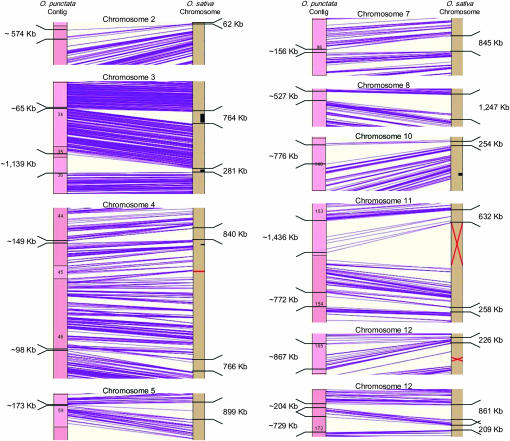
Figure 2.—
Alignment views with expansions/contractions of segments of the O. punctata genome against the collinear segment in O. sativa. The sizes of loci on O. punctata contigs were calculated using [1.27 kb/CB].
The regions containing size differences on contigs 36 and 34 (supplemental Figure 1 at http://www.genetics.org/supplemental/) mapped to physical gaps in the O. sativa reference sequence that were cytogenetically measured to be 112 and 500 kb (International Rice Genome Sequencing Project 2005), respectively. It is important to note that the estimated size differences for those regions rely on the accuracy of the physical gap size measurements in O. sativa. Contig 36 was created from three separate contigs (designated A, B, and C; supplemental Figure 1 at http://www.genetics.org/supplemental/) that were merged by 5 BES matches from contigs A and B (cutoff of 4e-17–4e-41; supplemental Figure 1 at http://www.genetics.org/supplemental/) and 14 BES matches from contigs B and C (cutoff of 1e-16–2e-42; supplemental Figure 1 at http://www.genetics.org/supplemental/). The expanded region in contig 36 was fully contained within contig A and was independent of any subsequent manual contig merge. In addition, the 9 BES alignments derived from contig A were in agreement with the alignments adjacent to them, which supports the integrity of contig 36. The contraction on contig 34 at the CB position of 1210–1260 was detected at the junction of a manual contig merge at 1221 CB (Table 2). Nevertheless, the five BAC clones at position 1157–1276 CB of contig 34, which span the contraction, as well as the manually contig-merged junction, were aligned onto the O. sativa sequence with an alignment length of 266–965 bp, a sequence identity of >91%, and an e-value of 1e-56 to 1e-242 (supplemental Table 1 at http://www.genetics.org/supplemental/). All five clones support the contraction, as the alignments of those clones span from 781 to 813 kb of the O. sativa reference sequence, which is ∼660 kb larger than the actual size of the clones.
These expansion/contraction regions aligned poorly between the species, with few or no clones contained within the region and dense coverage just outside of the region (Figure 1, red and black arrows; Figure 2). The estimated size of the expansions on the O. punctata map ranged from 512 kb for chromosome 2 to 858 kb for chromosome 3 (Table 2), while the size of the contractions on the O. punctata map ranged from 656 to 726 kb for chromosomes 12 and 5, respectively. Chromosomes 3 and 12 of O. punctata displayed both expansion and contraction of 2.7 Mb and 74 kb, respectively, as estimated by their relative positions on the O. sativa genome sequence. The expansion on chromosome 3 represents the largest difference among the seven expansions, and it is expected to be larger than measured due to the physical gap between contigs 35 and 36. The two expansions on chromosome 11 were detected in the pericentromeric region, contributing 1.3 Mb of excess sequence to O. punctata chromosome 11. Chromosome 4 contained two contractions, representing 1.4 Mb in total. All detected expansions and contractions (except those on contig 172) were detected within 1–5 Mb of the CentO repeat positions of O. sativa (International Rice Genome Sequencing Project 2005).
One inversion was detected on chromosome 8, representing ∼1.1 Mb of sequence in the O. sativa genome (Figure 3A). This inversion was detected by the alignment of 79 O. punctata BESs and, of those, 53 BESs aligned with an e-value of >1e-100, a sequence identity of >90%, and an alignment length of >400 bp (supplemental Table 2 at http://www.genetics.org/supplemental/). The position of the inversion on O. punctata was from 1820 CB of contig 111 to 784 CB of contig 112, corresponding to position 19.6–20.7 Mb of O. sativa chromosome 8 (Table 2). This inversion was further supported by confirming the order of the clones adjacent to the breakpoints in the O. punctata FPC map by FISH analysis (Figure 3B). The clones, OP__Ba0073K20 and OP__Ba0078K10, which contain the breakpoint for each end of the inversion, were detected in the alignments and were represented by both BESs for each clone on the O. sativa sequence (Figure 3A, red highlighted; Table 3).
Figure 3.—
Inversion between two species on chromosome 8. (A) O. punctata contigs 111 and 112 contain an inversion with respect to the O. sativa genome sequence. The inversion was detected by 79 BES alignments and two clones containing the breakpoints of the inversion were selected (red highlighted). (B) The confirmation of the inversion by FISH analysis. (Left) The clones selected for FISH analysis. (Right) FISH pattern of the clones on pachytene chromosome of O. punctata.
TABLE 3.
Breakpoint-containing clones for inversion and translocation between O. punctata and O. sativa genomes
| Alignment
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Clone | Contig | CB position | Chromosome | O. punctata BES | Start on O. sativa (bp) | End on O. sativa (bp) | e-Value | % identity |
| Inversion | a0073K20 | 111 | 1675–1820 | 8 | a0073K20f | 19517686 | 19518562 | 1.00E-215 | 95 |
| a0073K20r | 20703719 | 20704556 | 1.00E-162 | 93 | |||||
| a0078K10 | 112 | 770–911 | 8 | a0078K10f | 19667817 | 19668627 | 1.00E-182 | 96 | |
| a0078K10r | 20752956 | 20753518 | 1.00E-128 | 95 | |||||
| Transposition | a0092G08 | 75 | 760–896 | 6 | a0092G08f | 18450596 | 18451456 | 1.00E-118 | 93 |
| a0092G08r | 13002825 | 13004742 | 1.00E-156 | 94 | |||||
| a0090D18 | 76 | 12–176 | 6 | a0090D18f | 13174718 | 13175434 | 1.00E-182 | 94 | |
| a0090D18r | 18708349 | 18708663 | 1.00E-73 | 95 | |||||
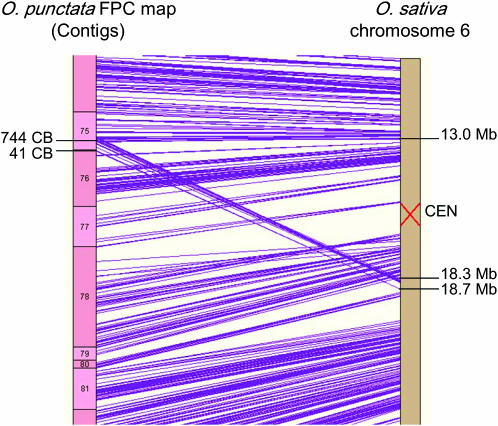
Approximately 400 kb of an O. punctata region on chromosome 6 was identified to represent an intrachromosomal transposition with respect to O. sativa (Figure 4). The position of this event was from 744 CB of contig 75 to 41 CB of contig 76, corresponding to 18.3–18.7 Mb of O. sativa chromosome 6 (Table 2). This transposition was detected by the alignment of 15 BES onto O. sativa chromosome 6 with a sequence identity of >91% and an alignment length of >300 bp (supplemental Table 3 at http://www.genetics.org/supplemental/). The translocation was confirmed by verifying the assembly of contig 75 and contig 76 using the overgo hybridization method described by Chen et al. (2002; data not shown). The clone OP__Ba0092G08 on contig 75 and OP__Ba0090D18 on contig 76 were determined to be breakpoint-containing clones (Table 3), and both ends of each clone aligned onto the O. sativa sequences with an e-value of <1e-73, the identity of >93%, and an aligned length of >300 bp.
Figure 4.—
Detailed view of an intrachromosomal transposition on chromosome 6. The 15 BESs from 744 CB of contig 75 to 41 CB of contig 76 were aligned to 18.3–18.7 Mb of O. sativa chromosome 6. The integrity of the contig assembly of the locus was confirmed by the overgo hybridization method described by Chen et al. (2002; data not shown).
DISCUSSION
Resources for studying the O.punctata genome:
The BES and FPC physical map of the O. punctata genome presented in this study are of a higher quality than any other previously reported resource with respect to the BES read length, BES/fingerprinting success ratio, genome coverage, and number of gaps in the FPC physical map. In addition, integration of the BES data into the FPC physical map evenly represents the O. punctata genome, covering ∼10.4% (±0.5%) of each chromosome.
We applied a modified SNaPshot fingerprinting method (Luo et al. 2003) and adapted it for high-throughput processing (see materials and methods). Due to this modification, we were able to fingerprint an 11× genome equivalent of O. punctata BAC clones, which covered 98% of the O. punctata genome with one BES tag/5.3 CB for all 12 chromosomes. The number of physical gaps over the entire genome was 164 equating to an average of 14 gaps per each chromosome. The accuracy of the FPC physical map assembly using the modified fingerprinting method was demonstrated by the high degree of collinearity with the O. sativa genome sequence, as well as by confirmation of a detected transposition and an inversion using overgo hybridization and FISH analysis (Figure 3B). The well-maintained collinearity displayed is expected due to the short divergence time between O. sativa and O. punctata and proved useful in editing the O. punctata FPC physical map, as well as in demonstrating the quality of the FPC assembly.
The O.punctata genome size:
The genome size of O. punctata has been reported as 22% larger (1.11 pg/2C; Uozu et al. 1997) or 3–5% smaller (0.88 pg/2C; Ammiraju et al. 2006) than that of O. sativa ssp. japonica cv Nipponbare (0.91 pg/2C; Uozu et al. 1997) on the basis of flow cytometric analysis. To estimate the physical size of FPC contigs, we determined the average band (CB unit) size derived from the O. punctata FPC physical map by estimating the size of 539 O. punctata BAC clones and applying the following formula: one CB unit = real size of the clone/CB unit of the clone. Consequently, the size of each chromosome, as well as the genome as a whole, was based on the constant of [1 CB unit = 1.27 kb] and the length of the FPC physical map in CB, an estimate that is independent of the previous reports by flow cytometry. The total FPC CB length of 329,435 (Table 1) converts to 418 Mb—our estimate of the size of the O. punctata genome covered by the FPC map. In addition, since 87% of the FPC physical map covered most of the O. sativa reference sequence (98%) or a larger portion of the O. sativa genome (93%) with estimated gaps (International Rice Genome Sequencing Project 2005), we estimate that the O. punctata genome is slightly larger than the O. sativa genome by ∼8% (∼30 Mb). The map also demonstrates that the size difference between O. punctata and O. sativa is not evenly distributed over the chromosomes, possibly reflecting dynamics that have affected each chromosome differently over evolutionary time (Table 1). One possible explanation for the genome size difference is the report that O. punctata centromeres have ∼2.5 times more CentO satellite DNA and a wider distribution of centromeric retrotransposon of rice sequence than O. sativa (Zhang et al. 2005). It warrants noting that our size estimate of the O. punctata genome may be an underestimate due to the presence of physical gaps between the contigs and the fact that the nuclear organizer region (Shishido et al. 2000) on the tip of chromosome 9S was not included in the FPC map. Therefore, although the chromosome 9 of O. punctata has a smaller observed size than O. sativa, the chromosome is actually larger than the corresponding O. sativa chromosome if the 6.95 Mb of rDNA on the O. sativa chromosome is excluded.
Comparison between the O. punctata and O.sativa genomes using the comparative FPC physical map:
O. punctata and O. sativa diverged ∼2 MYA (Ge et al. 2005) and, consequently, the collinearity between these two species is expected to be very high. As a result, we were able to generate a comparative physical map of the two species using the BES-tagged FPC physical map of O. punctata and the O. sativa reference sequence by sequence similarity. We found that overall collinearity between the species was well maintained and was mostly disrupted in centromeric and heterochromatic regions (Figure 1). This could be explained in part by more rapid divergence in the intergenic and repetitive regions as compared to more gene-rich regions (Bennetzen and Ma 2003).
The full genome-level macro-structural differences between O. punctata and O sativa have been illustrated in this study (Figure 2). Only ∼0.5 Mb of size difference between total expansions and contractions was observed at the macro level (>500 kb) in this study (Table 2), demonstrating that most of the contractions/expansions between the two species occur on a scale <500 kb. That the regions that did differ in size between the species had no or very few alignments indicates that these areas may represent regions of the chromosomes that are unique to that species. An alternate explanation would be that no alignments were detected due to stringent filtering of repeats by SyMAP (Soderlund et al. 2006). However, on the basis of the theory of 0.14 ± 0.06 structural mutations/chromosome/million years of divergence put forth by Paterson et al. (1996), we expect two to five structural mutations to have occurred between the O. punctata and O. sativa genomes over 2 million years of divergence. At least three times the expected number of structural mutations was detected between O. punctata and O. sativa, possibly due to the size threshold used for detecting the rearrangement. The timing of those events should be further investigated for clarifying whether the changes occurred concurrently with or subsequent to the divergence of the AA and BB genome groups. The map also suggests that there are many microlevel rearrangements between these two species that can account for the differentiation of O. punctata and O. sativa structurally as well functionally.
The degree of conservation for each chromosome, in terms of the alignment ratio of BES, varied from 31% (chromosome 12) to 53% (chromosome 3) uncorrelated with the variation in size. On the basis of the comparative analysis in this study, we estimated that chromosomes 6 and 12 are the most divergent between the species and that chromosome 3 is the most similar. It is interesting to note that chromosome 6 is known to possess one of the brown planthopper resistance genes (bph4; Kawaguchi et al. 2001), as well as one of the domestication genes of O. sativa, hd1 (Yano et al. 2000). Chromosome 12 is known to contain several of the brown planthopper resistance genes, such as bph1 (Hirabayashi and Ogawa 1995), bph2 (Murata et al. 1998), bph9 (Murata et al. 2001) bph10 (Ishii et al. 1994), and bph18(t) (Jena et al. 2005). The presence of resistance genes on these chromosomes may partially explain the higher levels of divergence relative to the rest of the genome as the resistant gene families tend to rapidly evolve through diversification (Ronald 1998).
The comparative map introduced here serves as a reference to develop the materials necessary to exploit the untapped genetic resource of O. punctata as a close relative to cultivated rice. The map provides the basis for positional cloning or introducing useful traits into cultivated rice by means of interspecific hybrids between cultivated rice and O. punctata, monosomic alien addition lines, introgression lines, and mapping populations. In addition, the well-aligned regions can be used to develop markers useful in integrating the physical and genetic maps of other important cereals. Finally, this comparative map not only elucidates several regions that differentiate the two genomes and warrant further investigation in an evolutionary context, but also will serve as a basis for future evolutionary studies on a finer scale. The comparative map will be updated as the study progresses and more anchoring information becomes available. Refinements of the physical map can be accessed at http://www.genome.arizona.edu/symap_punctata.
Acknowledgments
We thank Arizona Genomics Institute's BAC/EST Resource Center for providing a copy of the O. punctata BAC library, Paul Parker for help with BES generation, Rick Westerman for help with the trace file submission, Austin Shoemaker for help with Java graphics of SyMAP, Michele Braidotti for help with the SyMAP website, Andrea Zuccolo for providing repeat database and helpful discussion, and Jiming Jiang for supportive discussion. This work was supported by National Science Foundation grant 0321678 to R.A.W, S.A.J., and C.S.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. CW502583–CW509125, CW514009–539178, CW620733–CW624836, CW628185–633039, CW672722–CW676096, CW691361–692844, CW748472–CW754418, CW775494–CW778432, CW778534–CW784312, CW829027–CW834867, CW848277–CW850106, and CW935822–936338.
References
- Ahn, S., and S. D. Tanksley, 1993. Comparative linkage maps of rice and maize genomes. Proc. Natl. Acad. Sci. USA 90: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju, J. S., M. Luo, J. L. Goicoechea, W. Wang, D. Kudrna et al., 2006. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and J. Ma, 2003. The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr. Opin. Plant Biol. 6: 128–133. [DOI] [PubMed] [Google Scholar]
- Bonierbale, M. W., R. L. Plaisted and S. D. Tanksley, 1988. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J. E., M. A. Arias, R. Asher, J. A. Avise, R. T. Ball et al., 2005. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc. Natl. Acad. Sci. USA 102: 13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, C., R. Guyot, A. Zuccolo, B. Piegu and O. Panaud, 2007. RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res. 35: D66–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. S., P. SanMiguel and J. L. Bennetzen, 1998. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., G. Presting, W. B. Barbazuk, J. L. Goicoechea, B. Blackmon et al., 2002. An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., C. R. Buell, R. A. Wing, M. Gu and J. Jiang, 2001. Toward a cytological characterization of the rice genome. Genome Res. 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H. H., and M. H. Holmes, 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1094. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., 2005. Updating the ‘Crop Circle’. Curr. Opin. Plant Biol. 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Engler, F. W., J. Hatfield, W. Nelson and C. A. Soderlund, 2003. Locating sequence on FPC maps and selecting a minimal tiling path. Genome Res. 13: 2152–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendle and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Gale, M., and K. M. Devos, 1998. a Plant comparative genetics after 10 years. Science 282: 656–659. [DOI] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. b Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95: 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, S., Y.-L. Guo and Q.-H. Zhu, 2005. Molecular phylogeny and divergence of the rice tribe (Oryzeae), with special reference to the origin of the genus Oryza, pp. 40–44 in Rice Is Life: Scientific Perspectives for the 21st Century, edited by K. Toriyama, K. L. Heong and B. Hardy. IRRI, Philippines.
- Goff, S. A., D. Ricke, T. H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Han, B., and Y. Xue, 2003. Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 6: 134–138. [DOI] [PubMed] [Google Scholar]
- Hansson, B., A. Kawabe, S. Preuss, H. Kuittinen and D. Charlesworth, 2006. Comparative gene mapping in Arabidopsis lyrata chromosomes 1 and 2 and the corresponding A. thaliana chromosome 1: recombination rates, rearrangements and centromere location. Genet. Res. 87: 75–85. [DOI] [PubMed] [Google Scholar]
- Hirabayashi, H., and T. Ogawa, 1995. RFLP mapping of Bph-1 (brown planthopper resistance gene) in rice. Breed. Sci. 45: 369–371. [Google Scholar]
- Hulbert, S. H., T. E. Richter, J. D. Axtell and J. L. Bennetzen, 1990. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. USA 87: 4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium, 2001. A physical map of the human genome. Nature 409: 934–941. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Ishii, T., D. S. Brar, D. S. Multani and G. S. Khush, 1994. Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa. Genome 37: 217–221. [DOI] [PubMed] [Google Scholar]
- Jena, K. K., G. S. Khush and G. Kochert, 1994. Comparative RFLP mapping of a wild rice, Oryza officinalis, and cultivated rice, O. sativa. Genome 37: 382–389. [DOI] [PubMed] [Google Scholar]
- Jena, K. K., J. U. Jeung, J. H. Lee, H. C. Choi and D. S. Brar, 2005. High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 112: 288–297. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, M., K. Murata, T. Ishii, S. Takumi, N. Mori et al., 2001. Assignment of a brown planthopper (Nilaparvata lugens Stal) resistance gene bph4 to rice chromosome 6. Breed. Sci. 51: 13–18. [Google Scholar]
- Keller, B., and C. Feuillet, 2000. Colinearity and gene density in grass genomes. Trends Plant Sci. 5: 246–251. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, S. Ouyang et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389. [DOI] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M. A., T. A. Kucaba, N. L. Dietrich, E. D. Green, B. Brownstein et al., 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, G., K. M. Devos, Z. Wang and M. D. Gale, 1995. Cereal genome evolution: grasses, line up and form a circle. Curr. Biol. 5: 737–739. [DOI] [PubMed] [Google Scholar]
- Mozo, T., K. Dewar, P. Dunn, J. R. Ecker, S. Fischer et al., 1999. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat. Genet. 22: 271–275. [DOI] [PubMed] [Google Scholar]
- Murata, K., M. Fujiwara, C. Kaneda, S. Takumi, N. Mori et al., 1998. RFLP mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene bph2 of indica rice introgressed into a japonica breeding line ‘Norin-PL4.’ Genes Genet. Syst. 73: 359–364. [Google Scholar]
- Murata, K., M. Fujiwara, H. Murai, S. Takumi, N. Mori et al., 2001. Mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene Bph9 on the long arm of rice chromosome 12. Cereal Res. Commun. 29: 245–250. [Google Scholar]
- Nelson, W. M., A. K. Bharti, E. Butler, F. Wei, G. Fuks et al., 2005. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 139: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M. V., J. E. Dutchik, M. Y. Graham, G. M. Brodeur, C. Helms et al., 1986. Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. USA 83: 7826–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., T. H. Lan, K. P. Reischmann, C. Chang, Y. R. Lin et al., 1996. Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat. Genet. 14: 380–382. [DOI] [PubMed] [Google Scholar]
- Rice Chromosome 3 Sequencing Consortium, 2005. Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 15: 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, P. C., 1998. Resistance gene evolution. Curr. Opin. Plant Biol. 4: 294–298. [DOI] [PubMed] [Google Scholar]
- Salse, J., B. Piegu, R. Cooke and M. Delseny, 2004. New in silico insight into the synteny between rice (Oryza sativa L.) and maize (Zea mays L.) highlights reshuffling and identifies new duplications in the rice genome. Plant J. 38: 396–409. [DOI] [PubMed] [Google Scholar]
- Shishido, R., Y. Sano and K. Fukui, 2000. Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol. Gen. Genet. 263: 586–591. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., S. Humphray, A. Dunham and L. French, 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., W. Nelson, A. Shoemaker and A. Paterson, 2006. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 16: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe, K., H. Tsukazaki, H. Iketani, K. Hatakeyama, M. Kondo et al., 2006. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., R. Bernatzky, N. L. Lapitan and J. P. Prince, 1988. Conservation of gene repertoire but not gene order in pepper and tomato. Proc. Natl. Acad. Sci. USA 85: 6419–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms, L., R. Jimenez, M. Chase, D. Lavelle, L. McHale et al., 2006. Analyses of synteny between Arabidopsis thaliana and species in the Asteraceae reveal a complex network of small syntenic segments and major chromosomal rearrangements. Genetics 173: 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town, C. D., F. Cheung, R. Maiti, J. Crabtree, B. J. Haas et al., 2006. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozu, S., H. Ikehashi, N. Ohmido, H. Ohtsubo, E. Ohtsubo et al., 1997. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 35: 791–799. [DOI] [PubMed] [Google Scholar]
- Walling, J. G., J. C. Pires and S. A. Jackson, 2005. Preparation of samples for comparative studies of plant chromosomes using in situ hybridization methods, pp. 442–459 in Molecular Evolution: Producing the Biochemical Data, Part B. Elsevier, Amsterdam. [DOI] [PubMed]
- Yano, M., Y. Katayose, M. Ashikari, U. Yamanouchi, L. Monna et al., 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogeeswaran, K., A. Frary, T. L. York, A. Amenta, A. H. Lesser et al., 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., C. Yi, W. Bao, B. Liu, J. Cui et al., 2005. The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 139: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]